Calcitriol Promotes Differentiation of Glioma Stem-Like Cells and Increases Their Susceptibility to Temozolomide
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
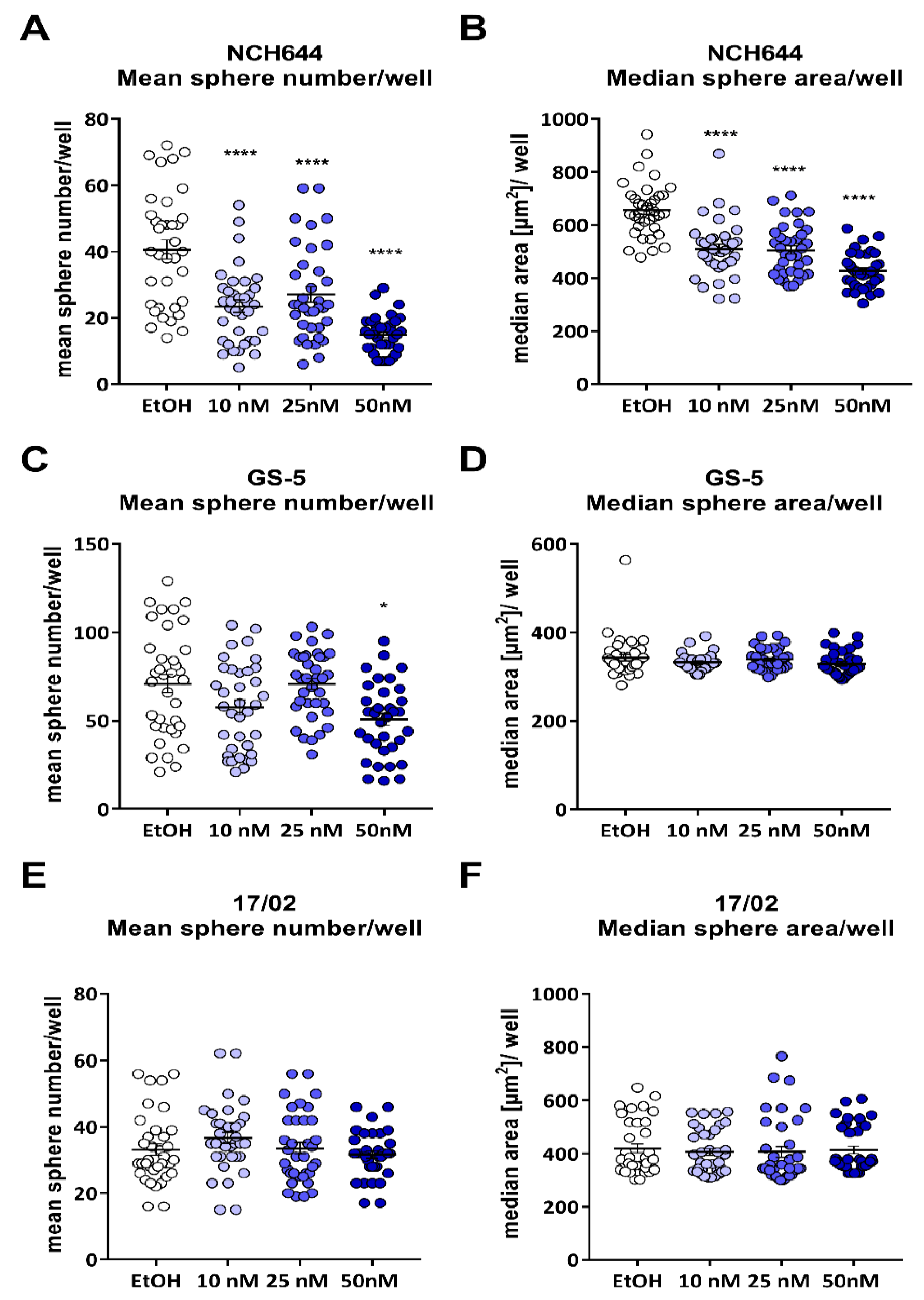
2.1. Calcitriol Reduces Stemness in GSCs In Vitro
2.2. Differential Activity of Calcitriol
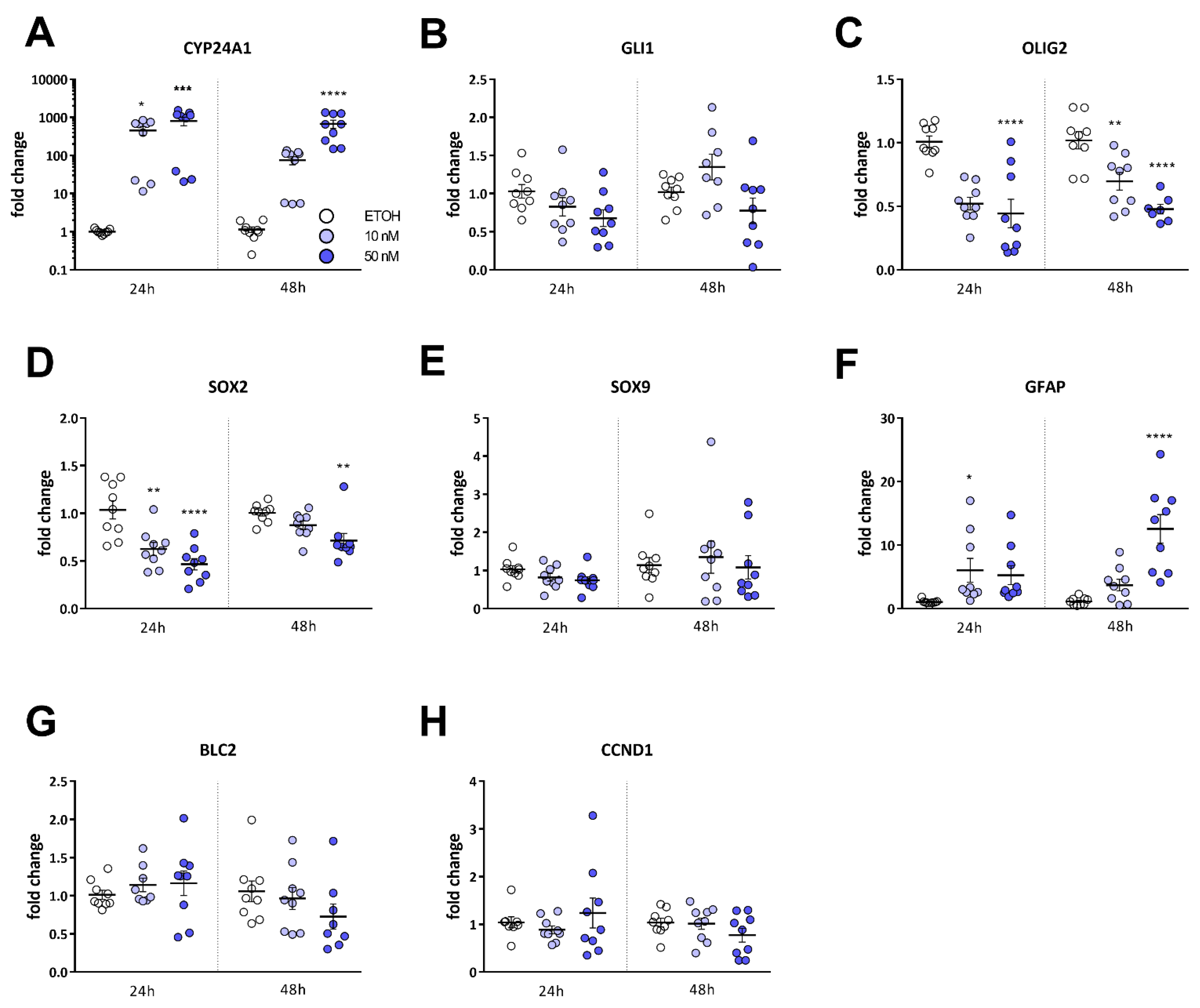
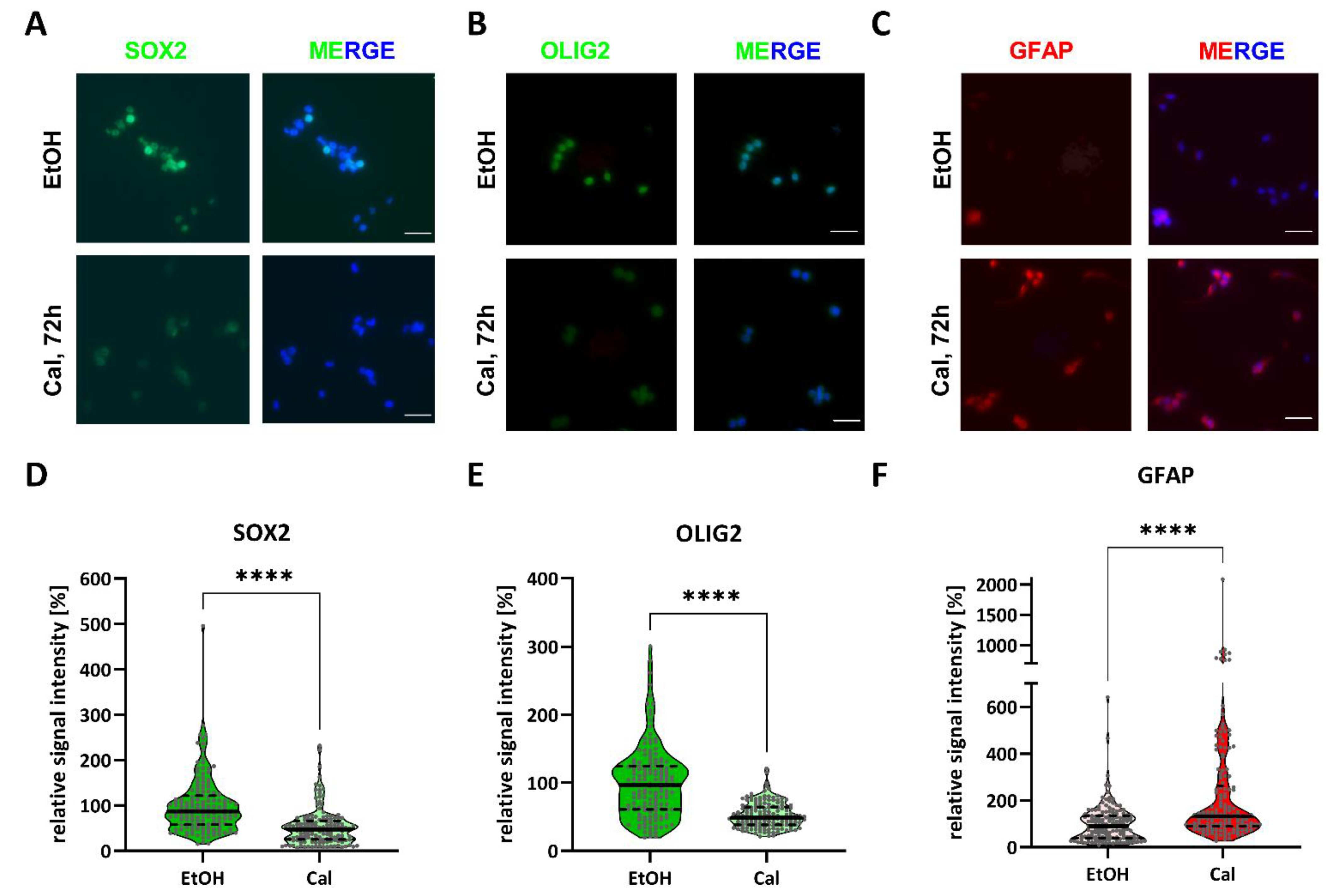
2.3. Calcitriol Reduces Stemness-Associated Marker Genes
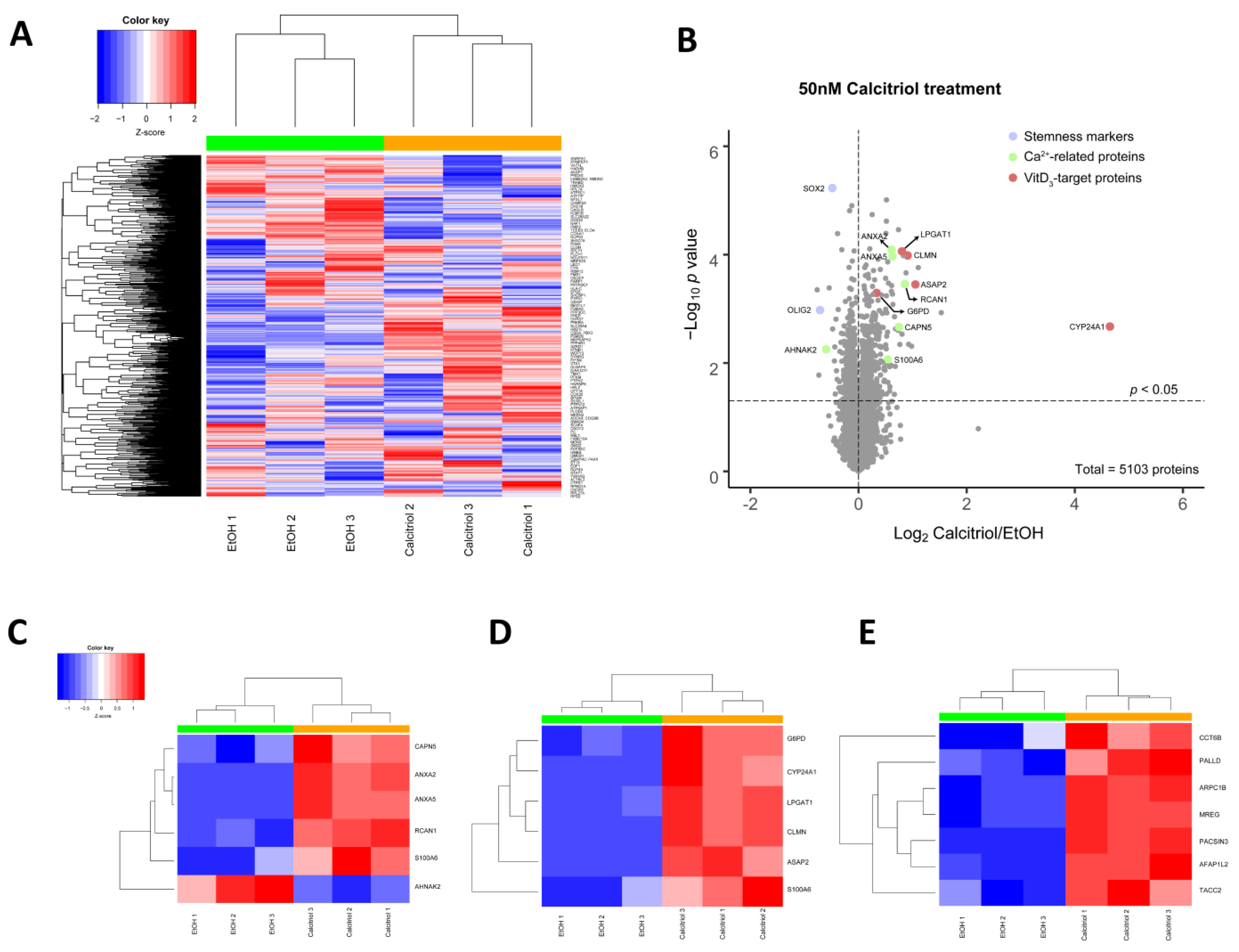
2.4. Alterations in Global Proteomic Profiles after Calcitriol Treatment
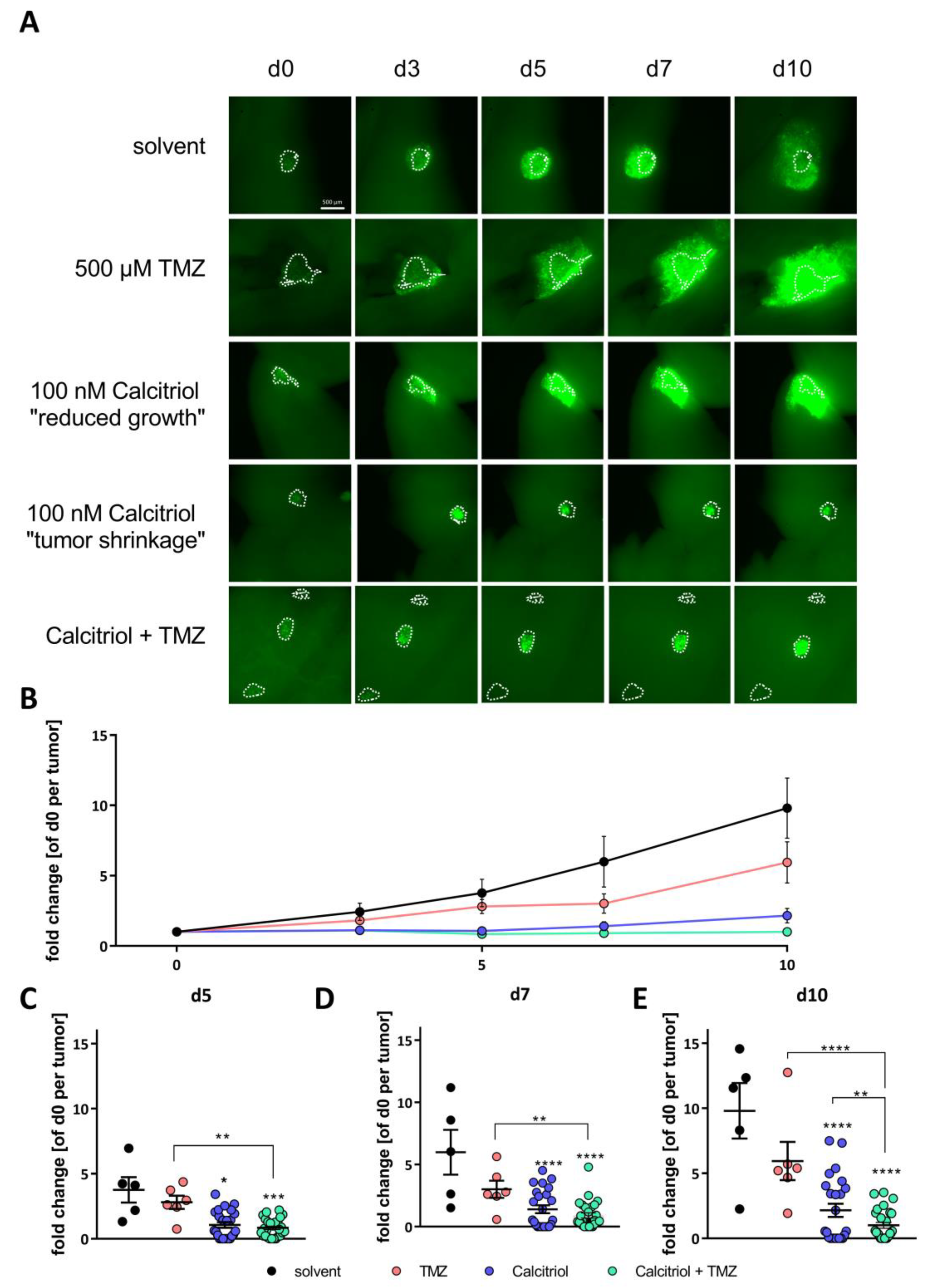
2.5. Calcitriol Reduces Tumor Growth Ex Vivo and Synergizes with TMZ
3. Discussion
4. Materials and Methods
4.1. Cells and Cell Culture
4.2. Compounds
4.3. Limiting Dilution Assay
4.4. Tagman-Based qRT-PCR
4.5. Immunofluorescence Staining
4.6. Proteomic Sample Preparation and Data Analyses
4.6.1. Sample Preparation for LC-MS2
4.6.2. Mass Spectrometry
4.6.3. Data Analysis
4.7. Lentiviral Transduction of GSCs
4.8. Adult Organotypic Slice Cultures and Ex Vivo Tumor Growth Assay
4.9. Statistics
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 world health organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed]
- Berger, T.R.; Wong, E.T. Tumor treating fields in neuro-oncology: Integration of alternating electric fields therapy into promising treatment strategies. Chin. Clin. Oncol. 2020, 9, 204. [Google Scholar] [CrossRef]
- Mittal, S.; Klinger, N.V.; Michelhaugh, S.K.; Barger, G.R.; Pannullo, S.C.; Juhasz, C. Alternating electric tumor treating fields for treatment of glioblastoma: Rationale, preclinical, and clinical studies. J. Neurosurg. 2018, 128, 414–421. [Google Scholar] [CrossRef] [PubMed]
- Giladi, M.; Weinberg, U.; Schneiderman, R.S.; Porat, Y.; Munster, M.; Voloshin, T.; Blatt, R.; Cahal, S.; Itzhaki, A.; Onn, A.; et al. Alternating electric fields (tumor-treating fields therapy) can improve chemotherapy treatment efficacy in non-small cell lung cancer both in vitro and in vivo. Semin. Oncol. 2014, 41 (Suppl. 6), S35–S41. [Google Scholar] [CrossRef] [PubMed]
- Rominiyi, O.; Vanderlinden, A.; Clenton, S.J.; Bridgewater, C.; Al-Tamimi, Y.; Collis, S.J. Tumour treating fields therapy for glioblastoma: Current advances and future directions. Br. J. Cancer 2021, 124, 697–709. [Google Scholar] [CrossRef] [PubMed]
- Butler, M.; Pongor, L.; Su, Y.T.; Xi, L.; Raffeld, M.; Quezado, M.; Trepel, J.; Aldape, K.; Pommier, Y.; Wu, J. MGMT status as a clinical biomarker in glioblastoma. Trends Cancer 2020, 6, 380–391. [Google Scholar] [CrossRef]
- Luwor, R.B.; Baradaran, B.; Taylor, L.E.; Iaria, J.; Nheu, T.V.; Amiry, N.; Hovens, C.M.; Wang, B.; Kaye, A.H.; Zhu, H.J. Targeting Stat3 and Smad7 to restore TGF-beta cytostatic regulation of tumor cells in vitro and in vivo. Oncogene 2013, 32, 2433–2441. [Google Scholar] [CrossRef] [PubMed]
- Bradshaw, A.; Wickremsekera, A.; Tan, S.T.; Peng, L.; Davis, P.F.; Itinteang, T. Cancer Stem Cell Hierarchy in Glioblastoma Multiforme. Front. Surg. 2016, 3, 21. [Google Scholar] [CrossRef]
- Soeda, A.; Park, M.; Lee, D.; Mintz, A.; Androutsellis-Theotokis, A.; McKay, R.D.; Engh, J.; Iwama, T.; Kunisada, T.; Kassam, A.B.; et al. Hypoxia promotes expansion of the CD133-positive glioma stem cells through activation of HIF-1alpha. Oncogene 2009, 28, 3949–3959. [Google Scholar] [CrossRef]
- Gilbertson, R.J.; Rich, J.N. Making a tumour’s bed: Glioblastoma stem cells and the vascular niche. Nat. Rev. Cancer 2007, 7, 733–736. [Google Scholar] [CrossRef] [PubMed]
- Hide, T.; Shibahara, I.; Kumabe, T. Novel concept of the border niche: Glioblastoma cells use oligodendrocytes progenitor cells (GAOs) and microglia to acquire stem cell-like features. Brain Tumor Pathol. 2019, 36, 63–73. [Google Scholar] [CrossRef]
- DeLuca, H.F. Overview of general physiologic features and functions of vitamin D. Am. J. Clin. Nutr. 2004, 80, 1689S–1696S. [Google Scholar] [CrossRef]
- Negri, M.; Gentile, A.; de Angelis, C.; Monto, T.; Patalano, R.; Colao, A.; Pivonello, R.; Pivonello, C. Vitamin D-Induced Molecular Mechanisms to Potentiate Cancer Therapy and to Reverse Drug-Resistance in Cancer Cells. Nutrients 2020, 12, 1798. [Google Scholar] [CrossRef] [PubMed]
- Christakos, S.; Ajibade, D.V.; Dhawan, P.; Fechner, A.J.; Mady, L.J. Vitamin D: Metabolism. Endocrinol. Metab. Clin. N. Am. 2010, 39, 243–253. [Google Scholar] [CrossRef]
- Deeb, K.K.; Trump, D.L.; Johnson, C.S. Vitamin D signalling pathways in cancer: Potential for anticancer therapeutics. Nat. Rev. Cancer 2007, 7, 684–700. [Google Scholar] [CrossRef]
- Bikle, D.D. The vitamin D receptor: A tumor suppressor in skin. Discov. Med. 2011, 11, 7–17. [Google Scholar]
- Hu, P.; Li, S.; Tian, N.; Wu, F.; Hu, Y.; Li, D.; Qi, Y.; Wei, Z.; Wei, Q.; Li, Y.; et al. Acidosis enhances the self-renewal and mitochondrial respiration of stem cell-like glioma cells through CYP24A1-mediated reduction of vitamin D. Cell Death Dis. 2019, 10, 25. [Google Scholar] [CrossRef]
- Takahashi, H.; Cornish, A.J.; Sud, A.; Law, P.J.; Kinnersley, B.; Ostrom, Q.T.; Labreche, K.; Eckel-Passow, J.E.; Armstrong, G.N.; Claus, E.B.; et al. Mendelian randomisation study of the relationship between vitamin D and risk of glioma. Sci. Rep. 2018, 8, 2339. [Google Scholar] [CrossRef] [PubMed]
- Zigmont, V.; Garrett, A.; Peng, J.; Seweryn, M.; Rempala, G.A.; Harris, R.; Holloman, C.; Gundersen, T.E.; Ahlbom, A.; Feychting, M.; et al. Association between Prediagnostic Serum 25-Hydroxyvitamin D Concentration and Glioma. Nutr. Cancer 2015, 67, 1120–1130. [Google Scholar] [CrossRef]
- Salomon, D.G.; Fermento, M.E.; Gandini, N.A.; Ferronato, M.J.; Arevalo, J.; Blasco, J.; Andres, N.C.; Zenklusen, J.C.; Curino, A.C.; Facchinetti, M.M. Vitamin D receptor expression is associated with improved overall survival in human glioblastoma multiforme. J. Neurooncol. 2014, 118, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Norlin, M. Effects of vitamin D in the nervous system: Special focus on interaction with steroid hormone signalling and a possible role in the treatment of brain cancer. J. Neuroendocrinol. 2020, 32, e12799. [Google Scholar] [CrossRef]
- Elmaci, I.; Ozpinar, A.; Ozpinar, A.; Perez, J.L.; Altinoz, M.A. From epidemiology and neurometabolism to treatment: Vitamin D in pathogenesis of glioblastoma Multiforme (GBM) and a proposal for Vitamin D + all-trans retinoic acid + Temozolomide combination in treatment of GBM. Metab. Brain Dis. 2019, 34, 687–704. [Google Scholar] [CrossRef] [PubMed]
- Maleklou, N.; Allameh, A.; Kazemi, B. Preparation, characterization and in vitro-targeted delivery of novel Apolipoprotein E-based nanoparticles to C6 glioma with controlled size and loading efficiency. J. Drug Target. 2016, 24, 348–358. [Google Scholar] [CrossRef] [PubMed]
- Bak, D.H.; Kang, S.H.; Choi, D.R.; Gil, M.N.; Yu, K.S.; Jeong, J.H.; Lee, N.S.; Lee, J.H.; Jeong, Y.G.; Kim, D.K.; et al. Autophagy enhancement contributes to the synergistic effect of vitamin D in temozolomide-based glioblastoma chemotherapy. Exp. Ther. Med. 2016, 11, 2153–2162. [Google Scholar] [CrossRef] [PubMed]
- Sui, A.; Xu, Y.; Pan, B.; Guo, T.; Wu, J.; Shen, Y.; Yang, J.; Guo, X. Histone demethylase KDM6B regulates 1,25-dihydroxyvitamin D3-induced senescence in glioma cells. J. Cell Physiol. 2019, 234, 17990–17998. [Google Scholar] [CrossRef]
- Trouillas, P.; Honnorat, J.; Bret, P.; Jouvet, A.; Gerard, J.P. Redifferentiation therapy in brain tumors: Long-lasting complete regression of glioblastomas and an anaplastic astrocytoma under long term 1-alpha-hydroxycholecalciferol. J. Neurooncol. 2001, 51, 57–66. [Google Scholar] [CrossRef]
- Campos, B.; Wan, F.; Farhadi, M.; Ernst, A.; Zeppernick, F.; Tagscherer, K.E.; Ahmadi, R.; Lohr, J.; Dictus, C.; Gdynia, G.; et al. Differentiation therapy exerts antitumor effects on stem-like glioma cells. Clin. Cancer Res. 2010, 16, 2715–2728. [Google Scholar] [CrossRef] [PubMed]
- Gunther, H.S.; Schmidt, N.O.; Phillips, H.S.; Kemming, D.; Kharbanda, S.; Soriano, R.; Modrusan, Z.; Meissner, H.; Westphal, M.; Lamszus, K. Glioblastoma-derived stem cell-enriched cultures form distinct subgroups according to molecular and phenotypic criteria. Oncogene 2008, 27, 2897–2909. [Google Scholar] [CrossRef] [PubMed]
- Linder, B.; Wehle, A.; Hehlgans, S.; Bonn, F.; Dikic, I.; Rodel, F.; Seifert, V.; Kogel, D. Arsenic Trioxide and (-)-Gossypol Synergistically Target Glioma Stem-Like Cells via Inhibition of Hedgehog and Notch Signaling. Cancers 2019, 11, 350. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.; Reyner, K.; Deleyrolle, L.; Millette, S.; Azari, H.; Day, B.W.; Stringer, B.W.; Boyd, A.W.; Johns, T.G.; Blot, V.; et al. Neurosphere and adherent culture conditions are equivalent for malignant glioma stem cell lines. Anat. Cell Biol. 2015, 48, 25–35. [Google Scholar] [CrossRef]
- Hu, Y.; Smyth, G.K. ELDA: Extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods 2009, 347, 70–78. [Google Scholar] [CrossRef]
- Nagpal, S.; Na, S.; Rathnachalam, R. Noncalcemic actions of vitamin D receptor ligands. Endocr. Rev. 2005, 26, 662–687. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Genssler, S.; Burger, M.C.; Zhang, C.; Oelsner, S.; Mildenberger, I.; Wagner, M.; Steinbach, J.P.; Wels, W.S. Dual targeting of glioblastoma with chimeric antigen receptor-engineered natural killer cells overcomes heterogeneity of target antigen expression and enhances antitumor activity and survival. Oncoimmunology 2016, 5, e1119354. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Burger, M.C.; Jennewein, L.; Genssler, S.; Schonfeld, K.; Zeiner, P.; Hattingen, E.; Harter, P.N.; Mittelbronn, M.; Tonn, T.; et al. ErbB2/HER2-Specific NK Cells for Targeted Therapy of Glioblastoma. J. Natl. Cancer Inst. 2016, 108. [Google Scholar] [CrossRef] [PubMed]
- Maire, C.L.; Fuh, M.M.; Kaulich, K.; Fita, K.D.; Stevic, I.; Heiland, D.H.; Welsh, J.A.; Jones, J.C.; Gorgens, A.; Ricklefs, T.; et al. Genome-wide methylation profiling of glioblastoma cell-derived extracellular vesicle DNA allows tumor classification. Neuro Oncol. 2021, 23, 1087–1099. [Google Scholar] [CrossRef] [PubMed]
- Gobin, M.; Nazarov, P.V.; Warta, R.; Timmer, M.; Reifenberger, G.; Felsberg, J.; Vallar, L.; Chalmers, A.J.; Herold-Mende, C.C.; Goldbrunner, R.; et al. A DNA Repair and cell-cycle gene expression signature in primary and recurrent glioblastoma: Prognostic value and clinical implications. Cancer Res. 2019, 79, 1226–1238. [Google Scholar] [CrossRef] [PubMed]
- Clement, V.; Sanchez, P.; De Tribolet, N.; Radovanovic, I.; Ruiz i Altaba, A. HEDGEHOG-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal, and tumorigenicity. Curr. Biol. 2007, 17, 165–172. [Google Scholar] [CrossRef]
- Bijlsma, M.F.; Spek, C.A.; Zivkovic, D.; van de Water, S.; Rezaee, F.; Peppelenbosch, M.P. Repression of smoothened by patched-dependent (pro-)vitamin D3 secretion. PLoS Biol. 2006, 4, e232. [Google Scholar] [CrossRef]
- Linder, B.; Weber, S.; Dittmann, K.; Adamski, J.; Hahn, H.; Uhmann, A. A Functional and Putative Physiological Role of Calcitriol in Patched1/Smoothened Interaction. J. Biol. Chem. 2015, 290, 19614–19628. [Google Scholar] [CrossRef]
- Remy, J.; Linder, B.; Weirauch, U.; Konovalova, J.; Marschalek, R.; Aigner, A.; Kogel, D. Inhibition of PIM1 blocks the autophagic flux to sensitize glioblastoma cells to ABT-737-induced apoptosis. Biochim. Biophys. Acta Mol. Cell Res. 2018, 1866, 175–189. [Google Scholar] [CrossRef] [PubMed]
- Lan, X.; Jorg, D.J.; Cavalli, F.M.G.; Richards, L.M.; Nguyen, L.V.; Vanner, R.J.; Guilhamon, P.; Lee, L.; Kushida, M.M.; Pellacani, D.; et al. Fate mapping of human glioblastoma reveals an invariant stem cell hierarchy. Nature 2017, 549, 227–232. [Google Scholar] [CrossRef] [PubMed]
- D’Arcy, M.S. Cell death: A review of the major forms of apoptosis, necrosis and autophagy. Cell Biol. Int. 2019, 43, 582–592. [Google Scholar] [CrossRef] [PubMed]
- Filatova, A.; Acker, T.; Garvalov, B.K. The cancer stem cell niche(s): The crosstalk between glioma stem cells and their microenvironment. Biochim. Biophys. Acta 2013, 1830, 2496–2508. [Google Scholar] [CrossRef]
- Ahmed, A.U.; Auffinger, B.; Lesniak, M.S. Understanding glioma stem cells: Rationale, clinical relevance and therapeutic strategies. Expert Rev. Neurother. 2013, 13, 545–555. [Google Scholar] [CrossRef]
- Gilbert, C.A.; Ross, A.H. Cancer stem cells: Cell culture, markers, and targets for new therapies. J. Cell Biochem. 2009, 108, 1031–1038. [Google Scholar] [CrossRef]
- Beier, D.; Hau, P.; Proescholdt, M.; Lohmeier, A.; Wischhusen, J.; Oefner, P.J.; Aigner, L.; Brawanski, A.; Bogdahn, U.; Beier, C.P. CD133(+) and CD133(−) glioblastoma-derived cancer stem cells show differential growth characteristics and molecular profiles. Cancer Res. 2007, 67, 4010–4015. [Google Scholar] [CrossRef]
- Bao, S.; Wu, Q.; McLendon, R.E.; Hao, Y.; Shi, Q.; Hjelmeland, A.B.; Dewhirst, M.W.; Bigner, D.D.; Rich, J.N. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 2006, 444, 756–760. [Google Scholar] [CrossRef]
- Bikle, D.D.; Elalieh, H.; Welsh, J.; Oh, D.; Cleaver, J.; Teichert, A. Protective role of vitamin D signaling in skin cancer formation. J. Steroid Biochem. Mol. Biol. 2013, 136, 271–279. [Google Scholar] [CrossRef]
- Emanuelsson, I.; Wikvall, K.; Friman, T.; Norlin, M. Vitamin D Analogues Tacalcitol and Calcipotriol Inhibit Proliferation and Migration of T98G Human Glioblastoma Cells. Basic Clin. Pharmacol. Toxicol. 2018, 123, 130–136. [Google Scholar] [CrossRef] [PubMed]
- Arensman, M.D.; Nguyen, P.; Kershaw, K.M.; Lay, A.R.; Ostertag-Hill, C.A.; Sherman, M.H.; Downes, M.; Liddle, C.; Evans, R.M.; Dawson, D.W. Calcipotriol Targets LRP6 to Inhibit Wnt Signaling in Pancreatic Cancer. Mol. Cancer Res. 2015, 13, 1509–1519. [Google Scholar] [CrossRef]
- Gorchs, L.; Ahmed, S.; Mayer, C.; Knauf, A.; Fernandez Moro, C.; Svensson, M.; Heuchel, R.; Rangelova, E.; Bergman, P.; Kaipe, H. The vitamin D analogue calcipotriol promotes an anti-tumorigenic phenotype of human pancreatic CAFs but reduces T cell mediated immunity. Sci. Rep. 2020, 10, 17444. [Google Scholar] [CrossRef]
- Segovia-Mendoza, M.; Diaz, L.; Gonzalez-Gonzalez, M.E.; Martinez-Reza, I.; Garcia-Quiroz, J.; Prado-Garcia, H.; Ibarra-Sanchez, M.J.; Esparza-Lopez, J.; Larrea, F.; Garcia-Becerra, R. Calcitriol and its analogues enhance the antiproliferative activity of gefitinib in breast cancer cells. J. Steroid Biochem. Mol. Biol. 2015, 148, 122–131. [Google Scholar] [CrossRef]
- Colston, K.W.; Chander, S.K.; Mackay, A.G.; Coombes, R.C. Effects of synthetic vitamin D analogues on breast cancer cell proliferation in vivo and in vitro. Biochem. Pharmacol. 1992, 44, 693–702. [Google Scholar] [CrossRef]
- Haussler, M.R.; Whitfield, G.K.; Haussler, C.A.; Hsieh, J.C.; Thompson, P.D.; Selznick, S.H.; Dominguez, C.E.; Jurutka, P.W. The nuclear vitamin D receptor: Biological and molecular regulatory properties revealed. J. Bone Miner. Res. 1998, 13, 325–349. [Google Scholar] [CrossRef]
- Li, W.; Wang, Q.L.; Liu, X.; Dong, S.H.; Li, H.X.; Li, C.Y.; Guo, L.S.; Gao, J.M.; Berger, N.A.; Li, L.; et al. Combined use of vitamin D3 and metformin exhibits synergistic chemopreventive effects on colorectal neoplasia in rats and mice. Cancer Prev. Res. 2015, 8, 139–148. [Google Scholar] [CrossRef]
- Santucci, K.L.; Snyder, K.K.; Baust, J.M.; Van Buskirk, R.G.; Mouraviev, V.; Polascik, T.J.; Gage, A.A.; Baust, J.G. Use of 1.25 alpha dihydroxyvitamin D3 as a cryosensitizing agent in a murine prostate cancer model. Prostate Cancer Prostatic Dis. 2011, 14, 97–104. [Google Scholar] [CrossRef]
- Dirkse, A.; Golebiewska, A.; Buder, T.; Nazarov, P.V.; Muller, A.; Poovathingal, S.; Brons, N.H.C.; Leite, S.; Sauvageot, N.; Sarkisjan, D.; et al. Stem cell-associated heterogeneity in Glioblastoma results from intrinsic tumor plasticity shaped by the microenvironment. Nat. Commun. 2019, 10, 1787. [Google Scholar] [CrossRef]
- Fijten, R.; de Nooijer, K.; Kutmon, M.; Pico, A.; Ehrhart, F.; Hanspers, K.; Willighagen, E.; Weitz, E. Vitamin D Receptor Pathway. Available online: https://www.wikipathways.org/index.php/Pathway:WP2877 (accessed on 27 May 2021).
- Khammissa, R.A.G.; Fourie, J.; Motswaledi, M.H.; Ballyram, R.; Lemmer, J.; Feller, L. The Biological Activities of Vitamin D and Its Receptor in Relation to Calcium and Bone Homeostasis, Cancer, Immune and Cardiovascular Systems, Skin Biology, and Oral Health. Biomed. Res. Int. 2018, 2018, 9276380. [Google Scholar] [CrossRef]
- Hirakawa, Y.; Nary, L.J.; Medh, R.D. Glucocorticoid evoked upregulation of RCAN1-1 in human leukemic CEM cells susceptible to apoptosis. J. Mol. Signal. 2009, 4, 6. [Google Scholar] [CrossRef][Green Version]
- Torac, E.; Gaman, L.; Atanasiu, V. The regulator of calcineurin (RCAN1) an important factor involved in atherosclerosis and cardiovascular diseases development. J. Med. Life 2014, 7, 481–487. [Google Scholar]
- Bikle, D.D.; Munson, S. 1,25-Dihydroxyvitamin D increases calmodulin binding to specific proteins in the chick duodenal brush border membrane. J. Clin. Investig. 1985, 76, 2312–2316. [Google Scholar] [CrossRef]
- Nimitphong, H.; Park, E.; Lee, M.J. Vitamin D regulation of adipogenesis and adipose tissue functions. Nutr. Res. Pract. 2020, 14, 553–567. [Google Scholar] [CrossRef]
- Coronas, V.; Terrie, E.; Deliot, N.; Arnault, P.; Constantin, B. Calcium Channels in Adult Brain Neural Stem Cells and in Glioblastoma Stem Cells. Front. Cell Neurosci. 2020, 14, 600018. [Google Scholar] [CrossRef]
- Robil, N.; Petel, F.; Kilhoffer, M.C.; Haiech, J. Glioblastoma and calcium signaling--analysis of calcium toolbox expression. Int. J. Dev. Biol. 2015, 59, 407–415. [Google Scholar] [CrossRef]
- Leclerc, C.; Haeich, J.; Aulestia, F.J.; Kilhoffer, M.C.; Miller, A.L.; Neant, I.; Webb, S.E.; Schaeffer, E.; Junier, M.P.; Chneiweiss, H.; et al. Calcium signaling orchestrates glioblastoma development: Facts and conjunctures. Biochim. Biophys. Acta 2016, 1863, 1447–1459. [Google Scholar] [CrossRef]
- Meyer, N.; Henkel, L.; Linder, B.; Zielke, S.; Tascher, G.; Trautmann, S.; Geisslinger, G.; Münch, C.; Fulda, S.; Tegeder, I.; et al. Autophagy activation, lipotoxicity and lysosomal membrane permeabilization synergize to promote pimozide- and loperamide-induced glioma cell death. Biorxiv 2020, 1–20. [Google Scholar] [CrossRef]
- Sapcariu, S.C.; Kanashova, T.; Weindl, D.; Ghelfi, J.; Dittmar, G.; Hiller, K. Simultaneous extraction of proteins and metabolites from cells in culture. MethodsX 2014, 1, 74–80. [Google Scholar] [CrossRef]
- Hosp, F.; Scheltema, R.A.; Eberl, H.C.; Kulak, N.A.; Keilhauer, E.C.; Mayr, K.; Mann, M. A Double-Barrel Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS) System to Quantify 96 Interactomes per Day. Mol. Cell Proteom. 2015, 14, 2030–2041. [Google Scholar] [CrossRef]
- Kelstrup, C.D.; Young, C.; Lavallee, R.; Nielsen, M.L.; Olsen, J.V. Optimized fast and sensitive acquisition methods for shotgun proteomics on a quadrupole orbitrap mass spectrometer. J. Proteome Res. 2012, 11, 3487–3497. [Google Scholar] [CrossRef]
- McAlister, G.C.; Nusinow, D.P.; Jedrychowski, M.P.; Wuhr, M.; Huttlin, E.L.; Erickson, B.K.; Rad, R.; Haas, W.; Gygi, S.P. MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal. Chem. 2014, 86, 7150–7158. [Google Scholar] [CrossRef] [PubMed]
- Paulo, J.A.; O’Connell, J.D.; Gygi, S.P. A Triple Knockout (TKO) Proteomics Standard for Diagnosing Ion Interference in Isobaric Labeling Experiments. J. Am. Soc. Mass Spectrom. 2016, 27, 1620–1625. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing, 4.0.2. ed.; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- RStudio Team. RStudio: Integrated Development for R. RStudio; PBC: Boston, MA, USA, 2020. [Google Scholar]
- Warnes, G.R.; Bolker, B.; Bonebakker, L.; Gentleman, R.; Huber, W.; Liaw, A.; Lumley, T.; Maechler, M.; Magnusson, A.; Moeller, S.; et al. Gplots: Various R Programming Tools for Plotting Data. CRAN Project. Maintainer Tal Galili tal.galili@gmail.com; License GPL-2. 2020. Available online: https://github.com/talgalili/gplots (accessed on 27 May 2021).
- Blighe, K.R.S.; Lewis, M. EnhancedVolcano: Publication-Ready Volcano Plots with Enhanced Colouring and Labeling, R Package Version 1.6.0.ed.; Bioconductor; 2020. Available online: https://github.com/kevinblighe/EnhancedVolcano (accessed on 27 May 2021).
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef]


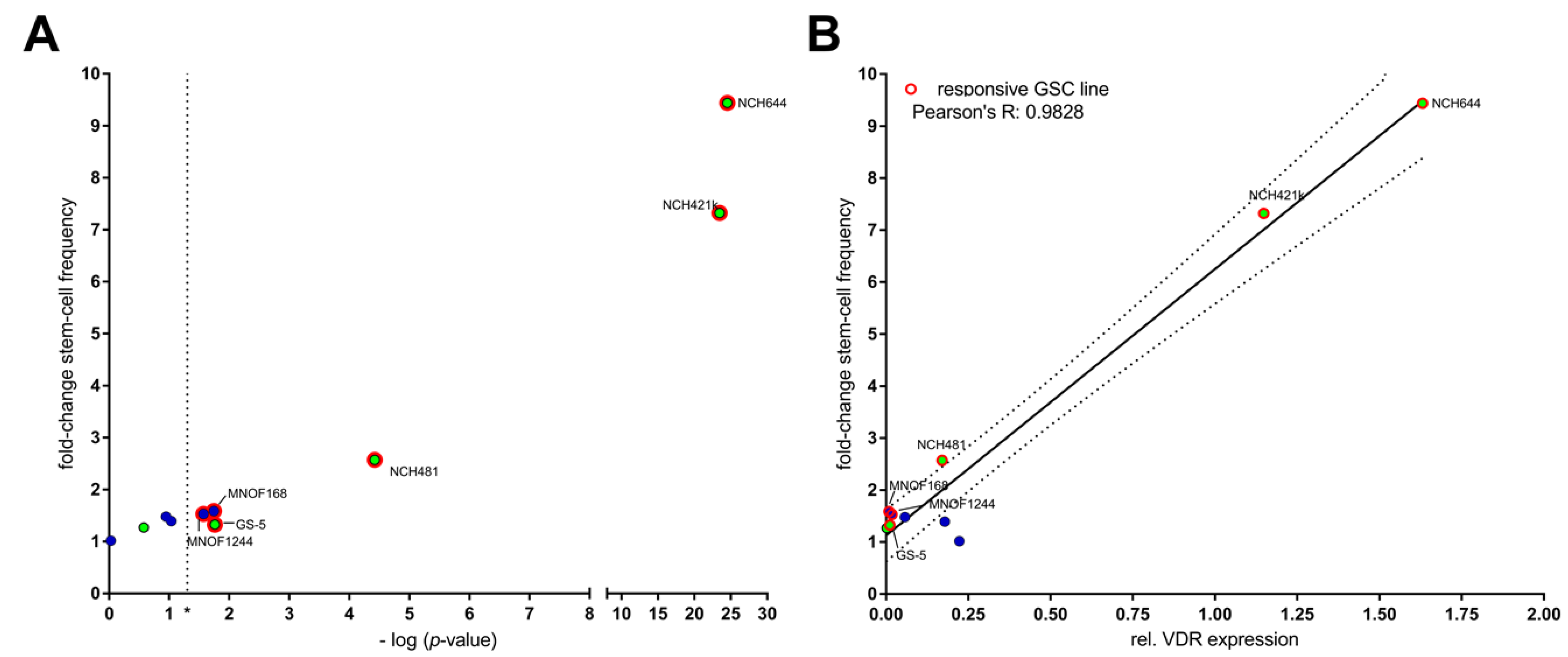

| Cell Line | Calcitriol [nM] | Stem-Cell Frequency [1/X] | p-Value (Chi2) | Calcipotriol [nM] | Stem-Cell Frequency [1/X] | p-Value (Chi2) |
|---|---|---|---|---|---|---|
| NCH644 | EtOH | 23.3 | DMSO | 50.6 | ||
| 10 | 81.6 | 1.46 × 10−15 | 100 | 80.5 | 0.00314 | |
| 25 | 119.1 | 3.01 ×10−25 | 500 | 104 | 5.05 × 10−6 | |
| 50 | 219.9 | 1.12 × 10−43 | 1000 | 129.5 | 4.58 × 10−9 | |
| GS-5 | EtOH | 188 | DMSO | 190 | ||
| 10 | 206 | 0.525 | 100 | 241 | 0.0861 | |
| 25 | 264 | 0.0245 | 500 | 232 | 0.140 | |
| 50 | 318 | 0.00061 | 1000 | 310 | 0.000556 | |
| 17/02 | EtOH | 153 | DMSO | 159 | ||
| 10 | 152 | 0.97 | 100 | 192 | 0.191 | |
| 25 | 159 | 0.81 | 500 | 209 | 0.0627 | |
| 50 | 198 | 0.101 | 1000 | 232 | 0.0109 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gerstmeier, J.; Possmayer, A.-L.; Bozkurt, S.; Hoffmann, M.E.; Dikic, I.; Herold-Mende, C.; Burger, M.C.; Münch, C.; Kögel, D.; Linder, B. Calcitriol Promotes Differentiation of Glioma Stem-Like Cells and Increases Their Susceptibility to Temozolomide. Cancers 2021, 13, 3577. https://doi.org/10.3390/cancers13143577
Gerstmeier J, Possmayer A-L, Bozkurt S, Hoffmann ME, Dikic I, Herold-Mende C, Burger MC, Münch C, Kögel D, Linder B. Calcitriol Promotes Differentiation of Glioma Stem-Like Cells and Increases Their Susceptibility to Temozolomide. Cancers. 2021; 13(14):3577. https://doi.org/10.3390/cancers13143577
Chicago/Turabian StyleGerstmeier, Julia, Anna-Lena Possmayer, Süleyman Bozkurt, Marina E. Hoffmann, Ivan Dikic, Christel Herold-Mende, Michael C. Burger, Christian Münch, Donat Kögel, and Benedikt Linder. 2021. "Calcitriol Promotes Differentiation of Glioma Stem-Like Cells and Increases Their Susceptibility to Temozolomide" Cancers 13, no. 14: 3577. https://doi.org/10.3390/cancers13143577
APA StyleGerstmeier, J., Possmayer, A.-L., Bozkurt, S., Hoffmann, M. E., Dikic, I., Herold-Mende, C., Burger, M. C., Münch, C., Kögel, D., & Linder, B. (2021). Calcitriol Promotes Differentiation of Glioma Stem-Like Cells and Increases Their Susceptibility to Temozolomide. Cancers, 13(14), 3577. https://doi.org/10.3390/cancers13143577









