Validation of a Point-of-Care Optical Coherence Tomography Device with Machine Learning Algorithm for Detection of Oral Potentially Malignant and Malignant Lesions
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
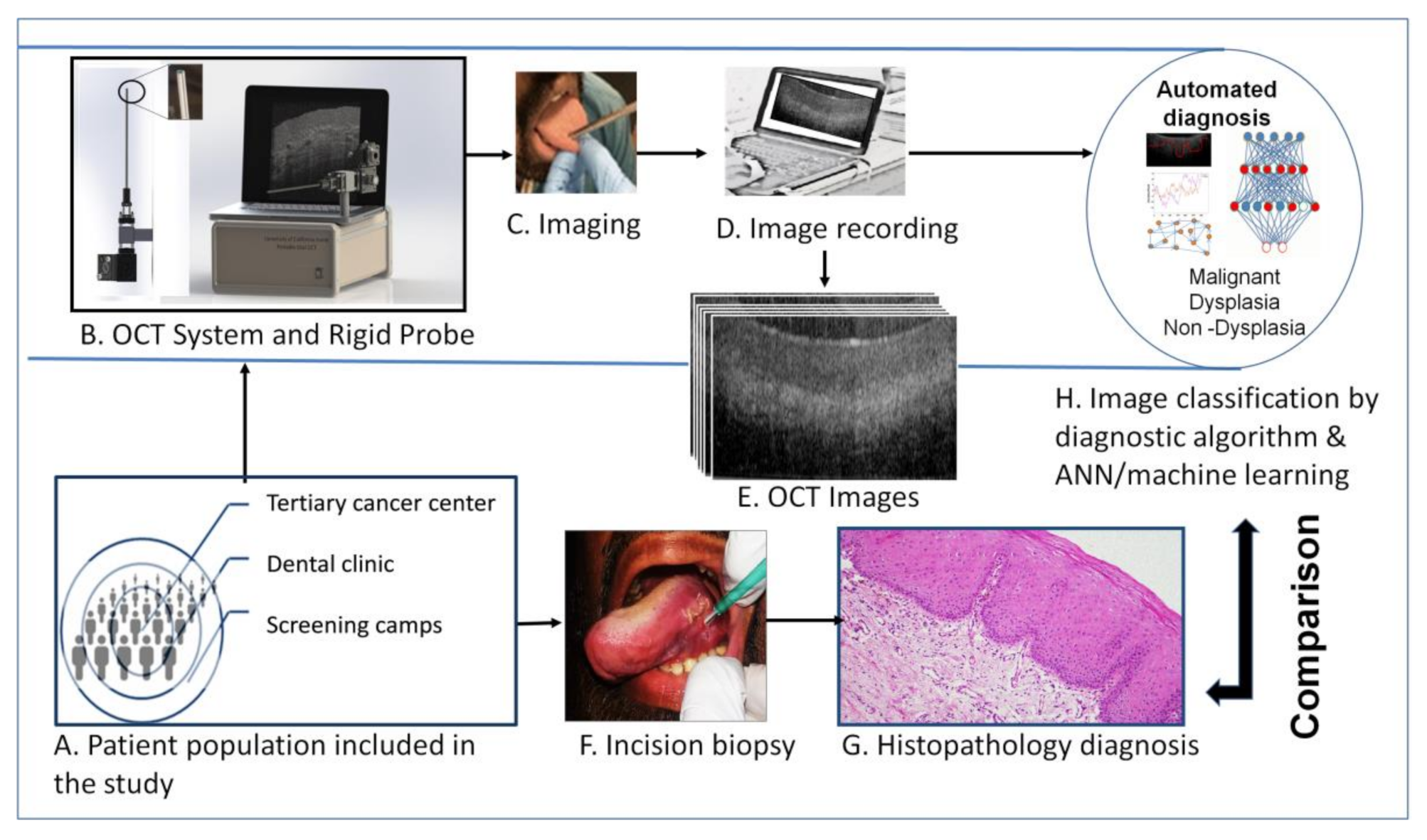
2.1. Study Population and Design
2.2. OCT System Design
2.3. Analysis of OCT Images
2.3.1. Algorithm-Score Based Analysis
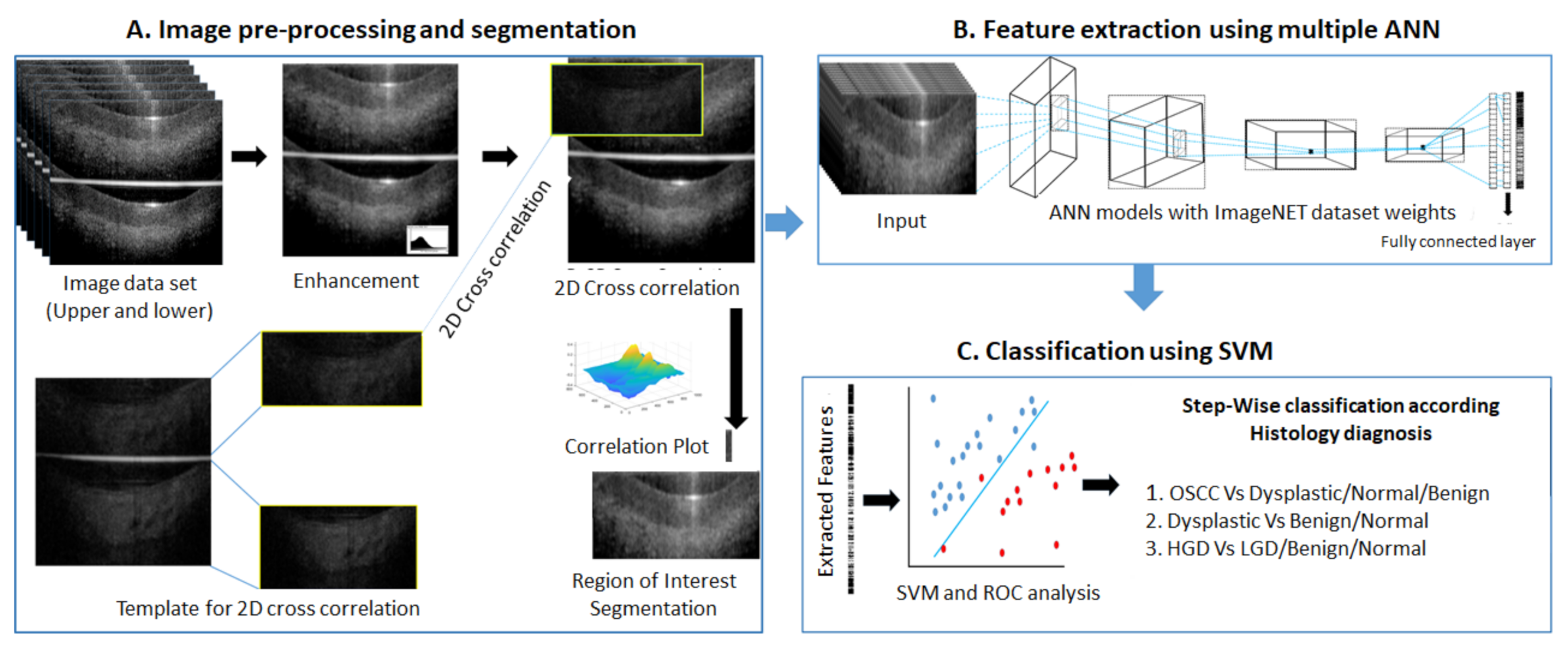
2.3.2. ANN-SVM Based Analysis
Image Data Preparation
Image Preprocessing
Feature Extraction by Deep Learning and SVM Model
2.4. Statistical Analysis
3. Results
3.1. Clinical and Demographic Details of Patients
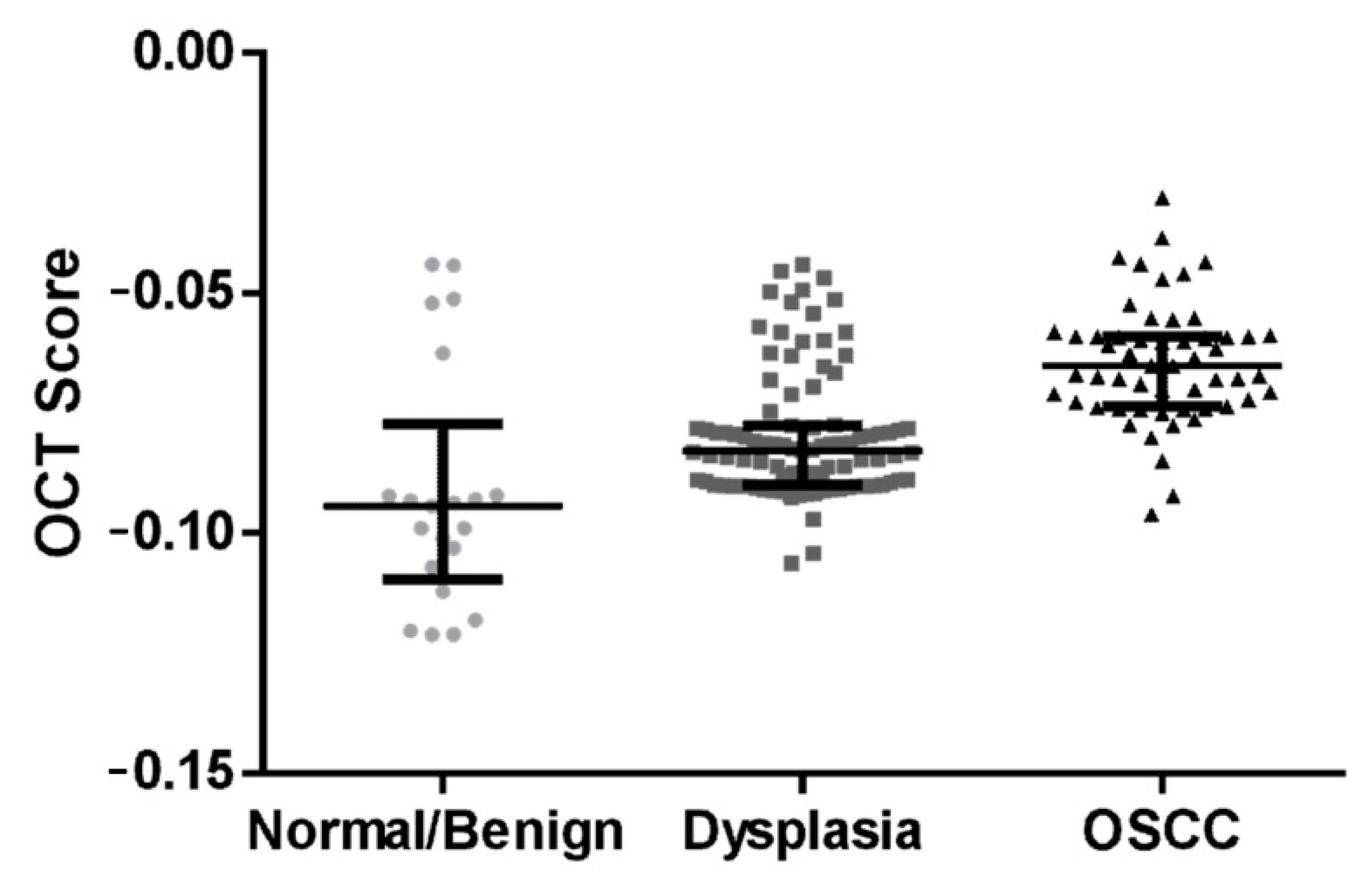
3.2. Algorithm Prediction Score Correlates with Histopathology Diagnosis
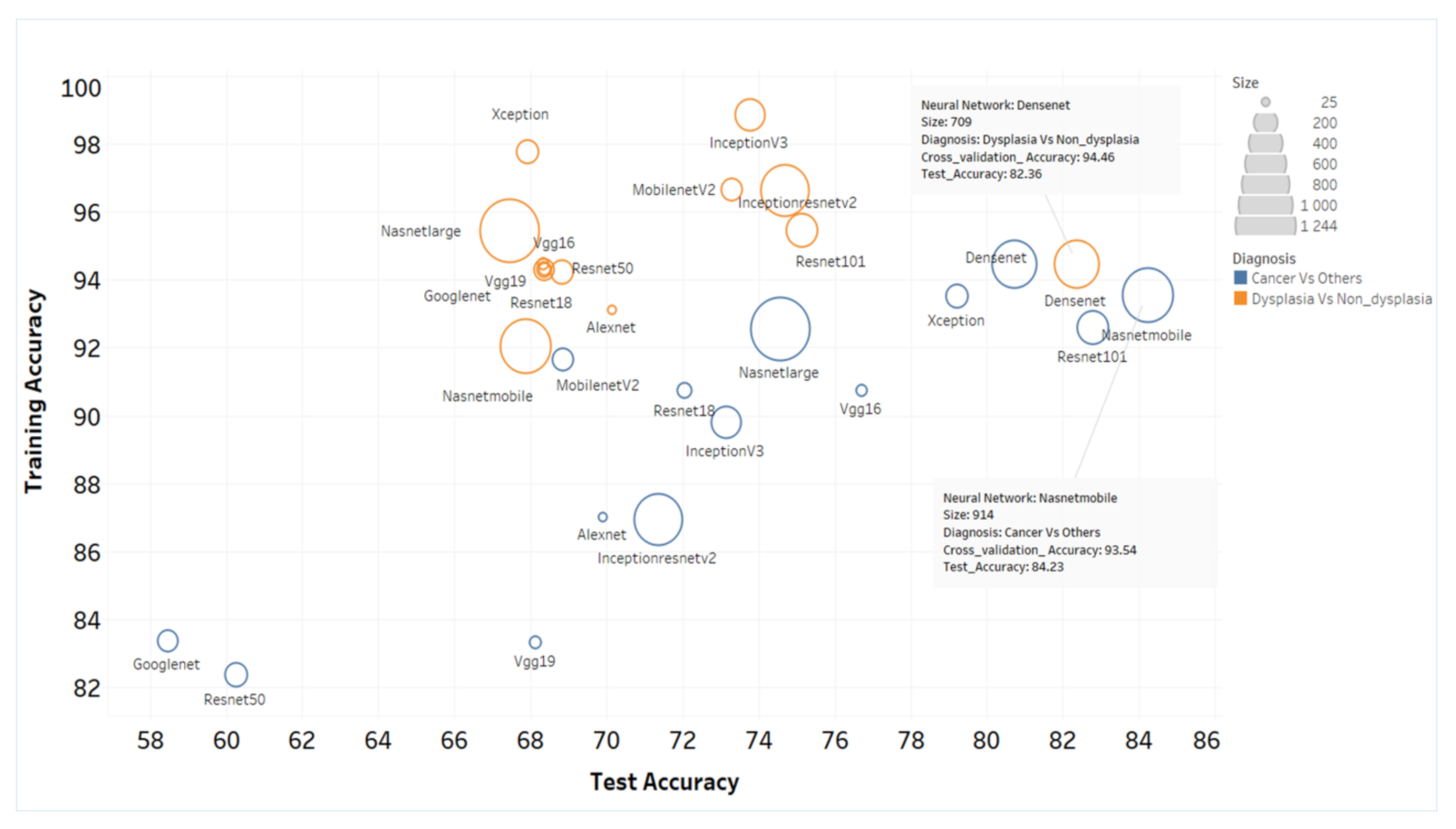
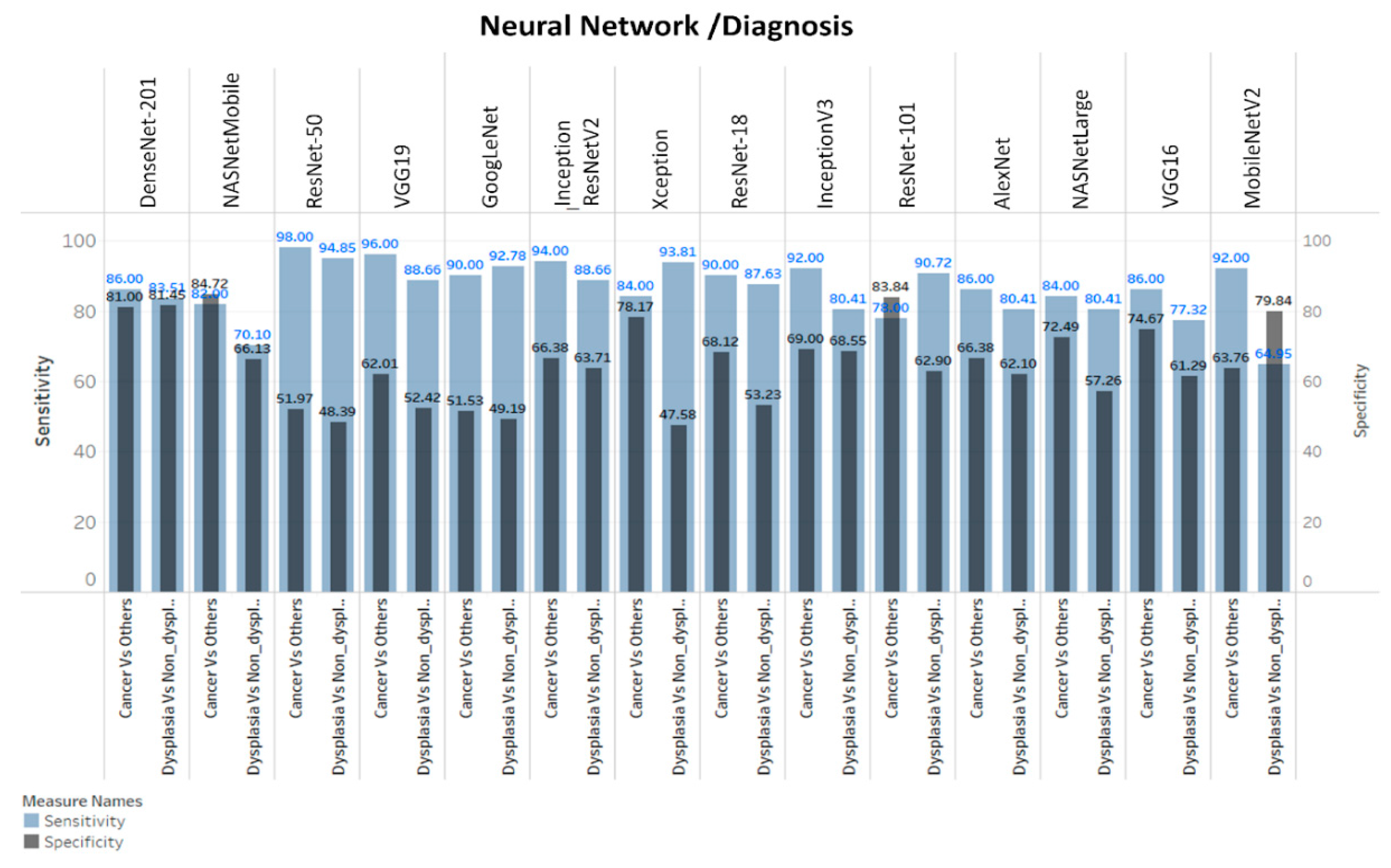
3.3. ANN-SVM Model Delineated Grades of Dysplasia
3.4. Clinical Application of OCT Device in Triaging Patients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Popli, G.; Bhat, S.; Mohan, S.; Sowdepalli, A.; Kumari, K. Oral cancer incidence trends in Delhi (1990–2014): An alarming scenario. South Asian J. Cancer 2019, 8, 116–119. [Google Scholar] [CrossRef] [PubMed]
- Globocan. Population Fact Sheets Globocan. Available online: https://gco.iarc.fr/today/online-analysis-map?v=2018&mode=population&mode_population=continents&population=900&populations=900&key=asr&sex=0&cancer=1&type=1&statistic=5&prevalence=0&population_group=0&ages_group%5B%5D=4&ages_group%5B%5D=17&nb_items=10&group_cancer=1&include_nmsc=1&include_nmsc_other=1&projection=natural-earth&color_palette=default&map_scale=quantile&map_nb_colors=7&continent=0&rotate=%255B10%252C0%255D (accessed on 24 August 2020).
- Sharma, S.; Satyanarayana, L.; Asthana, S.; Shivalingesh, K.K.; Goutham, B.S.; Ramachandra, S. Oral cancer statistics in India on the basis of first report of 29 population-based cancer registries. J. Oral Maxillofac. Pathol. JOMFP 2018, 22, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Borse, V.; Konwar, A.N.; Buragohain, P. Oral cancer diagnosis and perspectives in India. Sens. Int. 2020, 1, 100046. [Google Scholar] [CrossRef]
- Thavarool, S.B.; Muttath, G.; Nayanar, S.; Duraisamy, K.; Bhat, P.; Shringarpure, K.; Nayak, P.; Tripathy, J.P.; Thaddeus, A.; Philip, S.; et al. Improved survival among oral cancer patients: Findings from a retrospective study at a tertiary care cancer centre in rural Kerala, India. World J. Surg. Oncol. 2019, 17, 15. [Google Scholar] [CrossRef]
- Geum, D.H.; Roh, Y.C.; Yoon, S.Y.; Kim, H.G.; Lee, J.H.; Song, J.M.; Lee, J.Y.; Hwang, D.S.; Kim, Y.D.; Shin, S.H.; et al. The impact factors on 5-year survival rate in patients operated with oral cancer. J. Korean Assoc. Oral Maxillofac. Surg. 2013, 39, 207–216. [Google Scholar] [CrossRef] [Green Version]
- Bagan, J.; Sarrion, G.; Jimenez, Y. Oral cancer: Clinical features. Oral Oncol. 2010, 46, 414–417. [Google Scholar] [CrossRef]
- World Health Organization. Control of oral cancer in developing countries. A WHO meeting. Bull. World Health Org. 1984, 62, 817–830. [Google Scholar]
- Sankaranarayanan, R.; Ramadas, K.; Thomas, G.; Muwonge, R.; Thara, S.; Mathew, B.; Rajan, B.; Trivandrum Oral Cancer Screening Study, G. Effect of screening on oral cancer mortality in Kerala, India: A cluster-randomised controlled trial. Lancet 2005, 365, 1927–1933. [Google Scholar] [CrossRef]
- Elango, K.J.; Anandkrishnan, N.; Suresh, A.; Iyer, S.K.; Ramaiyer, S.K.; Kuriakose, M.A. Mouth self-examination to improve oral cancer awareness and early detection in a high-risk population. Oral Oncol. 2011, 47, 620–624. [Google Scholar] [CrossRef]
- Epstein, J.B.; Guneri, P.; Boyacioglu, H.; Abt, E. The limitations of the clinical oral examination in detecting dysplastic oral lesions and oral squamous cell carcinoma. Tex. Dent. J. 2013, 130, 410–424. [Google Scholar] [CrossRef]
- Epstein, J.B.; Silverman, S., Jr.; Epstein, J.D.; Lonky, S.A.; Bride, M.A. Analysis of oral lesion biopsies identified and evaluated by visual examination, chemiluminescence and toluidine blue. Oral Oncol. 2008, 44, 538–544. [Google Scholar] [CrossRef] [PubMed]
- Jitender, S.; Sarika, G.; Varada, H.R.; Omprakash, Y.; Mohsin, K. Screening for oral cancer. J. Exp. Ther. Oncol. 2016, 11, 303–307. [Google Scholar]
- Chakraborty, D.; Natarajan, C.; Mukherjee, A. Advances in oral cancer detection. Adv. Clin. Chem. 2019, 91, 181–200. [Google Scholar] [CrossRef] [PubMed]
- Aumann, S.; Donner, S.; Fischer, J.; Muller, F. Optical Coherence Tomography (OCT): Principle and Technical Realization. In High Resolution Imaging in Microscopy and Ophthalmology: New Frontiers in Biomedical Optics; Bille, J.F., Ed.; Springer: Cham, Switzerland, 2019; pp. 59–85. [Google Scholar] [CrossRef] [Green Version]
- Prestin, S.; Rothschild, S.I.; Betz, C.S.; Kraft, M. Measurement of epithelial thickness within the oral cavity using optical coherence tomography. Head Neck 2012, 34, 1777–1781. [Google Scholar] [CrossRef]
- Jerjes, W.; Hamdoon, Z.; Yousif, A.A.; Al-Rawi, N.H.; Hopper, C. Epithelial tissue thickness improves optical coherence tomography’s ability in detecting oral cancer. Photodiagn. Photodyn. Ther. 2019, 28, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Stasio, D.D.; Lauritano, D.; Iquebal, H.; Romano, A.; Gentile, E.; Lucchese, A. Measurement of Oral Epithelial Thickness by Optical Coherence Tomography. Diagnostics 2019, 9, 90. [Google Scholar] [CrossRef] [Green Version]
- Hamdoon, Z.; Jerjes, W.; Upile, T.; McKenzie, G.; Jay, A.; Hopper, C. Optical coherence tomography in the assessment of suspicious oral lesions: An immediate ex vivo study. Photodiagn. Photodyn. Ther. 2013, 10, 17–27. [Google Scholar] [CrossRef]
- Reddy, R.S.; Sai Praveen, K.N. Optical coherence tomography in oral cancer: A transpiring domain. J. Cancer Res. Ther. 2017, 13, 883–888. [Google Scholar] [CrossRef]
- Sharma, P.; Verma, Y.; Sahu, K.; Kumar, S.; Varma, A.V.; Kumawat, J.; Gupta, P.K. Human ex-vivo oral tissue imaging using spectral domain polarization sensitive optical coherence tomography. Lasers Med. Sci. 2017, 32, 143–150. [Google Scholar] [CrossRef]
- Heidari, A.E.; Sunny, S.P.; James, B.L.; Lam, T.M.; Tran, A.V.; Yu, J.; Ramanjinappa, R.D.; Uma, K.; Birur, P.; Suresh, A. Optical Coherence Tomography as an Oral Cancer Screening Adjunct in a Low Resource Settings. IEEE J. Sel. Top. Quantum Electron. 2018, 25, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Sunny, S.P.; Agarwal, S.; James, B.L.; Heidari, E.; Muralidharan, A.; Yadav, V.; Pillai, V.; Shetty, V.; Chen, Z.; Hedne, N.; et al. Intra-operative point-of-procedure delineation of oral cancer margins using optical coherence tomography. Oral Oncol. 2019, 92, 12–19. [Google Scholar] [CrossRef]
- Söllinger, D.; Trung, P.; Uhl, A. Non-reference image quality assessment and natural scene statistics to counter biometric sensor spoofing. IET Biom. 2018, 7, 314–324. [Google Scholar] [CrossRef]
- Harris, R.I.D. Testing for unit roots using the augmented Dickey-Fuller test. Econ. Lett. 1992, 38, 381–386. [Google Scholar] [CrossRef]
- de Jong, R.M.; Amsler, C.; Schmidt, P. A robust version of the KPSS test based on indicators. J. Econom. 2007, 137, 311–333. [Google Scholar] [CrossRef]
- Hajian-Tilaki, K. Sample size estimation in diagnostic test studies of biomedical informatics. J. Biomed. Inform. 2014, 48, 193–204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmmed, R.; Swakshar, A.S.; Hossain, M.F.; Rafiq, M.A. Classification of tumors and it stages in brain MRI using support vector machine and artificial neural network. In Proceedings of the International Conference on Electrical, Computer and Communication Engineering (ECCE), Cox’s Bazar, Bangladesh, 16–18 February 2017; pp. 229–234. [Google Scholar]
- Pande, P.; Shrestha, S.; Park, J.; Gimenez-Conti, I.; Brandon, J.; Applegate, B.E.; Jo, J.A. Automated analysis of multimodal fluorescence lifetime imaging and optical coherence tomography data for the diagnosis of oral cancer in the hamster cheek pouch model. Biomed. Opt. Express 2016, 7, 2000–2015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heidari, A.E.; Pham, T.T.; Ifegwu, I.; Burwell, R.; Armstrong, W.B.; Tjoson, T.; Whyte, S.; Giorgioni, C.; Wang, B.; Wong, B.J.F.; et al. The use of optical coherence tomography and convolutional neural networks to distinguish normal and abnormal oral mucosa. J. Biophotonics 2020, 13, e201900221. [Google Scholar] [CrossRef] [PubMed]
- Sunny, S.; Baby, A.; James, B.L.; Balaji, D.; Aparna, N.V.; Rana, M.H.; Gurpur, P.; Skandarajah, A.; D’Ambrosio, M.; Ramanjinappa, R.D.; et al. A smart tele-cytology point-of-care platform for oral cancer screening. PLoS ONE 2019, 14, e0224885. [Google Scholar] [CrossRef] [Green Version]
- Fakurnejad, S.; van Keulen, S.; Nishio, N.; Engelen, M.; van den Berg, N.S.; Lu, G.; Birkeland, A.; Baik, F.; Colevas, A.D.; Rosenthal, E.L.; et al. Fluorescence molecular imaging for identification of high-grade dysplasia in patients with head and neck cancer. Oral Oncol. 2019, 97, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Xiao, X.; Wu, W.; Shen, X.; Zhou, Z.; Liu, W.; Shi, L. Cytological study of DNA content and nuclear morphometric analysis for aid in the diagnosis of high-grade dysplasia within oral leukoplakia. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2017, 124, 280–285. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.H.; Wu, C.H.; Chen, Y.F.; Yeh, Y.C.; Lin, B.H.; Chang, K.W.; Lai, P.Y.; Hou, M.C.; Lu, C.L.; Kuo, W.C. Combination of structural and vascular optical coherence tomography for differentiating oral lesions of mice in different carcinogenesis stages. Biomed. Opt. Express 2018, 9, 1461–1476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogien, J.; Daures, A.; Cazalas, M.; Perrot, J.-L.; Dubois, A. Line-field confocal optical coherence tomography for three-dimensional skin imaging. Front. Optoelectron. 2020, 13, 381–392. [Google Scholar] [CrossRef]

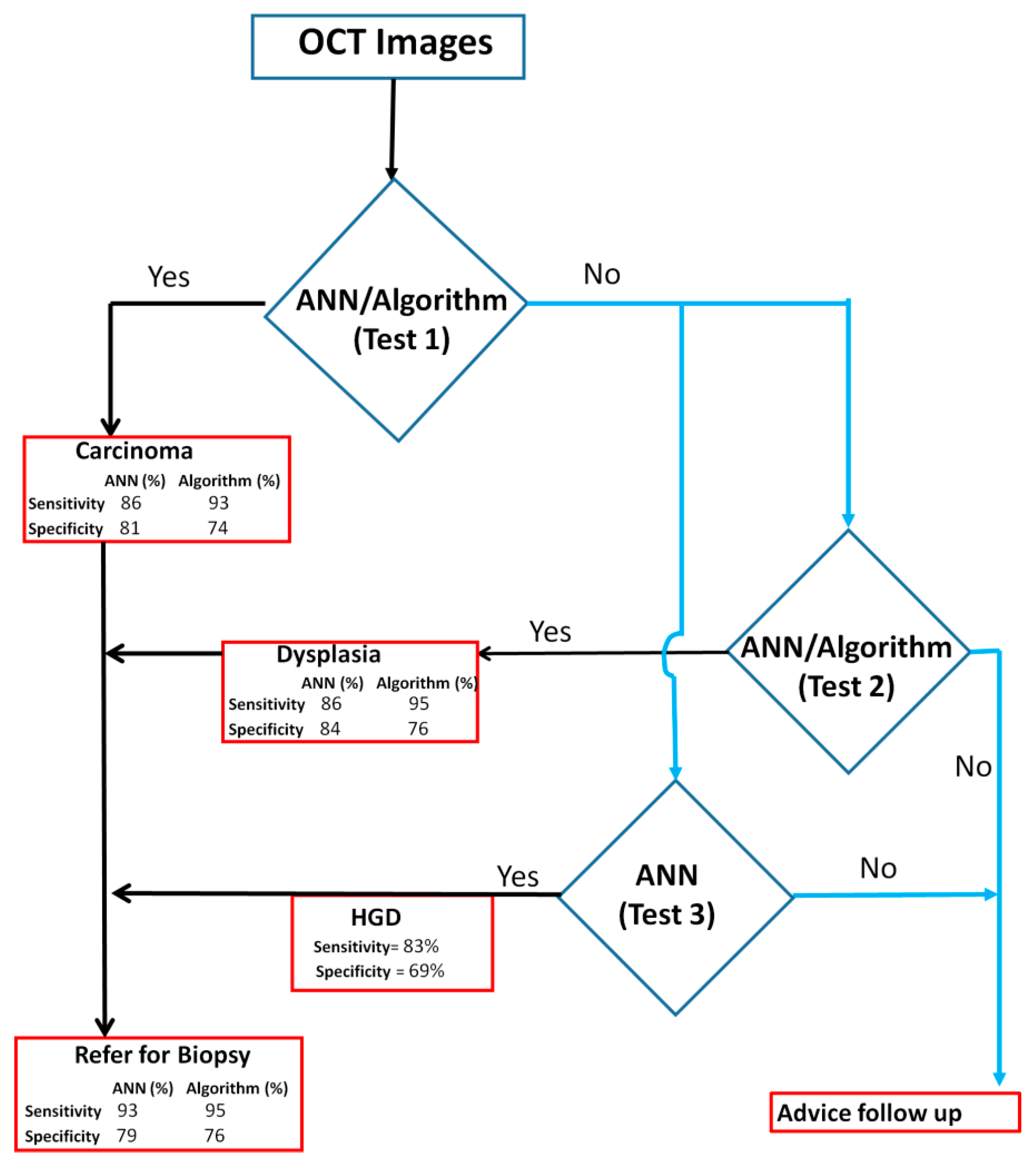
| Method | Diagnosis | Sensitivity (TP/(TP + FN)) | Specificity (TN/(TN + FP)) | PPV | NPV |
|---|---|---|---|---|---|
| Algorithm-Score | OSCC Vs Dysplasia/Benign/Normal | 93(51/55) | 74(87/117) | 63 | 96 |
| Dysplasia VsBenign/Normal | 95(91/96) | 76(16/21) | 95 | 76 | |
| DensNet-201-SVM | OSCC Vs Dysplasia/Benign/Normal | 86(43/50) | 81(179/221) | 51 | 96 |
| Dysplasia Vs Benign/Normal | 84(81/97) | 82(101/124) | 78 | 86 | |
| Inception-ResNet-v2-SVM | HGD Vs LGD/Benign/Normal | 83(63/75) | 69(100/146) | 58 | 89 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
James, B.L.; Sunny, S.P.; Heidari, A.E.; Ramanjinappa, R.D.; Lam, T.; Tran, A.V.; Kankanala, S.; Sil, S.; Tiwari, V.; Patrick, S.; et al. Validation of a Point-of-Care Optical Coherence Tomography Device with Machine Learning Algorithm for Detection of Oral Potentially Malignant and Malignant Lesions. Cancers 2021, 13, 3583. https://doi.org/10.3390/cancers13143583
James BL, Sunny SP, Heidari AE, Ramanjinappa RD, Lam T, Tran AV, Kankanala S, Sil S, Tiwari V, Patrick S, et al. Validation of a Point-of-Care Optical Coherence Tomography Device with Machine Learning Algorithm for Detection of Oral Potentially Malignant and Malignant Lesions. Cancers. 2021; 13(14):3583. https://doi.org/10.3390/cancers13143583
Chicago/Turabian StyleJames, Bonney Lee, Sumsum P. Sunny, Andrew Emon Heidari, Ravindra D. Ramanjinappa, Tracie Lam, Anne V. Tran, Sandeep Kankanala, Shiladitya Sil, Vidya Tiwari, Sanjana Patrick, and et al. 2021. "Validation of a Point-of-Care Optical Coherence Tomography Device with Machine Learning Algorithm for Detection of Oral Potentially Malignant and Malignant Lesions" Cancers 13, no. 14: 3583. https://doi.org/10.3390/cancers13143583
APA StyleJames, B. L., Sunny, S. P., Heidari, A. E., Ramanjinappa, R. D., Lam, T., Tran, A. V., Kankanala, S., Sil, S., Tiwari, V., Patrick, S., Pillai, V., Shetty, V., Hedne, N., Shah, D., Shah, N., Chen, Z.-p., Kandasarma, U., Raghavan, S. A., Gurudath, S., ... Kuriakose, M. A. (2021). Validation of a Point-of-Care Optical Coherence Tomography Device with Machine Learning Algorithm for Detection of Oral Potentially Malignant and Malignant Lesions. Cancers, 13(14), 3583. https://doi.org/10.3390/cancers13143583








