Regulation of Metabolic Reprogramming by Long Non-Coding RNAs in Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
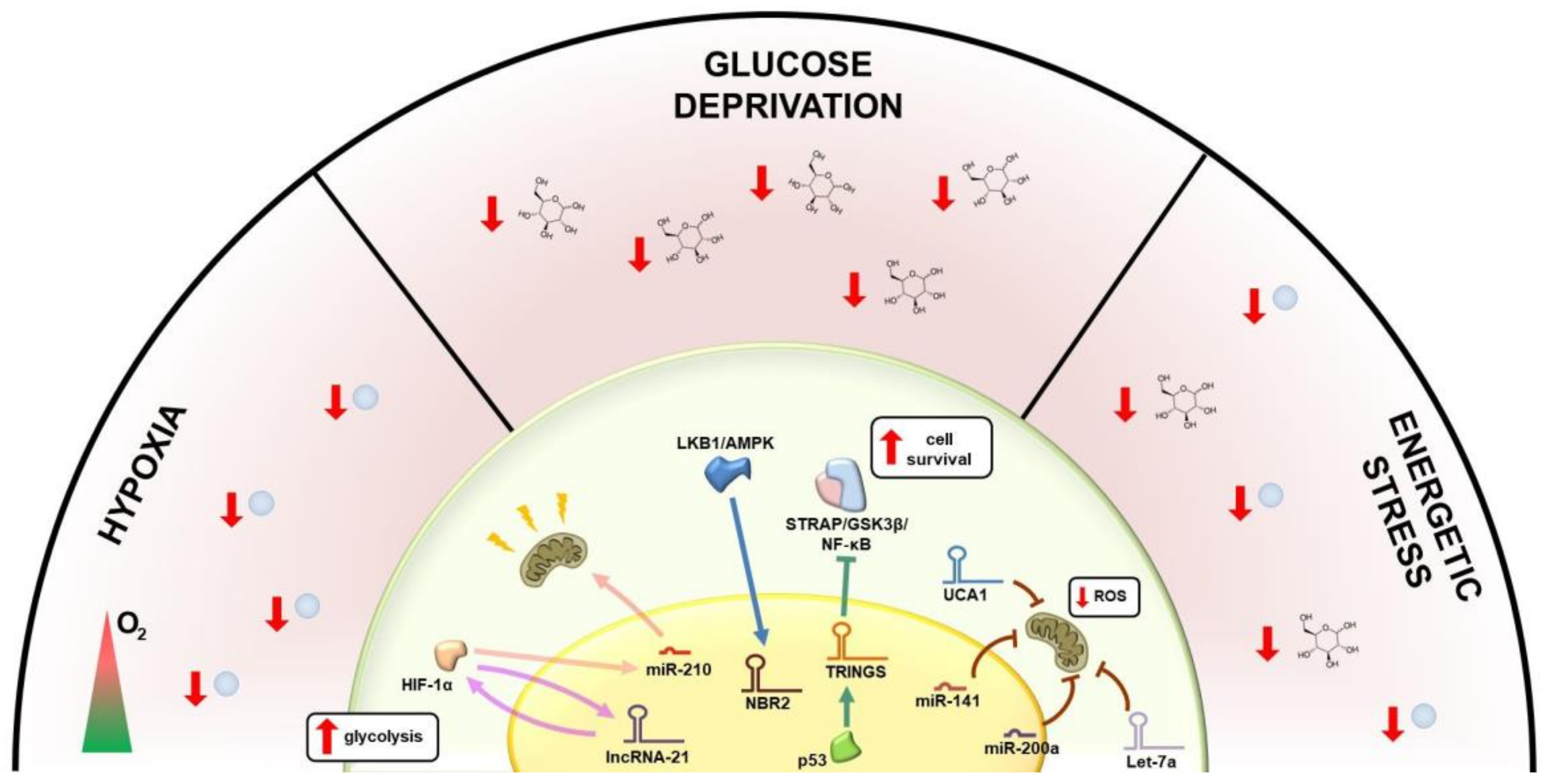
2. Metabolic Regulation of lncRNAs
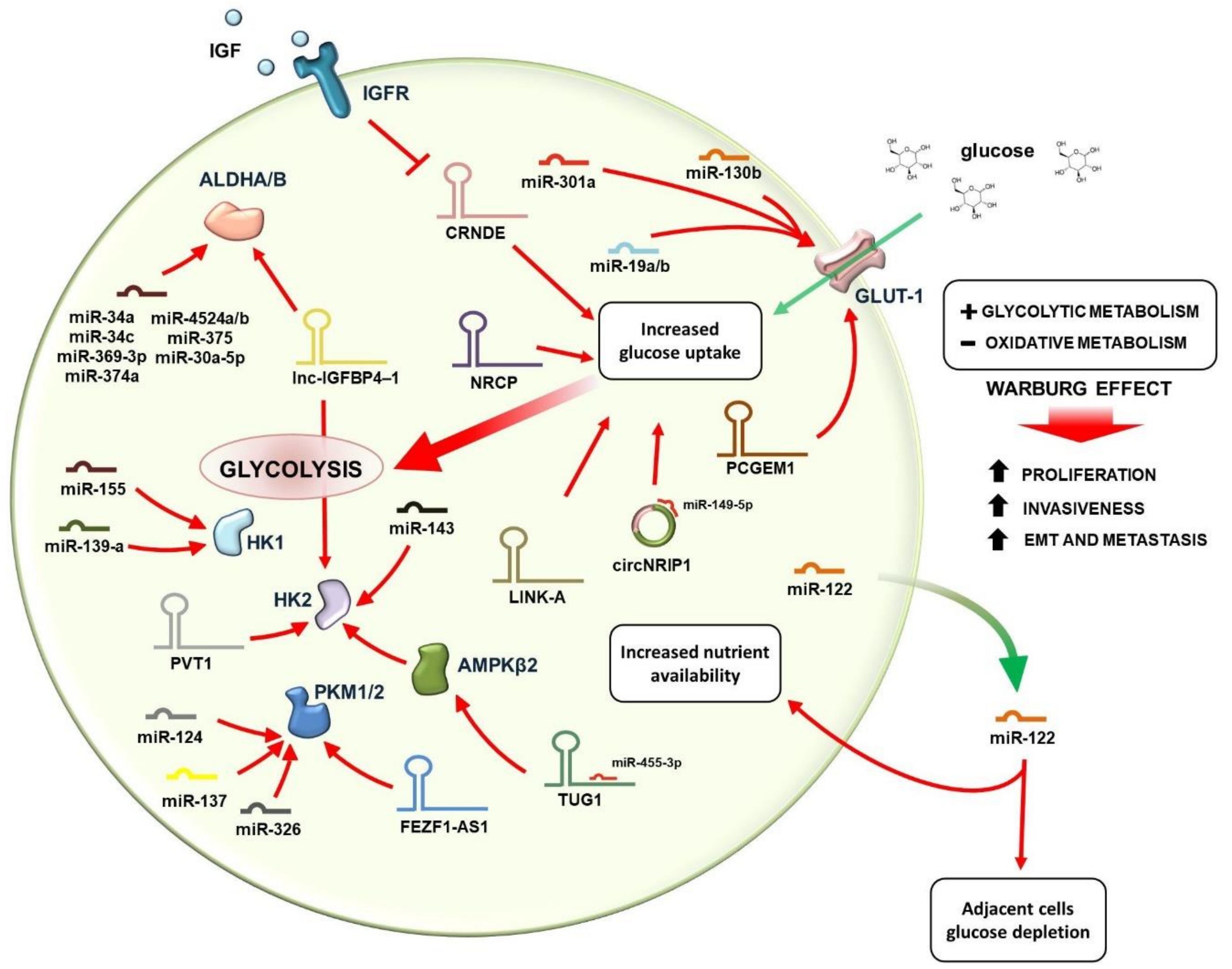
3. Regulation of Glucose Metabolism by lncRNAs
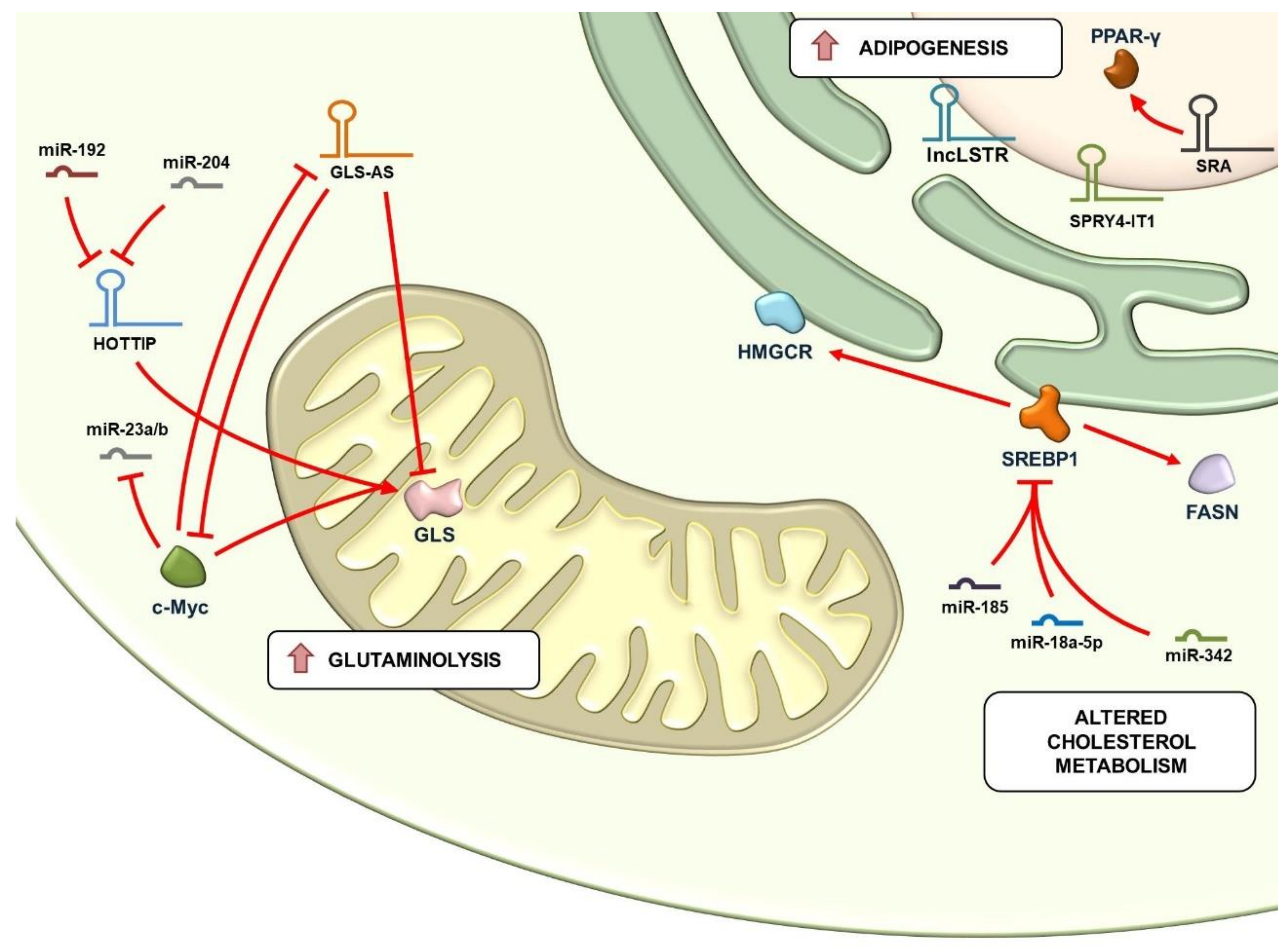
4. Regulation of Glutaminolysis and Mitochondrial Metabolism by lncRNAs
5. Regulation of Lipid Metabolism by lncRNAs
6. lncRNAs as Biomarkers and Therapeutic Targets
7. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ACC | Acetyl-CoA carboxylase |
| ACLY | adenosine triphosphate citrate lyase |
| ACSL1 | acyl-CoA synthetase long-chain family member 1 |
| AMPK | AMP-activated protein kinase |
| AMPKβ2 | adenosine monophosphate-activated protein kinase subunit β2 |
| AMPKβ2 | adenosine monophosphate-activated protein kinase subunit β2 |
| CDK6 | cyclin-dependent kinase 6 |
| CELF2 | CUGBP Elav-like family member 2 |
| CRNDE | Colorectal Neoplasia Differentially Expressed |
| dsRNAs | double-strand RNAs |
| EMT | epithelial-mesenchymal transition |
| ES | Ewing sarcoma |
| FASN | Fatty acid synthase |
| FILNC1 | FoxO-induced long non-coding RNA 1 |
| GLS | Glutaminase |
| GLS-AS | nuclear-located antisense lncRNA of glutaminase |
| GLUTs | Glucose transporters |
| GMAN | Gastric cancer metastasis associated |
| HCC | Hepatocellular carcinoma |
| HIF-1 α | Hypoxia-inducible factor-1 α |
| HIF-1α | Hypoxia-inducible factor 1-alpha |
| HIFAL | HIF-1α anti-sense lncRNA |
| HK2 | Hexokinase 2 |
| HKs | Hexokinases |
| HMGCR | 3-hydroxy-3-methyl-glutaryl CoA reductase) |
| HOTAIR | HOX Transcript Antisense Intergenic RNA |
| HULC | Highly Upregulated in Liver Cancer lncRNA |
| IGF | insulin-like growth factor |
| IGFBP4–1 | insulin-like growth factor binding protein 4–1 |
| LATS2 | large tumor suppressor 2 |
| LDH | lactate dehydrogenase |
| lincRNAs | long intergenic non-coding RNA |
| LIPA | lipase A |
| LKB1 | liver kinase B1 |
| lncRNAs | long non-coding RNA |
| MALAT1 | Metastasis Associated Lung Adenocarcinoma Transcript 1 |
| MEG3 | maternally expressed gene 3 |
| MMP-2 | matrix metalloproteinase-2 |
| NATs | natural antisense transcripts |
| NBR2 | neighbor of BRCA1 gene 2 |
| NRCP | ceruloplasmin lncRNA |
| PDAC | pancreatic ductal adenocarcinoma |
| PDK1 | pyruvate dehydrogenase lipoamide kinase isozyme 1 |
| PKM | Pyruvate kinase |
| PLD | phospholipase D |
| sncRNAs | small non-coding RNAs |
| SREBP1 | sterol regulatory element binding protein |
| TGIF2 | transforming growth factor-β-induced factor homeobox 2 |
| TRINGS | Tp53-regulated inhibitor of necrosis under glucose starvation |
| T-UCR | transcribed ultraconserved region |
| TUG1 | taurine up-regulated gene 1 |
References
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef] [PubMed]
- Anastasiadou, E.; Jacob, L.S.; Slack, F.J. Non-coding RNA networks in cancer. Nat. Rev. Cancer 2018, 18, 5–18. [Google Scholar] [CrossRef] [PubMed]
- Lu, P.; Li, M.; Zhang, D.; Jiang, W. Lnc-ing pluripotency maintenance and early differentiation in human pluripotent stem cells. FASEB J. 2021, 35, e21438. [Google Scholar] [CrossRef] [PubMed]
- Murillo-Maldonado, J.M.; Riesgo-Escovar, J.R. The various and shared roles of lncRNAs during development. Dev. Dyn. 2019, 248, 1059–1069. [Google Scholar] [CrossRef] [Green Version]
- Froberg, J.E.; Yang, L.; Lee, J.T. Guided by RNAs: X-inactivation as a model for lncRNA function. J. Mol. Biol. 2013, 425, 3698–3706. [Google Scholar] [CrossRef] [Green Version]
- Barlow, D.P.; Bartolomei, M.S. Genomic imprinting in mammals. Cold Spring Harb. Perspect. Biol. 2014, 6. [Google Scholar] [CrossRef] [Green Version]
- Wei, J.W.; Huang, K.; Yang, C.; Kang, C.S. Non-coding RNAs as regulators in epigenetics (Review). Oncol. Rep. 2017, 37, 3–9. [Google Scholar] [CrossRef] [Green Version]
- Han Li, C.; Chen, Y. Small and Long Non-Coding RNAs: Novel Targets in Perspective Cancer Therapy. Curr. Genom. 2015, 16, 319–326. [Google Scholar] [CrossRef] [Green Version]
- Tian, Y.; Ma, R.; Sun, Y.; Liu, H.; Zhang, H.; Sun, Y.; Liu, L.; Li, Y.; Song, L.; Gao, P. SP1-activated long noncoding RNA lncRNA GCMA functions as a competing endogenous RNA to promote tumor metastasis by sponging miR-124 and miR-34a in gastric cancer. Oncogene 2020, 39, 4854–4868. [Google Scholar] [CrossRef]
- Yu, Y.; Gao, F.; He, Q.; Li, G.; Ding, G. lncRNA UCA1 Functions as a ceRNA to Promote Prostate Cancer Progression via Sponging miR143. Mol. Ther. Nucleic Acids 2020, 19, 751–758. [Google Scholar] [CrossRef]
- Beltrán-Anaya, F.O.; Cedro-Tanda, A.; Hidalgo-Miranda, A.; Romero-Cordoba, S.L. Insights into the Regulatory Role of Non-coding RNAs in Cancer Metabolism. Front. Physiol. 2016, 7, 342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Sun, X. Non-Coding RNAs Operate in the Crosstalk Between Cancer Metabolic Reprogramming and Metastasis. Front. Oncol. 2020, 10, 810. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Croce, C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer 2006, 6, 857–866. [Google Scholar] [CrossRef] [PubMed]
- Amodio, N.; Stamato, M.A.; Juli, G.; Morelli, E.; Fulciniti, M.; Manzoni, M.; Taiana, E.; Agnelli, L.; Cantafio, M.E.G.; Romeo, E.; et al. Drugging the lncRNA MALAT1 via LNA gapmeR ASO inhibits gene expression of proteasome subunits and triggers anti-multiple myeloma activity. Leukemia 2018, 32, 1948–1957. [Google Scholar] [CrossRef] [Green Version]
- Mozdarani, H.; Ezzatizadeh, V.; Rahbar Parvaneh, R. The emerging role of the long non-coding RNA HOTAIR in breast cancer development and treatment. J. Transl. Med. 2020, 18, 152. [Google Scholar] [CrossRef] [Green Version]
- Hung, C.L.; Wang, L.Y.; Yu, Y.L.; Chen, H.W.; Srivastava, S.; Petrovics, G.; Kung, H.J. A long noncoding RNA connects c-Myc to tumor metabolism. Proc. Natl. Acad. Sci. USA 2014, 111, 18697–18702. [Google Scholar] [CrossRef] [Green Version]
- Shankaraiah, R.C.; Veronese, A.; Sabbioni, S.; Negrini, M. Non-coding RNAs in the reprogramming of glucose metabolism in cancer. Cancer Lett. 2018, 419, 167–174. [Google Scholar] [CrossRef]
- Sun, H.; Huang, Z.; Sheng, W.; Xu, M.D. Emerging roles of long non-coding RNAs in tumor metabolism. J. Hematol. Oncol. 2018, 11, 106. [Google Scholar] [CrossRef]
- Al Tameemi, W.; Dale, T.P.; Al-Jumaily, R.M.K.; Forsyth, N.R. Hypoxia-Modified Cancer Cell Metabolism. Front. Cell Dev. Biol. 2019, 7, 4. [Google Scholar] [CrossRef] [Green Version]
- Yang, F.; Zhang, H.; Mei, Y.; Wu, M. Reciprocal regulation of HIF-1α and lincRNA-p21 modulates the Warburg effect. Mol. Cell 2014, 53, 88–100. [Google Scholar] [CrossRef] [Green Version]
- Khan, M.R.; Xiang, S.; Song, Z.; Wu, M. The p53-inducible long noncoding RNA TRINGS protects cancer cells from necrosis under glucose starvation. EMBO J. 2017, 36, 3483–3500. [Google Scholar] [CrossRef]
- Tang, J.; Yan, T.; Bao, Y.; Shen, C.; Yu, C.; Zhu, X.; Tian, X.; Guo, F.; Liang, Q.; Liu, Q.; et al. LncRNA GLCC1 promotes colorectal carcinogenesis and glucose metabolism by stabilizing c-Myc. Nat. Commun. 2019, 10, 3499. [Google Scholar] [CrossRef]
- Chen, Z.; Li, Y.; Zhang, H.; Huang, P.; Luthra, R. Hypoxia-regulated microRNA-210 modulates mitochondrial function and decreases ISCU and COX10 expression. Oncogene 2010, 29, 4362–4368. [Google Scholar] [CrossRef] [Green Version]
- Benard, G.; Bellance, N.; Jose, C.; Melser, S.; Nouette-Gaulain, K.; Rossignol, R. Multi-site control and regulation of mitochondrial energy production. Biochim. Biophys. Acta 2010, 1797, 698–709. [Google Scholar] [CrossRef] [Green Version]
- Ebi, H.; Sato, T.; Sugito, N.; Hosono, Y.; Yatabe, Y.; Matsuyama, Y.; Yamaguchi, T.; Osada, H.; Suzuki, M.; Takahashi, T. Counterbalance between RB inactivation and miR-17-92 overexpression in reactive oxygen species and DNA damage induction in lung cancers. Oncogene 2009, 28, 3371–3379. [Google Scholar] [CrossRef] [Green Version]
- Mateescu, B.; Batista, L.; Cardon, M.; Gruosso, T.; de Feraudy, Y.; Mariani, O.; Nicolas, A.; Meyniel, J.P.; Cottu, P.; Sastre-Garau, X.; et al. miR-141 and miR-200a act on ovarian tumorigenesis by controlling oxidative stress response. Nat. Med. 2011, 17, 1627–1635. [Google Scholar] [CrossRef]
- Serguienko, A.; Grad, I.; Wennerstrøm, A.B.; Meza-Zepeda, L.A.; Thiede, B.; Stratford, E.W.; Myklebost, O.; Munthe, E. Metabolic reprogramming of metastatic breast cancer and melanoma by let-7a microRNA. Oncotarget 2015, 6, 2451–2465. [Google Scholar] [CrossRef] [Green Version]
- Ma, X.; Li, C.; Sun, L.; Huang, D.; Li, T.; He, X.; Wu, G.; Yang, Z.; Zhong, X.; Song, L.; et al. Lin28/let-7 axis regulates aerobic glycolysis and cancer progression via PDK1. Nat. Commun. 2014, 5, 5212. [Google Scholar] [CrossRef] [Green Version]
- Li, H.J.; Li, X.; Pang, H.; Pan, J.J.; Xie, X.J.; Chen, W. Long non-coding RNA UCA1 promotes glutamine metabolism by targeting miR-16 in human bladder cancer. Jpn. J. Clin. Oncol. 2015, 45, 1055–1063. [Google Scholar] [CrossRef] [Green Version]
- Kamarajugadda, S.; Stemboroski, L.; Cai, Q.; Simpson, N.E.; Nayak, S.; Tan, M.; Lu, J. Glucose oxidation modulates anoikis and tumor metastasis. Mol. Cell. Biol. 2012, 32, 1893–1907. [Google Scholar] [CrossRef] [Green Version]
- Ito, H.; Duxbury, M.; Zinner, M.J.; Ashley, S.W.; Whang, E.E. Glucose transporter-1 gene expression is associated with pancreatic cancer invasiveness and MMP-2 activity. Surgery 2004, 136, 548–556. [Google Scholar] [CrossRef]
- Ellis, B.C.; Graham, L.D.; Molloy, P.L. CRNDE, a long non-coding RNA responsive to insulin/IGF signaling, regulates genes involved in central metabolism. Biochim. Biophys. Acta 2014, 1843, 372–386. [Google Scholar] [CrossRef] [Green Version]
- Lin, A.; Li, C.; Xing, Z.; Hu, Q.; Liang, K.; Han, L.; Wang, C.; Hawke, D.H.; Wang, S.; Zhang, Y.; et al. The LINK-A lncRNA activates normoxic HIF1α signalling in triple-negative breast cancer. Nat. Cell Biol. 2016, 18, 213–224. [Google Scholar] [CrossRef]
- Fong, M.Y.; Zhou, W.; Liu, L.; Alontaga, A.Y.; Chandra, M.; Ashby, J.; Chow, A.; O’Connor, S.T.; Li, S.; Chin, A.R.; et al. Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat. Cell Biol. 2015, 17, 183–194. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Wang, S.; Wang, H.; Cao, J.; Huang, X.; Chen, Z.; Xu, P.; Sun, G.; Xu, J.; Lv, J.; et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol. Cancer 2019, 18, 20. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Han, S.; Lei, K.; Chang, X.; Wang, K.; Li, Z.; Liu, J. Anti-Warburg effect of rosmarinic acid via miR-155 in colorectal carcinoma cells. Eur. J. Cancer Prev. 2016, 25, 481–489. [Google Scholar] [CrossRef]
- Hua, S.; Lei, L.; Deng, L.; Weng, X.; Liu, C.; Qi, X.; Wang, S.; Zhang, D.; Zou, X.; Cao, C.; et al. miR-139-5p inhibits aerobic glycolysis, cell proliferation, migration, and invasion in hepatocellular carcinoma via a reciprocal regulatory interaction with ETS1. Oncogene 2018, 37, 1624–1636. [Google Scholar] [CrossRef]
- Peschiaroli, A.; Giacobbe, A.; Formosa, A.; Markert, E.K.; Bongiorno-Borbone, L.; Levine, A.J.; Candi, E.; D’Alessandro, A.; Zolla, L.; Finazzi Agrò, A.; et al. miR-143 regulates hexokinase 2 expression in cancer cells. Oncogene 2013, 32, 797–802. [Google Scholar] [CrossRef]
- Siu, M.K.Y.; Jiang, Y.X.; Wang, J.J.; Leung, T.H.Y.; Han, C.Y.; Tsang, B.K.; Cheung, A.N.Y.; Ngan, H.Y.S.; Chan, K.K.L. Hexokinase 2 Regulates Ovarian Cancer Cell Migration, Invasion and Stemness via FAK/ERK1/2/MMP9/NANOG/SOX9 Signaling Cascades. Cancers 2019, 11, 813. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.H.; Wu, M.H.; Huang, Y.H.; Yeh, C.T.; Cheng, M.L.; Chi, H.C.; Tsai, C.Y.; Chung, I.H.; Chen, C.Y.; Lin, K.H. Taurine up-regulated gene 1 functions as a master regulator to coordinate glycolysis and metastasis in hepatocellular carcinoma. Hepatology 2018, 67, 188–203. [Google Scholar] [CrossRef]
- Chen, J.; Yu, Y.; Li, H.; Hu, Q.; Chen, X.; He, Y.; Xue, C.; Ren, F.; Ren, Z.; Li, J.; et al. Long non-coding RNA PVT1 promotes tumor progression by regulating the miR-143/HK2 axis in gallbladder cancer. Mol. Cancer 2019, 18, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fabani, M.M.; Gait, M.J. miR-122 targeting with LNA/2’-O-methyl oligonucleotide mixmers, peptide nucleic acids (PNA), and PNA-peptide conjugates. RNA 2008, 14, 336–346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; Zhao, X.; Zhou, Y.; Hu, Y. miR-124, miR-137 and miR-340 regulate colorectal cancer growth via inhibition of the Warburg effect. Oncol. Rep. 2012, 28, 1346–1352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bian, Z.; Zhang, J.; Li, M.; Feng, Y.; Wang, X.; Yao, S.; Jin, G.; Du, J.; Han, W.; Yin, Y.; et al. LncRNA-FEZF1-AS1 Promotes Tumor Proliferation and Metastasis in Colorectal Cancer by Regulating PKM2 Signaling. Clin. Cancer Res. 2018, 24, 4808–4819. [Google Scholar] [CrossRef] [Green Version]
- Kefas, B.; Comeau, L.; Erdle, N.; Montgomery, E.; Amos, S.; Purow, B. Pyruvate kinase M2 is a target of the tumor-suppressive microRNA-326 and regulates the survival of glioma cells. Neuro-oncology 2010, 12, 1102–1112. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Wang, H.; Liu, A.; Fang, C.; Hao, J.; Wang, Z. Lactate dehydrogenase A negatively regulated by miRNAs promotes aerobic glycolysis and is increased in colorectal cancer. Oncotarget 2015, 6, 19456–19468. [Google Scholar] [CrossRef]
- Li, L.; Kang, L.; Zhao, W.; Feng, Y.; Liu, W.; Wang, T.; Mai, H.; Huang, J.; Chen, S.; Liang, Y.; et al. miR-30a-5p suppresses breast tumor growth and metastasis through inhibition of LDHA-mediated Warburg effect. Cancer Lett. 2017, 400, 89–98. [Google Scholar] [CrossRef]
- Isozaki, Y.; Hoshino, I.; Nohata, N.; Kinoshita, T.; Akutsu, Y.; Hanari, N.; Mori, M.; Yoneyama, Y.; Akanuma, N.; Takeshita, N.; et al. Identification of novel molecular targets regulated by tumor suppressive miR-375 induced by histone acetylation in esophageal squamous cell carcinoma. Int. J. Oncol. 2012, 41, 985–994. [Google Scholar] [CrossRef]
- Kinoshita, T.; Nohata, N.; Yoshino, H.; Hanazawa, T.; Kikkawa, N.; Fujimura, L.; Chiyomaru, T.; Kawakami, K.; Enokida, H.; Nakagawa, M.; et al. Tumor suppressive microRNA-375 regulates lactate dehydrogenase B in maxillary sinus squamous cell carcinoma. Int. J. Oncol. 2012, 40, 185–193. [Google Scholar] [CrossRef]
- Yang, B.; Zhang, L.; Cao, Y.; Chen, S.; Cao, J.; Wu, D.; Chen, J.; Xiong, H.; Pan, Z.; Qiu, F.; et al. Overexpression of lncRNA IGFBP4-1 reprograms energy metabolism to promote lung cancer progression. Mol. Cancer 2017, 16, 154. [Google Scholar] [CrossRef]
- Sato, H.; Sakaeda, M.; Ishii, J.; Kashiwagi, K.; Shimoyamada, H.; Okudela, K.; Tajiri, M.; Ohmori, T.; Ogura, T.; Woo, T.; et al. Insulin-like growth factor binding protein-4 gene silencing in lung adenocarcinomas. Pathol. Int. 2011, 61, 19–27. [Google Scholar] [CrossRef]
- Price, W.A.; Moats-Staats, B.M.; Stiles, A.D. Insulin-like growth factor-I (IGF-I) regulates IGFBP-3 and IGFBP-4 by multiple mechanisms in A549 human adenocarcinoma cells. Am. J. Respir. Cell Mol. Biol. 1995, 13, 466–476. [Google Scholar] [CrossRef]
- Noll, K.; Wegmann, B.R.; Havemann, K.; Jaques, G. Insulin-like growth factors stimulate the release of insulin-like growth factor-binding protein-3 (IGFBP-3) and degradation of IGFBP-4 in nonsmall cell lung cancer cell lines. J. Clin. Endocrinol. Metab. 1996, 81, 2653–2662. [Google Scholar] [CrossRef] [Green Version]
- Xiao, Z.D.; Han, L.; Lee, H.; Zhuang, L.; Zhang, Y.; Baddour, J.; Nagrath, D.; Wood, C.G.; Gu, J.; Wu, X.; et al. Energy stress-induced lncRNA FILNC1 represses c-Myc-mediated energy metabolism and inhibits renal tumor development. Nat. Commun. 2017, 8, 783. [Google Scholar] [CrossRef]
- Hu, J.; Locasale, J.W.; Bielas, J.H.; O’Sullivan, J.; Sheahan, K.; Cantley, L.C.; Vander Heiden, M.G.; Vitkup, D. Heterogeneity of tumor-induced gene expression changes in the human metabolic network. Nat. Biotechnol. 2013, 31, 522–529. [Google Scholar] [CrossRef]
- Dupuy, F.; Tabariès, S.; Andrzejewski, S.; Dong, Z.; Blagih, J.; Annis, M.G.; Omeroglu, A.; Gao, D.; Leung, S.; Amir, E.; et al. PDK1-Dependent Metabolic Reprogramming Dictates Metastatic Potential in Breast Cancer. Cell Metab. 2015, 22, 577–589. [Google Scholar] [CrossRef] [Green Version]
- Ge, Y.; Yan, X.; Jin, Y.; Yang, X.; Yu, X.; Zhou, L.; Han, S.; Yuan, Q.; Yang, M. MiRNA-192 [corrected] and miRNA-204 Directly Suppress lncRNA HOTTIP and Interrupt GLS1-Mediated Glutaminolysis in Hepatocellular Carcinoma. PLoS Genet. 2015, 11, e1005726. [Google Scholar] [CrossRef]
- Deng, S.J.; Chen, H.Y.; Zeng, Z.; Deng, S.; Zhu, S.; Ye, Z.; He, C.; Liu, M.L.; Huang, K.; Zhong, J.X.; et al. Nutrient Stress-Dysregulated Antisense lncRNA GLS-AS Impairs GLS-Mediated Metabolism and Represses Pancreatic Cancer Progression. Cancer Res. 2019, 79, 1398–1412. [Google Scholar] [CrossRef]
- Zhang, N.; Zhang, H.; Liu, Y.; Su, P.; Zhang, J.; Wang, X.; Sun, M.; Chen, B.; Zhao, W.; Wang, L.; et al. SREBP1, targeted by miR-18a-5p, modulates epithelial-mesenchymal transition in breast cancer via forming a co-repressor complex with Snail and HDAC1/2. Cell Death Differ. 2019, 26, 843–859. [Google Scholar] [CrossRef]
- Zheng, F.; Chen, J.; Zhang, X.; Wang, Z.; Chen, J.; Lin, X.; Huang, H.; Fu, W.; Liang, J.; Wu, W.; et al. The HIF-1alpha antisense long non-coding RNA drives a positive feedback loop of HIF-1alpha mediated transactivation and glycolysis. Nat. Commun. 2021, 12, 1341. [Google Scholar] [CrossRef]
- Xing, Z.; Zhang, Y.; Liang, K.; Yan, L.; Xiang, Y.; Li, C.; Hu, Q.; Jin, F.; Putluri, V.; Putluri, N.; et al. Expression of Long Noncoding RNA YIYA Promotes Glycolysis in Breast Cancer. Cancer Res. 2018, 78, 4524–4532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, W.; Xia, J.; Xie, S.; Zou, R.; Pan, S.; Wang, Z.W.; Assaraf, Y.G.; Zhu, X. Long non-coding RNAs as a determinant of cancer drug resistance: Towards the overcoming of chemoresistance via modulation of lncRNAs. Drug Resist. Updat. 2020, 50, 100683. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Hou, M.S.; Zhan, Y.; Shen, X.B.; Xue, H.Y. MALAT1 promotes cisplatin resistance in cervical cancer by activating the PI3K/AKT pathway. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 7653–7659. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.C.; Zhang, J.Q.; Jiang, X.L.; Hua, Y.Y.; Xie, S.W.; Qin, Y.A.; Yang, Y.J. LncRNA CRNDE facilitates epigenetic suppression of CELF2 and LATS2 to promote proliferation, migration and chemoresistance in hepatocellular carcinoma. Cell Death Dis. 2020, 11, 676. [Google Scholar] [CrossRef]
- Zhu, L.L.; Wu, Z.; Li, R.K.; Xing, X.; Jiang, Y.S.; Li, J.; Wang, Y.H.; Hu, L.P.; Wang, X.; Qin, W.T.; et al. Deciphering the genomic and lncRNA landscapes of aerobic glycolysis identifies potential therapeutic targets in pancreatic cancer. Int. J. Biol. Sci. 2021, 17, 107–118. [Google Scholar] [CrossRef]
- Ginn, S.L.; Alexander, I.E.; Edelstein, M.L.; Abedi, M.R.; Wixon, J. Gene therapy clinical trials worldwide to 2012 an update. J. Gene. Med. 2013, 15, 65–77. [Google Scholar] [CrossRef]
- Pedram Fatemi, R.; Salah-Uddin, S.; Modarresi, F.; Khoury, N.; Wahlestedt, C.; Faghihi, M.A. Screening for Small-Molecule Modulators of Long Noncoding RNA-Protein Interactions Using AlphaScreen. J. Biomol. Screen 2015, 20, 1132–1141. [Google Scholar] [CrossRef] [Green Version]
- Mercatelli, N.; Fortini, D.; Palombo, R.; Paronetto, M.P. Small molecule inhibition of Ewing sarcoma cell growth via targeting the long non coding RNA HULC. Cancer Lett. 2020, 469, 111–123. [Google Scholar] [CrossRef]
- Gong, N.; Teng, X.; Li, J.; Liang, X.J. Antisense Oligonucleotide-Conjugated Nanostructure-Targeting lncRNA MALAT1 Inhibits Cancer Metastasis. ACS Appl. Mater. Interfaces 2019, 11, 37–42. [Google Scholar] [CrossRef]
- Ren, Y.; Li, R.Q.; Cai, Y.R.; Xia, T.; Yang, M.; Xu, F.J. Effective Codelivery of lncRNA and pDNA by Pullulan-Based Nanovectors for Promising Therapy of Hepatocellular Carcinoma. Adv. Funct. Mater. 2016, 26, 7314–7325. [Google Scholar] [CrossRef]
- Zhuo, W.; Liu, Y.; Li, S.; Guo, D.; Sun, Q.; Jin, J.; Rao, X.; Li, M.; Sun, M.; Jiang, M.; et al. Long Noncoding RNA GMAN, Up-regulated in Gastric Cancer Tissues, Is Associated With Metastasis in Patients and Promotes Translation of Ephrin A1 by Competitively Binding GMAN-AS. Gastroenterology 2019, 156, 676–691. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sellitto, A.; Pecoraro, G.; Giurato, G.; Nassa, G.; Rizzo, F.; Saggese, P.; Martinez, C.A.; Scafoglio, C.; Tarallo, R. Regulation of Metabolic Reprogramming by Long Non-Coding RNAs in Cancer. Cancers 2021, 13, 3485. https://doi.org/10.3390/cancers13143485
Sellitto A, Pecoraro G, Giurato G, Nassa G, Rizzo F, Saggese P, Martinez CA, Scafoglio C, Tarallo R. Regulation of Metabolic Reprogramming by Long Non-Coding RNAs in Cancer. Cancers. 2021; 13(14):3485. https://doi.org/10.3390/cancers13143485
Chicago/Turabian StyleSellitto, Assunta, Giovanni Pecoraro, Giorgio Giurato, Giovanni Nassa, Francesca Rizzo, Pasquale Saggese, Cesar A. Martinez, Claudio Scafoglio, and Roberta Tarallo. 2021. "Regulation of Metabolic Reprogramming by Long Non-Coding RNAs in Cancer" Cancers 13, no. 14: 3485. https://doi.org/10.3390/cancers13143485
APA StyleSellitto, A., Pecoraro, G., Giurato, G., Nassa, G., Rizzo, F., Saggese, P., Martinez, C. A., Scafoglio, C., & Tarallo, R. (2021). Regulation of Metabolic Reprogramming by Long Non-Coding RNAs in Cancer. Cancers, 13(14), 3485. https://doi.org/10.3390/cancers13143485






