TBC1D9: An Important Modulator of Tumorigenesis in Breast Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Public BC Datasets Analysis
2.2. Breast Tissue Samples Selection and Tissue Microarray (TMA) Preparation
2.3. Immunohistochemistry (IHC) Analysis and Scoring
2.4. Cell Lines
2.5. RNA Extraction and Quantitative Real-Time PCR (qPCR) Analysis
2.6. Lentiviral Production and Infection
2.7. Proliferation
2.8. Migration Assay
2.9. Three-Dimensional Spheroid Formation Assay
2.10. CAM Assay
2.11. Statistical Analysis
3. Results
3.1. Low TBC1D9 Expression Correlates with Poor Survival Outcomes
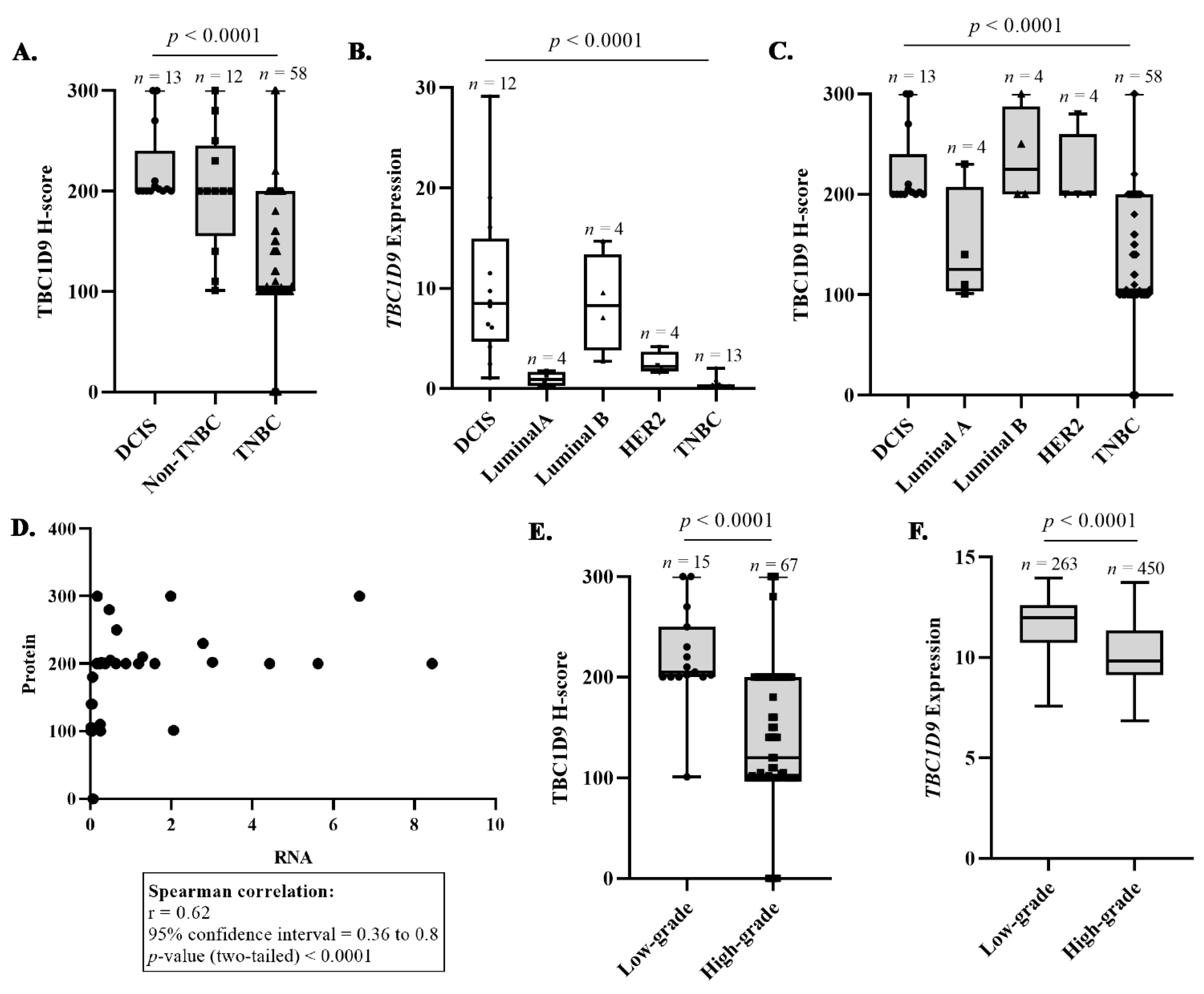
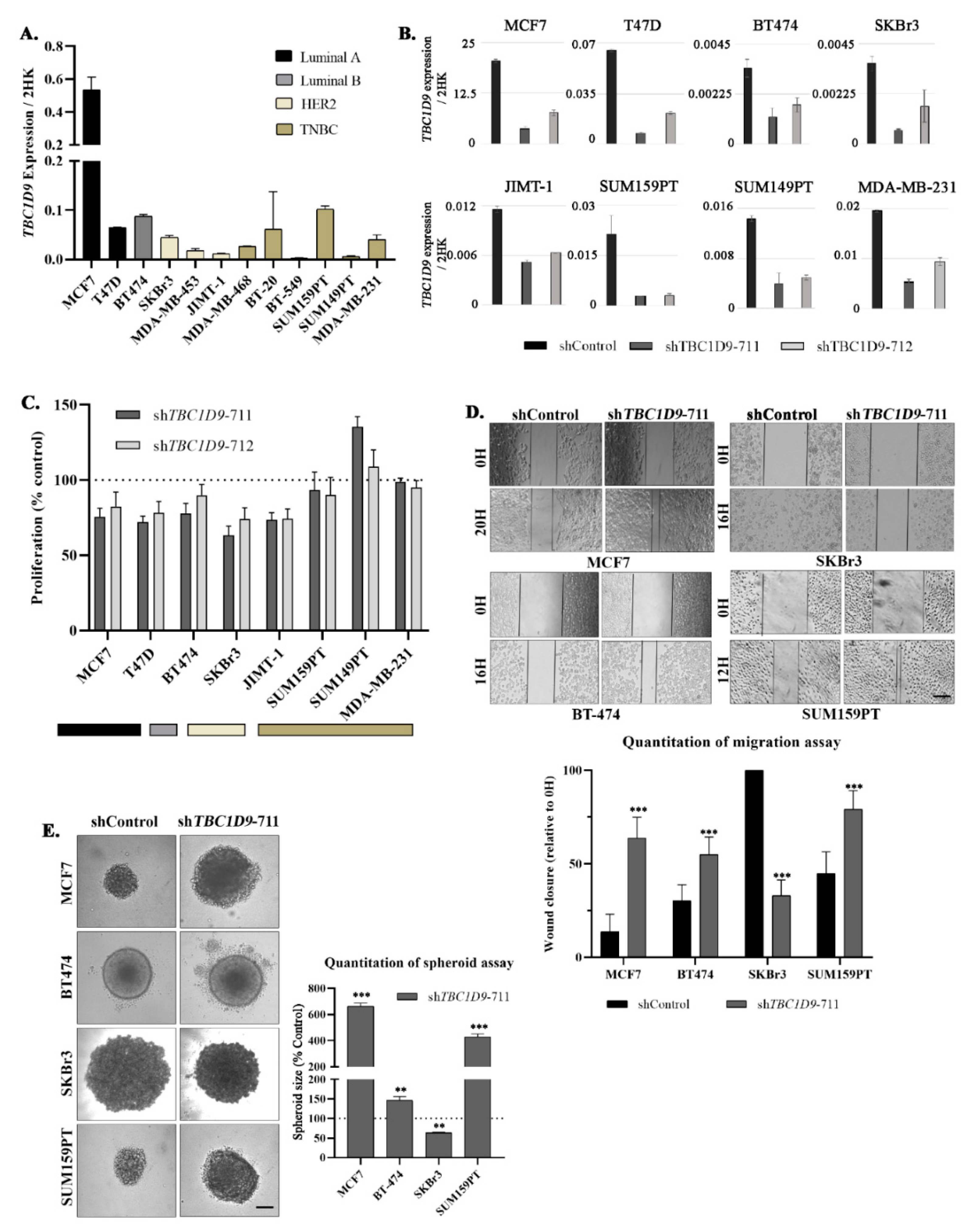
3.2. Confirmation That TBC1D9 Can Differentiate TNBC from Other BCs
3.3. TBC1D9 Expression Inversely Correlates with Grade and Tumor Size
3.4. TBC1D9 KD Increases Cell Migration and 3D Spheroid Formation in Luminal and TNBC Cell Lines
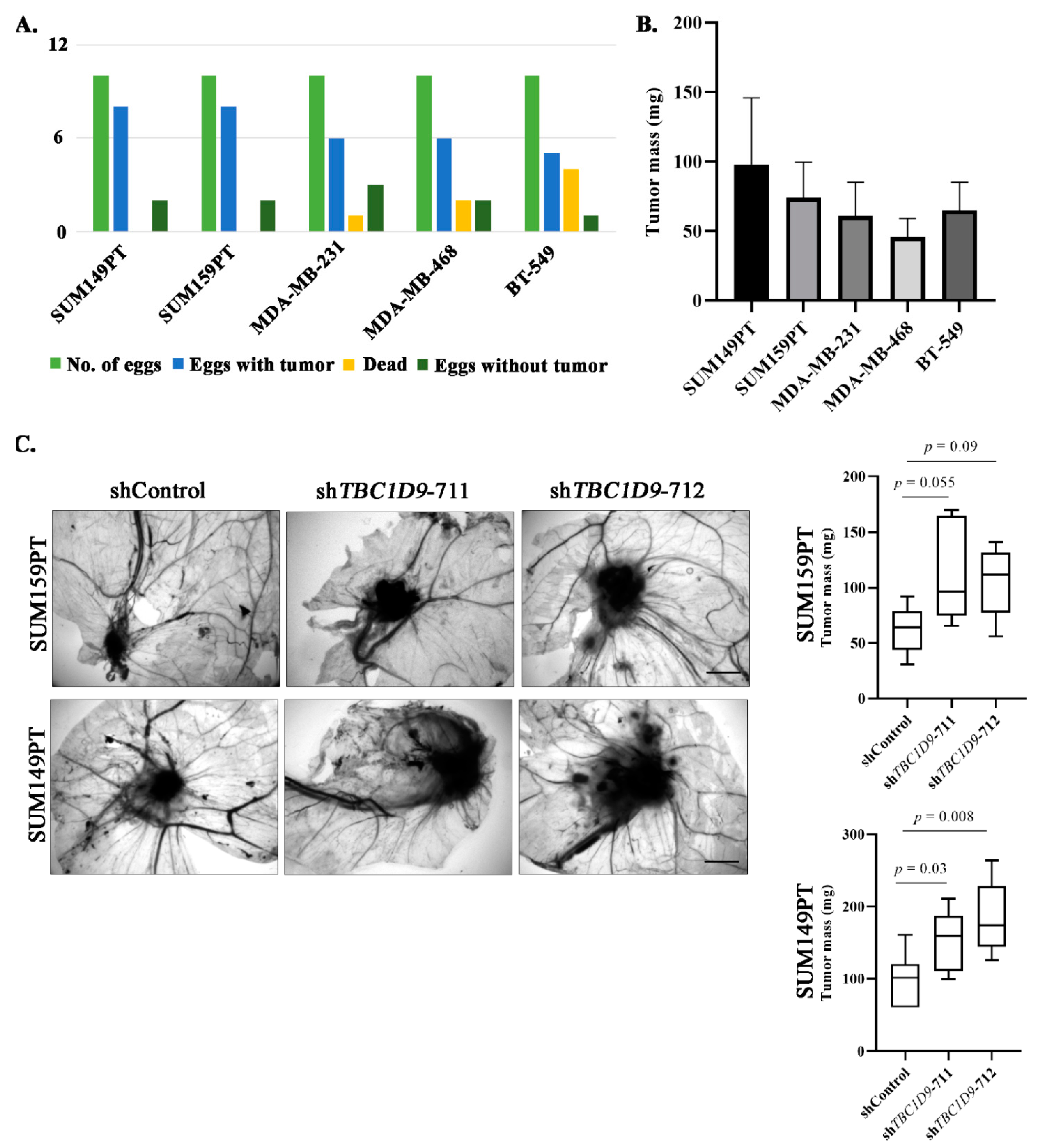
3.5. TBC1D9 KD Increases Tumor Growth of BC Cell Lines In Vivo
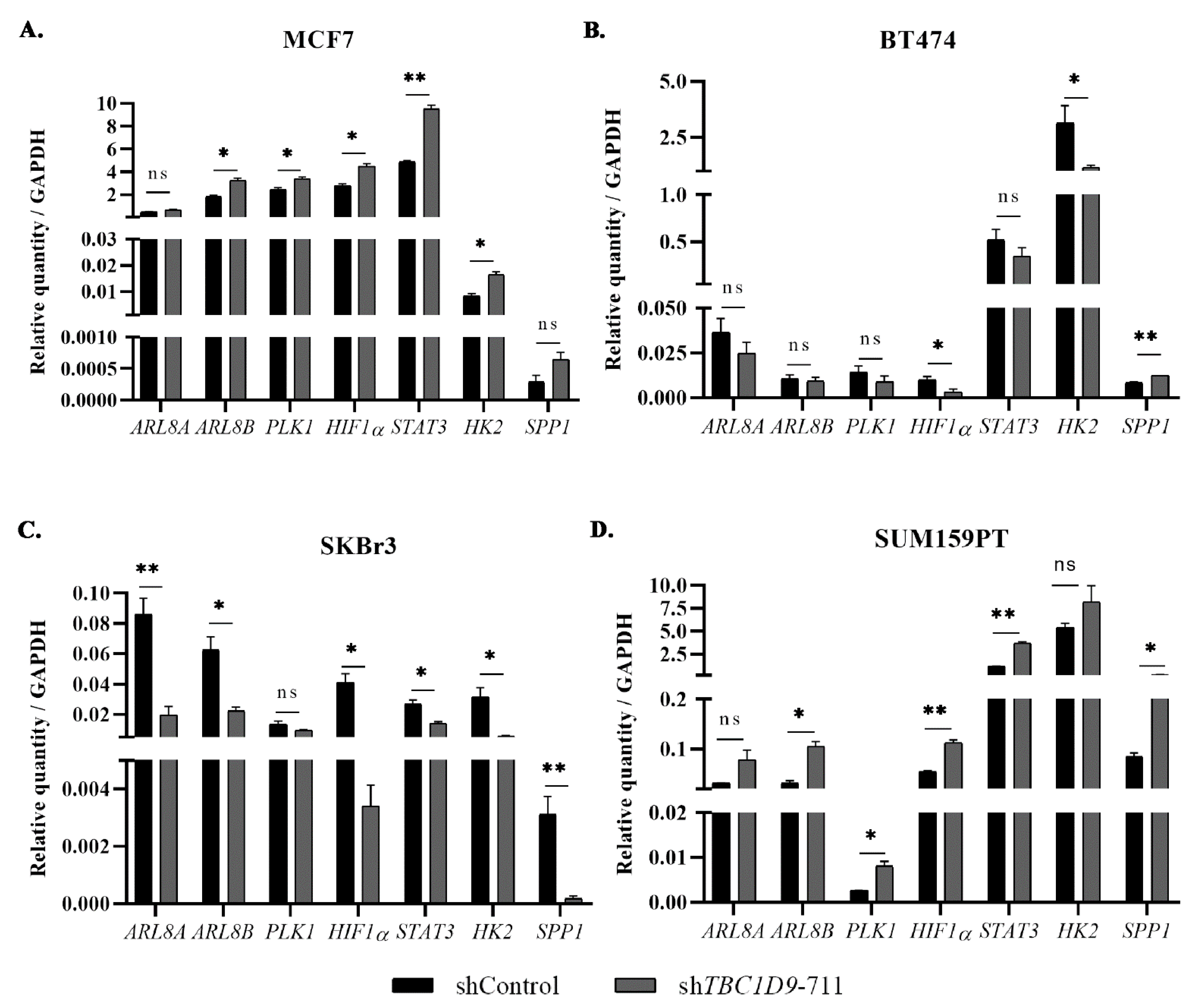
3.6. The Effect of TBC1D9 on BC Phenotypes Is Mediated by Genes Involved in Tumor Growth and Migration
4. Discussion
5. Conclusions
6. Future Perspectives
- It would be interesting to understand why TBC1D9 behaves differently in HER2-positive BC cell lines and how this information could be used in targeting HER2-positive BC.
- Is the difference in the altered oncogenic signaling upon KD of TBC1D9 in luminal A and luminal B due to the presence of HER2 in luminal B subtypes?
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- American Cancer Society. Breast Cancer Facts & Figures 2019–2020; American Cancer Society, Inc.: Atlanta, GA, USA, 2019. [Google Scholar]
- Saadatmand, S.; Bretveld, R.; Siesling, S.; A Tilanus-Linthorst, M.M. Influence of tumour stage at breast cancer detection on survival in modern times: Population based study in 173,797 patients. BMJ 2015, 351, h4901. [Google Scholar] [CrossRef] [Green Version]
- Kothari, C.; Diorio, C.; Durocher, F. Gene signatures of breast cancer development and the potential for novel targeted treatments. Pharmacogenomics 2020, 21, 157–161. [Google Scholar] [CrossRef] [Green Version]
- Al-Mahmood, S.; Sapiezynski, J.; Garbuzenko, O.B.; Minko, T. Metastatic and triple-negative breast cancer: Challenges and treatment options. Drug Deliv. Transl. Res. 2018, 8, 1483–1507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoeferlin, L.A.; Chalfant, C.E.; Park, M.A. Challenges in the Treatment of Triple Negative and HER2-Overexpressing Breast Cancer. J. Surg. Sci. 2013, 1, 3–7. [Google Scholar]
- Lehmann, B.D.; Bauer, J.A.; Chen, X.; Sanders, M.E.; Chakravarthy, A.B.; Shyr, Y.; Pietenpol, J.A. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Investig. 2011, 121, 2750–2767. [Google Scholar] [CrossRef] [Green Version]
- Kothari, C.; Osseni, M.A.; Agbo, L.; Ouellette, G.; Déraspe, M.; LaViolette, F.; Corbeil, J.; Lambert, J.-P.; Diorio, C.; Durocher, F. Machine learning analysis identifies genes differentiating triple negative breast cancers. Sci. Rep. 2020, 10, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Nozawa, T.; Sano, S.; Minowa-Nozawa, A.; Toh, H.; Nakajima, S.; Murase, K.; Aikawa, C.; Nakagawa, I. TBC1D9 regulates TBK1 activation through Ca2+ signaling in selective autophagy. Nat. Commun. 2020, 11, 770. [Google Scholar] [CrossRef] [PubMed]
- Andres, A.S.; Brock, G.N.; Wittliff, J.L. Interrogating differences in expression of targeted gene sets to predict breast cancer outcome. BMC Cancer 2013, 13, 326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, W.-Y.; Yang, T.-H.O.; Anastassiou, D. Biomolecular Events in Cancer Revealed by Attractor Metagenes. PLoS Comput. Biol. 2013, 9, e1002920. [Google Scholar] [CrossRef] [Green Version]
- Cyprian, F.S.; Akhtar, S.; Gatalica, Z.; Vranic, S. Targeted immunotherapy with a checkpoint inhibitor in combination with chemotherapy: A new clinical paradigm in the treatment of triple-negative breast cancer. Bosn. J. Basic Med Sci. 2019, 19, 227–233. [Google Scholar] [CrossRef] [Green Version]
- Schmid, P.; Adams, S.; Rugo, H.S.; Schneeweiss, A.; Barrios, C.H.; Iwata, H.; Diéras, V.; Hegg, R.; Im, S.A.; Shaw Wright, G.; et al. Atezolizumab and Nab-Paclitaxel in Advanced Triple-Negative Breast Cancer. N. Engl. J. Med. 2018, 379, 2108–2121. [Google Scholar] [CrossRef]
- Park, S.-J.; Yoon, B.-H.; Kim, S.-Y. GENT2: An updated gene expression database for normal and tumor tissues. BMC Med. Genom. 2019, 12, 101. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.V.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Györffy, B.; Lanczky, A.; Eklund, A.C.; Denkert, C.; Budczies, J.; Li, Q.; Szallasi, Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1809 patients. Breast Cancer Res. Treat. 2009, 123, 725–731. [Google Scholar] [CrossRef] [Green Version]
- Clemenceau, A.; Hanna, M.; Ennour-Idrissi, K.; Burguin, A.; Diorio, C.; Durocher, F. Secreted Frizzled-Related Protein 1 as a Biomarker against Incomplete Age-Related Lobular Involution and Microcalcifications’ Development. Cancers 2020, 12, 2693. [Google Scholar] [CrossRef] [PubMed]
- Burguin, A.; Furrer, D.; Ouellette, G.; Jacob, S.; Diorio, C.; Durocher, F. Trastuzumab effects depend on HER2 phosphorylation in HER2-negative breast cancer cell lines. PLoS ONE 2020, 15, e0234991. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Koo, J.S.; Park, H.S.; Kim, J.-H.; Choi, S.-Y.; Lee, J.H.; Park, B.-W.; Lee, K.S. Expression of androgen receptors in primary breast cancer. Ann. Oncol. 2010, 21, 488–492. [Google Scholar] [CrossRef] [PubMed]
- Detre, S.; Jotti, G.S.; Dowsett, M. A “quickscore” method for immunohistochemical semiquantitation: Validation for oestrogen receptor in breast carcinomas. J. Clin. Pathol. 1995, 48, 876–878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dowsett, M.; Nielsen, T.O.; A’Hern, R.; Bartlett, J.; Coombes, R.C.; Cuzick, J.; Ellis, M.; Henry, N.L.; Hugh, J.C.; Lively, T.; et al. Assessment of Ki67 in Breast Cancer: Recommendations from the International Ki67 in Breast Cancer Working Group. J. Natl. Cancer Inst. 2011, 103, 1656–1664. [Google Scholar] [CrossRef] [Green Version]
- Pouliot, M.-C.; Kothari, C.; Joly-Beauparlant, C.; Labrie, Y.; Ouellette, G.; Simard, J.; Droit, A.; Durocher, F. Transcriptional signature of lymphoblastoid cell lines of BRCA1, BRCA2 and non-BRCA1/2 high risk breast cancer families. Oncotarget 2017, 8, 78691–78712. [Google Scholar] [CrossRef] [Green Version]
- Kothari, C.; Ouellette, G.; Labrie, Y.; Jacob, S.; Diorio, C.; Durocher, F. Identification of a gene signature for different stages of breast cancer development that could be used for early diagnosis and specific therapy. Oncotarget 2018, 9, 37407–37420. [Google Scholar] [CrossRef] [Green Version]
- Luu-The, V.; Paquet, N.; Calvo, E.; Cumps, J. Improved real-time RT-PCR method for high-throughput measurements using second derivative calculation and double correction. Biotechniques 2005, 38, 287–293. [Google Scholar] [CrossRef]
- Kothari, C.; Clemenceau, A.; Ouellette, G.; Ennour-Idrissi, K.; Michaud, A.; Diorio, C.; Durocher, F. Is Carboxypeptidase B1 a Prognostic Marker for Ductal Carcinoma In Situ? Cancers 2021, 13, 1726. [Google Scholar] [CrossRef]
- Herrmann, A.; Moss, D.; Sée, V. The Chorioallantoic Membrane of the Chick Embryo to Assess Tumor Formation and Metastasis. Methods Mol. Biol. 2016, 1464, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Provencher, L.; Diorio, C.; Hogue, J.-C.; Doyle, C.; Jacob, S. Does breast cancer tumor size really matter that much? Breast 2012, 21, 682–685. [Google Scholar] [CrossRef] [PubMed]
- Nahed, A.S.; Shaimaa, M.Y. Ki-67 as a prognostic marker according to breast cancer molecular subtype. Cancer Biol. Med. 2016, 13, 496–504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, X.; Miao, H.; Jing, B.; Pan, Q.; Zhang, H.; Chen, Y.; Zhang, D.; Liang, Z.; Wen, Z.; Li, M. Claudin-4 controls the proliferation, apoptosis, migration and in vivo growth of MCF-7 breast cancer cells. Oncol. Rep. 2015, 34, 681–690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiwa, L.S.; Van Diest, P.J.; Hoefnagel, L.D.; Wesseling, J.; Wesseling, P.; Dutch Distant Breast Cancer Metastases Consortium; Moelans, C.B. Upregulation of Claudin-4, CAIX and GLUT-1 in distant breast cancer metastases. BMC Cancer 2014, 14, 864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Todd, M.C.; Petty, H.M.; King, J.; Marshall, B.N.P.; Sheller, R.A.; Cuevas, M.E. Overexpression and delocalization of claudin-3 protein in MCF-7 and MDA-MB-415 breast cancer cell lines. Oncol. Lett. 2015, 10, 156–162. [Google Scholar] [CrossRef] [Green Version]
- Kwon, M.J. Emerging Roles of Claudins in Human Cancer. Int. J. Mol. Sci. 2013, 14, 18148–18180. [Google Scholar] [CrossRef] [Green Version]
- Jang, M.H.; Kang, H.J.; Jang, K.S.; Paik, S.S.; Kim, W.S. Clinicopathological analysis of CD44 and CD24 expression in invasive breast cancer. Oncol. Lett. 2016, 12, 2728–2733. [Google Scholar] [CrossRef] [Green Version]
- Kristiansen, G.; Winzer, K.-J.; Mayordomo, E.; Bellach, J.; Schlüns, K.; Denkert, C.; Dahl, E.; Pilarsky, C.; Altevogt, P.; Guski, H.; et al. CD24 expression is a new prognostic marker in breast cancer. Clin. Cancer Res. 2003, 9, 4906–4913. [Google Scholar] [PubMed]
- Singletary, S.E.; Allred, C.; Ashley, P.; Bassett, L.W.; Berry, D.; Bland, K.I.; Borgen, P.I.; Clark, G.; Edge, S.B.; Hayes, D.F.; et al. Revision of the American Joint Committee on Cancer Staging System for Breast Cancer. J. Clin. Oncol. 2002, 20, 3628–3636. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Apple, S.; Zhao, H.; Song, J.; Lee, M.; Luo, W.; Wu, X.; Chung, D.; Pietras, R.J.; Chang, H.R. CD24 Expression and differential resistance to chemotherapy in triple-negative breast cancer. Oncotarget 2017, 8, 38294–38308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Zhang, Q.; Sun, C.; Huang, Y.; Zhang, J.; Wang, Q. Clinical relevance of ARF/ARL family genes and oncogenic function of ARL4C in endometrial cancer. Biomed. Pharmacother. 2020, 125, 110000. [Google Scholar] [CrossRef]
- Zhou, B.; Guo, R. Genomic and regulatory characteristics of significant transcription factors in colorectal cancer metastasis. Sci. Rep. 2018, 8, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Suarez-Arroyo, I.; Feliz-Mosquea, Y.R.; Pérez-Laspiur, J.; Arju, R.; Giashuddin, S.; Maldonado-Martínez, G.; Cubano, L.A.; Schneider, R.; Martinez-Montemayor, M. The proteome signature of the inflammatory breast cancer plasma membrane identifies novel molecular markers of disease. Am. J. Cancer Res. 2016, 6, 1720–1740. [Google Scholar]
- Montaudon, E.; Nikitorowicz-Buniak, J.; Sourd, L.; Morisset, L.; EL Botty, R.; Huguet, L.; Dahmani, A.; Painsec, P.; Nemati, F.; Vacher, S.; et al. PLK1 inhibition exhibits strong anti-tumoral activity in CCND1-driven breast cancer metastases with acquired palbociclib resistance. Nat. Commun. 2020, 11, 4053. [Google Scholar] [CrossRef]
- Giordano, A.; Liu, Y.; Armeson, K.; Park, Y.; Ridinger, M.; Erlander, M.; Reuben, J.; Britten, C.; Kappler, C.; Yeh, E.; et al. Polo-like kinase 1 (Plk1) inhibition synergizes with taxanes in triple negative breast cancer. PLoS ONE 2019, 14, e0224420. [Google Scholar] [CrossRef]
- Ueda, A.; Oikawa, K.; Fujita, K.; Ishikawa, A.; Sato, E.; Ishikawa, T.; Kuroda, M.; Kanekura, K. Therapeutic potential of PLK1 inhibition in triple-negative breast cancer. Lab. Investig. 2019, 99, 1275–1286. [Google Scholar] [CrossRef]
- Wu, P.-H.; Onodera, Y.; Giaccia, A.J.; Le, Q.-T.; Shimizu, S.; Shirato, H.; Nam, J.-M. Lysosomal trafficking mediated by Arl8b and BORC promotes invasion of cancer cells that survive radiation. Commun. Biol. 2020, 3, 1–15. [Google Scholar] [CrossRef]
- Dykes, S.S.; Gray, A.L.; Coleman, D.T.; Saxena, M.; Stephens, C.A.; Carroll, J.L.; Pruitt, K.; Cardelli, J.A. The Arf-like GTPase Arl8b is essential for three-dimensional invasive growth of prostate cancer in vitro and xenograft formation and growth in vivo. Oncotarget 2016, 7, 31037–31052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castello, L.M.; Raineri, D.; Salmi, L.; Clemente, N.; Vaschetto, R.; Quaglia, M.; Garzaro, M.; Gentilli, S.; Navalesi, P.; Canta-luppi, V.; et al. Osteopontin at the Crossroads of Inflammation and Tumor Progression. Available online: https://www.hindawi.com/journals/mi/2017/4049098/ (accessed on 3 February 2021).
- Rodrigues, L.R.; Teixeira, J.; Schmitt, F.; Paulsson, M.; Lindmark-Mänsson, H. The Role of Osteopontin in Tumor Progression and Metastasis in Breast Cancer. Cancer Epidemiol. Biomark. Prev. 2007, 16, 1087–1097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eremo, A.G.; Lagergren, K.; Othman, L.; Montgomery, S.; Andersson, G.; Tina, E. Evaluation of SPP1/osteopontin expression as predictor of recurrence in tamoxifen treated breast cancer. Sci. Rep. 2020, 10, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.-H.; Qin, L.; Li, X. Role of STAT3 signaling pathway in breast cancer. Cell Commun. Signal. 2020, 18, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Segatto, I.; Baldassarre, G.; Belletti, B. STAT3 in Breast Cancer Onset and Progression: A Matter of Time and Context. Int. J. Mol. Sci. 2018, 19, 2818. [Google Scholar] [CrossRef] [Green Version]
- Qin, J.-J.; Yan, L.; Zhang, J.; Zhang, W.-D. STAT3 as a potential therapeutic target in triple negative breast cancer: A systematic review. J. Exp. Clin. Cancer Res. 2019, 38, 1–16. [Google Scholar] [CrossRef]
- Li, Y.; Wang, Y.; Shi, Z.; Liu, J.; Zheng, S.; Yang, J.; Liu, Y.; Yang, Y.; Chang, F.; Yu, W. Clinicopathological and Prognostic Role of STAT3/p-STAT3 in Breast Cancer Patients in China: A Meta-Analysis. Sci. Rep. 2019, 9, 11243. [Google Scholar] [CrossRef]
- Patra, K.C.; Wang, Q.; Bhaskar, P.T.; Miller, L.; Wang, Z.; Wheaton, W.; Chandel, N.; Laakso, M.; Muller, W.J.; Allen, E.L.; et al. Hexokinase 2 Is Required for Tumor Initiation and Maintenance and Its Systemic Deletion Is Therapeutic in Mouse Models of Cancer. Cancer Cell 2013, 24, 213–228. [Google Scholar] [CrossRef] [Green Version]
- Wu, Z.; Wu, J.; Zhao, Q.; Fu, S.; Jin, J. Emerging roles of aerobic glycolysis in breast cancer. Clin. Transl. Oncol. 2020, 22, 631–646. [Google Scholar] [CrossRef]
- Zhang, T.; Zhu, X.; Wu, H.; Jiang, K.; Zhao, G.; Shaukat, A.; Deng, G.; Qiu, C. Targeting the ROS/PI3K/AKT/HIF-1α/HK2 axis of breast cancer cells: Combined administration of Polydatin and 2-Deoxy-d-glucose. J. Cell. Mol. Med. 2019, 23, 3711–3723. [Google Scholar] [CrossRef] [PubMed]
- De Francesco, E.M.; Maggiolini, M.; Musti, A.M. Crosstalk between Notch, HIF-1α and GPER in Breast Cancer EMT. Int. J. Mol. Sci. 2018, 19, 2011. [Google Scholar] [CrossRef] [Green Version]
- Dunn, L.K.; Mohammad, K.S.; Fournier, P.; McKenna, C.R.; Davis, H.W.; Niewolna, M.; Peng, X.H.; Chirgwin, J.M.; Guise, T.A. Hypoxia and TGF-β Drive Breast Cancer Bone Metastases through Parallel Signaling Pathways in Tumor Cells and the Bone Microenvironment. PLoS ONE 2009, 4, e6896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, J.; Wang, X.; Ran, L.; Song, J.; Luo, R.; Wang, Y. Hypoxia-Inducible Factor 1α Regulates the Transforming Growth Factor β1/SMAD Family Member 3 Pathway to Promote Breast Cancer Progression. J. Breast Cancer 2018, 21, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Samanta, D.; Lu, H.; Bullen, J.W.; Zhang, H.; Chen, I.; He, X.; Semenza, G.L. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m6A-demethylation of NANOG mRNA. Proc. Natl. Acad. Sci. USA 2016, 113, E2047–E2056. [Google Scholar] [CrossRef] [Green Version]

| All BCs (n = 83) | IDC (n = 70) | TNBC (n = 58) | Non-TNBC Including DCIS (n = 25) | Non-TNBC (n = 12) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Markers | Rho | p-Value | Rho | p-Value | Rho | p-Value | Rho | p-Value | Rho | p-Value |
| Tumor Size (mm) | −0.25 | 0.02 | −0.22 | 0.06 | −0.14 | 0.29 | −0.49 | 0.01 | −0.80 | 0.01 |
| Ki-67 percent staining | −0.29 | 0.008 | −0.11 | 0.34 | 0.03 | 0.83 | −0.04 | 0.85 | −0.04 | 0.52 |
| CD24 H-score | 0.20 | 0.07 | 0.27 | 0.02 | 0.36 | 0.006 | −0.09 | 0.66 | −0.09 | 0.85 |
| Claudin 3 H-score | 0.08 | 0.48 | 0.10 | 0.41 | 0.18 | 0.18 | 0.25 | 0.24 | 0.25 | 0.53 |
| Claudin 4 H-score | −0.11 | 0.34 | −0.02 | 0.84 | 0.04 | 0.76 | −0.15 | 0.46 | −0.15 | 0.83 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kothari, C.; Clemenceau, A.; Ouellette, G.; Ennour-Idrissi, K.; Michaud, A.; C.-Gaudreault, R.; Diorio, C.; Durocher, F. TBC1D9: An Important Modulator of Tumorigenesis in Breast Cancer. Cancers 2021, 13, 3557. https://doi.org/10.3390/cancers13143557
Kothari C, Clemenceau A, Ouellette G, Ennour-Idrissi K, Michaud A, C.-Gaudreault R, Diorio C, Durocher F. TBC1D9: An Important Modulator of Tumorigenesis in Breast Cancer. Cancers. 2021; 13(14):3557. https://doi.org/10.3390/cancers13143557
Chicago/Turabian StyleKothari, Charu, Alisson Clemenceau, Geneviève Ouellette, Kaoutar Ennour-Idrissi, Annick Michaud, René C.-Gaudreault, Caroline Diorio, and Francine Durocher. 2021. "TBC1D9: An Important Modulator of Tumorigenesis in Breast Cancer" Cancers 13, no. 14: 3557. https://doi.org/10.3390/cancers13143557
APA StyleKothari, C., Clemenceau, A., Ouellette, G., Ennour-Idrissi, K., Michaud, A., C.-Gaudreault, R., Diorio, C., & Durocher, F. (2021). TBC1D9: An Important Modulator of Tumorigenesis in Breast Cancer. Cancers, 13(14), 3557. https://doi.org/10.3390/cancers13143557






