Systematic Roadmap for Cancer Drug Screening Using Zebrafish Embryo Xenograft Cancer Models: Melanoma Cell Line as a Case Study
Abstract
Simple Summary
Abstract
1. Introduction
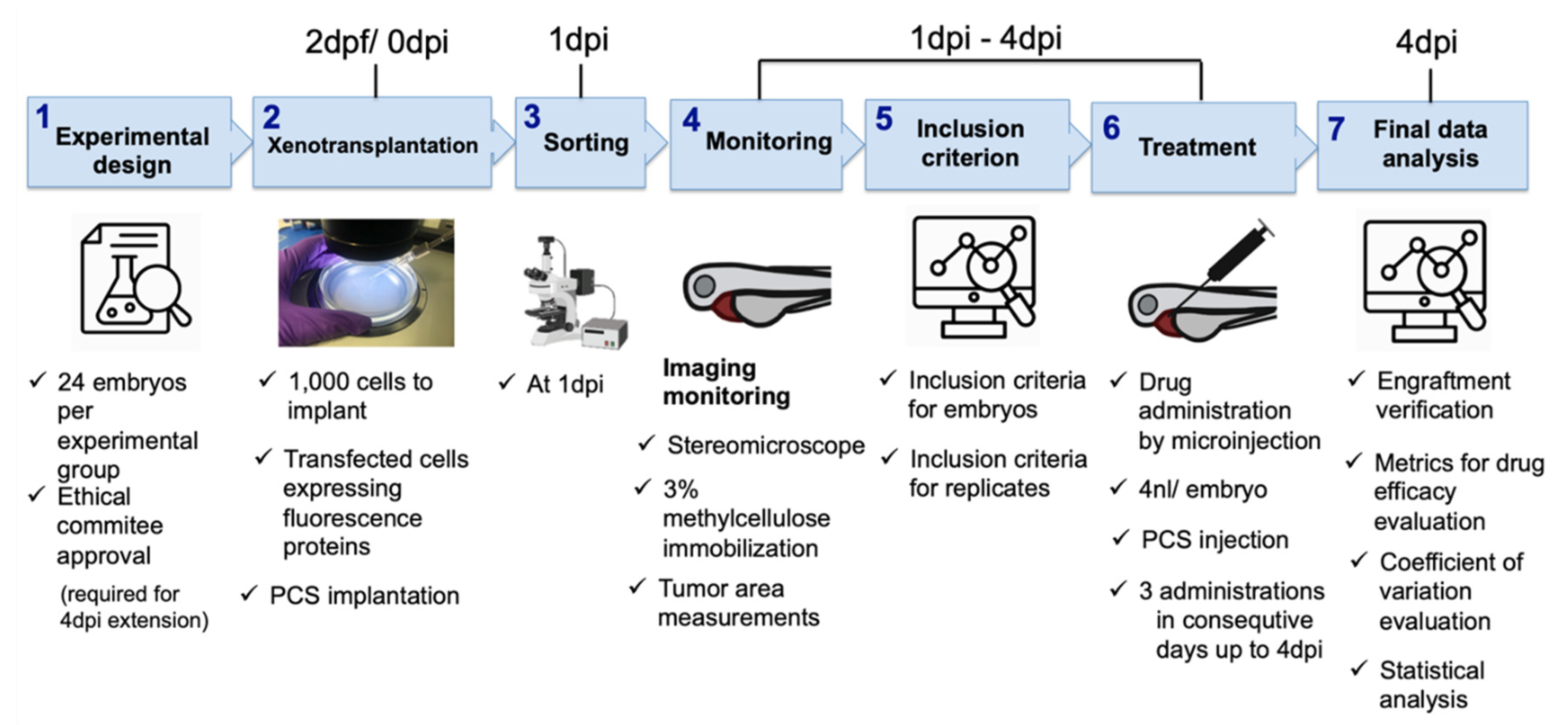
2. Materials and Methods
2.1. Image Analysis
2.2. RNA/DNA Genetic Extraction and Reverse Transcription
2.3. Primers Quality Test
2.4. qPCR Standard Curves
2.5. Zebrafish Maintenance and Egg Collection
2.6. Cell line and Culture Methods
2.7. Xenotransplantation and Injection Site Experiment
2.8. Engraftment Definition and Experimental Inclusion Criteria
2.8.1. Biodistribution and Dissemination Assay
2.8.2. Drug
2.8.3. In Vitro Cell Viability Assay (MTS)
2.8.4. Maximum Tolerated Dose (MTD) Assay
2.8.5. In Vivo Drug Efficacy Assays
2.8.6. Metrics Definition for Efficacy Assays
2.8.7. Statistical Methods
3. Results
3.1. Monitoring Approaches
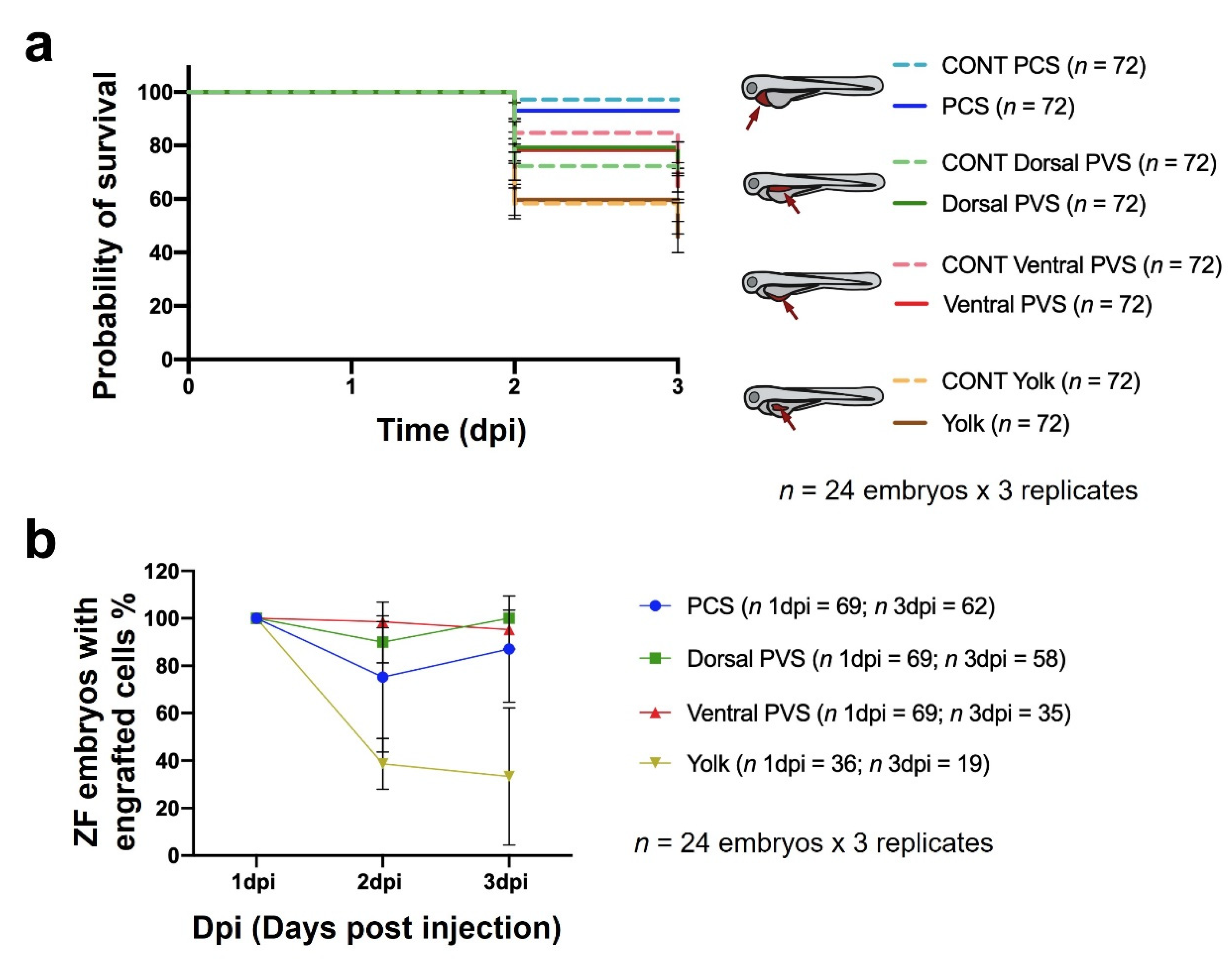
3.2. Injection Site Assay
3.3. Compound Administration and Biodistribution Assay
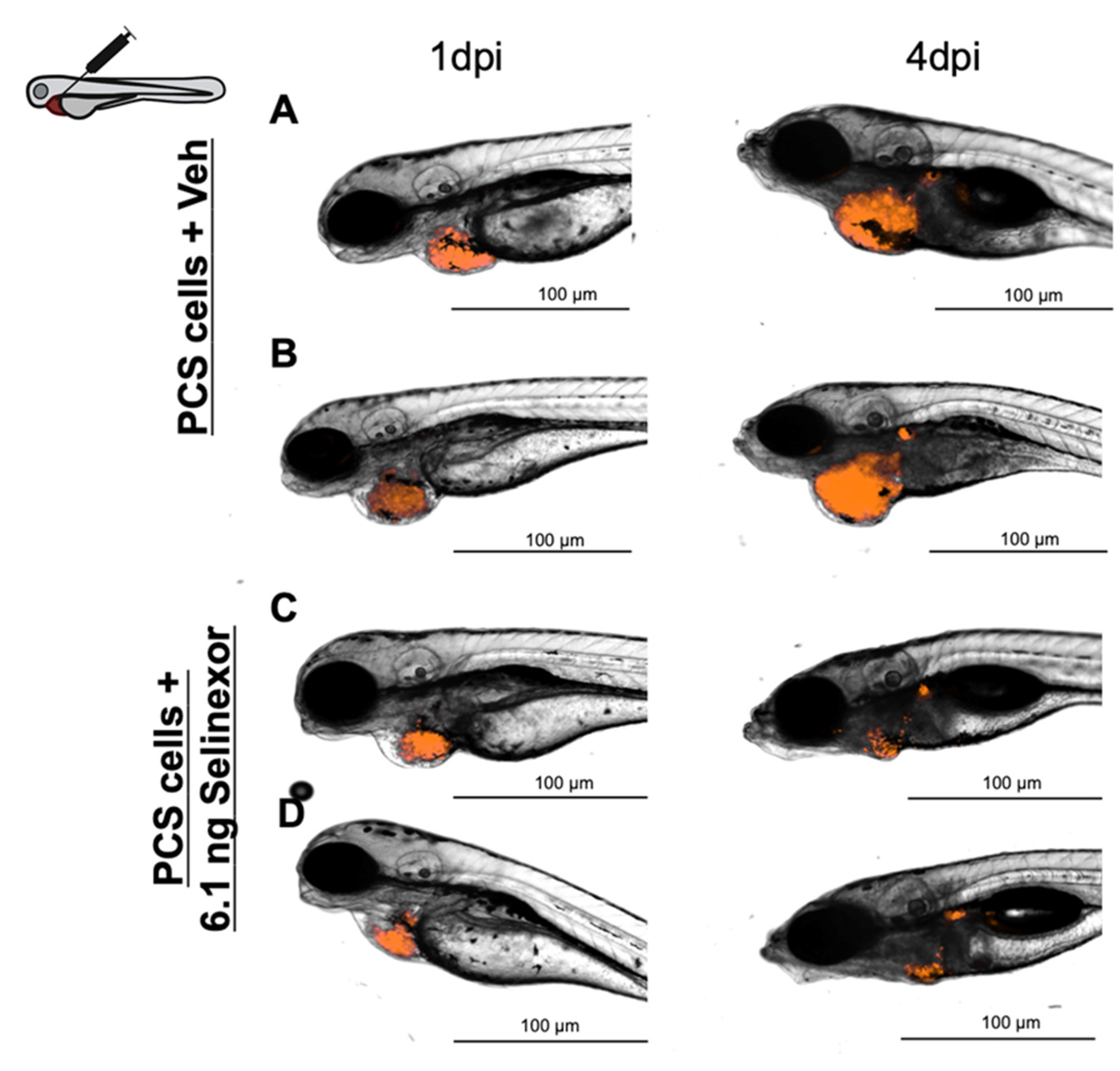
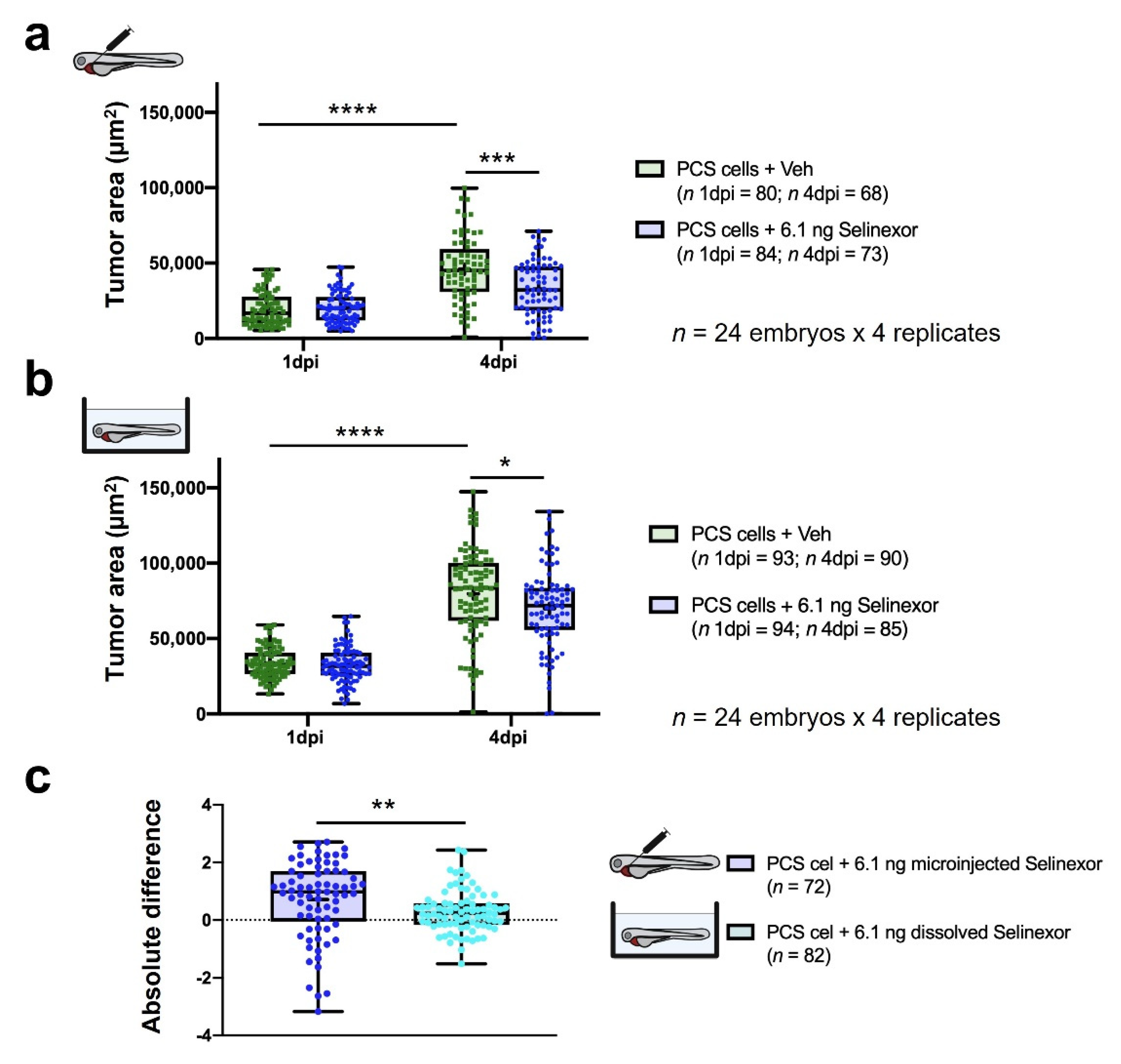
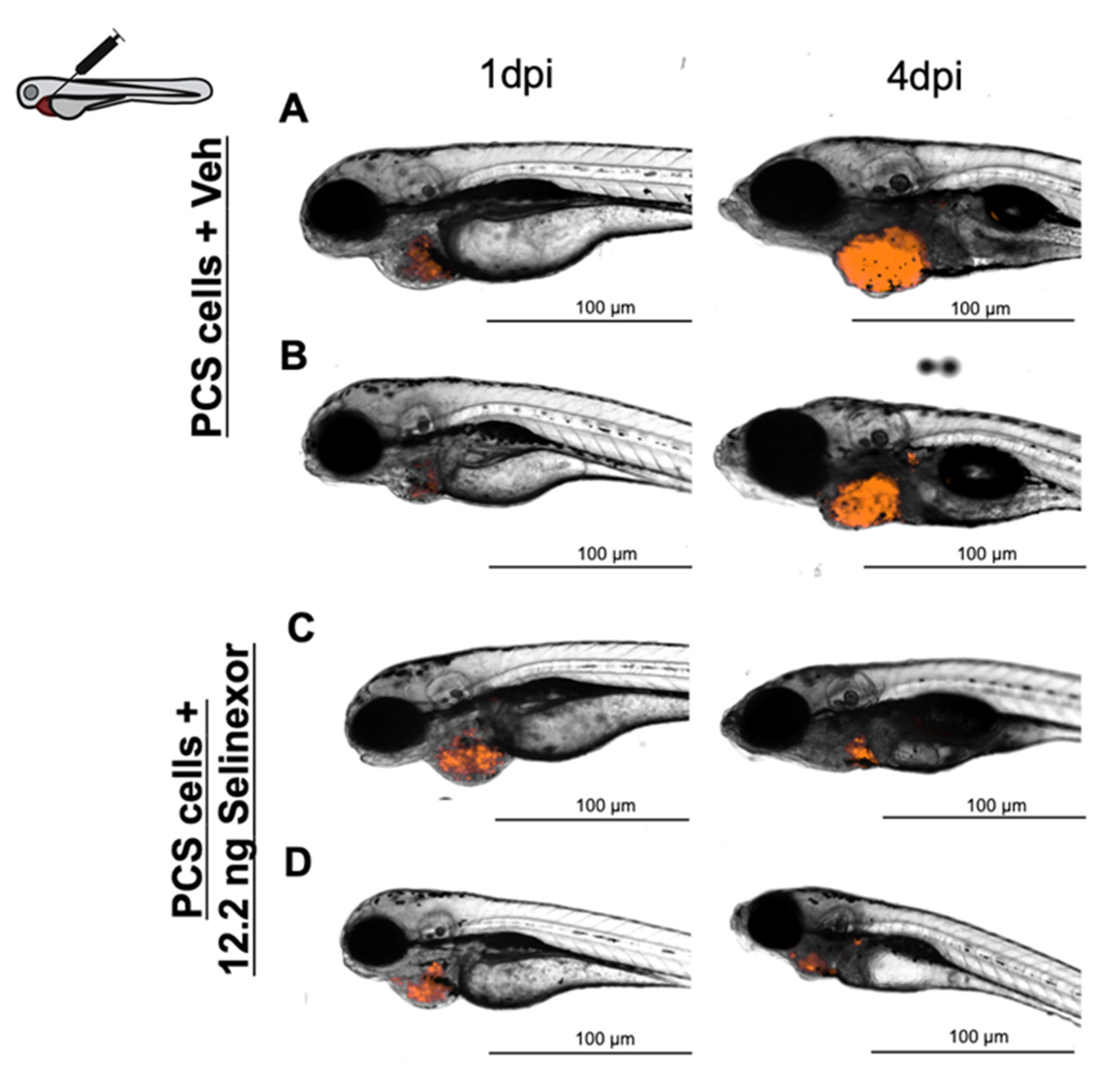
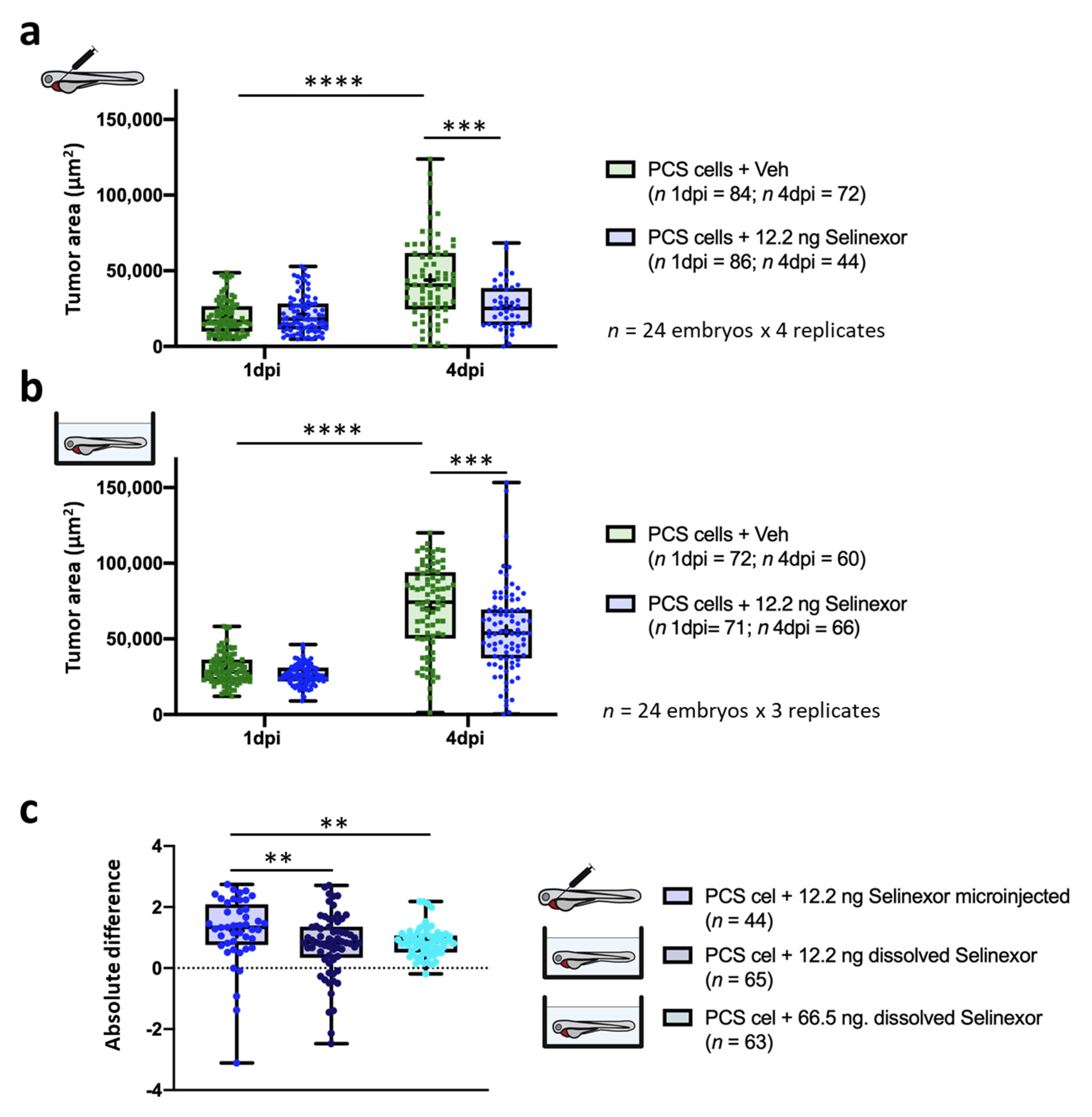
3.4. In Vivo Efficacy Assays
4. Discussion
5. Conclusions
- (1)
- Experimental design. According to our results, twenty-four embryos per experimental group and three independent replicates were enough to see statistically significant results on the final day of the experiment and increased sensitivity to detect smaller antitumoral effects precluding false negatives. Our experimental setup was extended to 4 dpi, which had a clear added value and had a statistically significant impact on the efficacy assessment window and the DTG metric; therefore, ethical approval was required.
- (2)
- Cancer cell xenotransplantation. A total of 1000 cells was sufficient to detect tumor engraftment by imaging, and our results discourage the use of the yolk as the site of implantation. Conversely, the PCS was identified as the best site, as it yielded higher rates of cancer cell engraftment and was less harmful than the other sites, as determined by the engrafted embryos showing higher survival rates.
- (3)
- Sorting. At 1 dpi, sorting should be carefully performed using a fluorescence stereomicroscope to select properly microinjected embryos that exhibit cells at only the correct location and homogeneous tumor masses.
- (4)
- Tumor monitoring. As reported above, gDNA qPCR is a less time-consuming technique that provides some advantages for only ventral PVS implantation. However, due to its detrimental impact on statistical power (requires pools of embryos) and due to the PCS being selected as the site of tumor implantation, fluorescence imaging is the proposed tumor monitoring technique.
- (5)
- User-independent inclusion criteria. We aimed to propose a systematic screening roadmap, including a user-independent decision-making process, to minimize variability and maximize reproducibility. Therefore, we defined assay-based user-independent criteria, including the inclusion threshold (IT), to the efficacy assessment of only embryos that presented with a TA at 1 dpi that was higher than the IT, which was key for decision making.
- (6)
- Treatment. According to our results, compound administration by direct intratumoral inoculation is the best approach to treat engrafted embryos and for reliable efficacy assessments. As the experimental setup was extended to 4 dpi, compounds were administered for three consecutive days.
- (7)
- Final data analysis. Only embryos meeting the inclusion criteria (IT) were considered for efficacy assessment and decision making.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fior, R.; Póvoa, V.; Mendes, R.V.; Carvalho, T.; Gomes, A.; Figueiredo, N.; Ferreira, M.G. Single-cell functional and chemosensitive profiling of combinatorial colorectal therapy in zebrafish xenografts. Proc. Natl. Acad. Sci. USA 2017, 114, E8234–E8243. [Google Scholar] [CrossRef]
- Letrado, P.; De Miguel, I.; Lamberto, I.; Díez-Martínez, R.; Oyarzabal, J. Zebrafish: Speeding Up the Cancer Drug Discovery Process. Cancer Res. 2018, 78, 6048–6058. [Google Scholar] [CrossRef]
- van der Ent, W.; Jochemsen, A.G.; Teunisse, A.F.; Krens, S.G.; Szuhai, K.; Spaink, H.P.; Hogendoorn, P.C.; Snaar-Jagalska, B.E. Ewing sarcoma inhibition by disruption of EWSR1-FLI1 transcriptional activity and reactivation of p53. J. Pathol. 2014, 233, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Brunson, D.; Tang, Q.; Do, D.; Iftimia, N.A.; Moore, J.C.; Hayes, M.N.; Welker, A.M.; Garcia, E.G.; Dubash, T.D.; et al. Visualizing Engrafted Human Cancer and Therapy Responses in Immunodeficient Zebrafish. Cell 2019, 177, 1903–1914. [Google Scholar] [CrossRef] [PubMed]
- Huiting, L.N.; Laroche, F.; Feng, H. The Zebrafish as a Tool to Cancer Drug Discovery. Austin J. Pharmacol. Ther. 2015, 3, 1069. [Google Scholar]
- Póvoa, V.; de Almeida, C.R.; Maia-Gil, M.; Sobral, D.; Domingues, M.; Martinez-Lopez, M.; Fuzeta, M.D.A.; Silva, C.; Grosso, A.R.; Fior, R. Innate immune evasion revealed in a colorectal zebrafish xenograft model. Nat. Commun. 2021, 12, 1–15. [Google Scholar] [CrossRef]
- Jin, W.; Zhou, L.; Yan, B.; Yan, L.; Liu, F.; Tong, P.; Yu, W.; Dong, X.; Xie, L.; Zhang, J.; et al. Theabrownin triggersDNAdamage to suppress human osteosarcoma U2OScells by activating p53 signalling pathway. J. Cell. Mol. Med. 2018, 22, 4423–4436. [Google Scholar] [CrossRef]
- Xu, W.; Foster, B.A.; Richards, M.; Bondioli, K.R.; Shah, G.; Green, C.C. Characterization of prostate cancer cell progression in zebrafish xenograft model. Int. J. Oncol. 2017, 52, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Avci, M.E.; Keskus, A.; Targen, S.; Isilak, M.E.; Ozturk, M.; Atalay, R.C.; Adams, M.M.; Konu, Ö. Development of a novel zebrafish xenograft model in ache mutants using liver cancer cell lines. Sci. Rep. 2018, 8, 1570. [Google Scholar] [CrossRef] [PubMed]
- Wertman, J.; Veinotte, C.J.; Dellaire, G.; Berman, J.N. The Zebrafish Xenograft Platform: Evolution of a Novel Cancer Model and Preclinical Screening Tool. Adv. Exp. Med. Biol. 2016, 916, 289–314. [Google Scholar] [CrossRef]
- Lieschke, G.J.; Currie, P. Animal models of human disease: Zebrafish swim into view. Nat. Rev. Genet. 2007, 8, 353–367. [Google Scholar] [CrossRef]
- Zhang, F.; Qin, W.; Zhang, J.-P.; Hu, C.-Q. Antibiotic Toxicity and Absorption in Zebrafish Using Liquid Chromatography-Tandem Mass Spectrometry. PLoS ONE 2015, 10, e0124805. [Google Scholar] [CrossRef]
- Fda, C.; Purdie, F.P. Waiver of In Vivo Bioavailability and Bioequivalence Studies for Immediate-Release Solid Oral Dosage Forms Based on a Biopharmaceutics Classification System Guidance for Industry; U.S. Food and Drug Administraiton: Silver Spring, MD, USA, 2017.
- Ku, M.S. Use of the Biopharmaceutical Classification System in Early Drug Development. AAPS J. 2008, 10, 208–212. [Google Scholar] [CrossRef]
- Vliegenthart, A.D.B.; Tucker, C.S.; Del Pozo, J.; Dear, J.W. Zebrafish as model organisms for studying drug-induced liver injury. Br. J. Clin. Pharmacol. 2014, 78, 1217–1227. [Google Scholar] [CrossRef]
- Pugach, E.K.; Li, P.; White, R.; Zon, L. Retro-orbital Injection in Adult Zebrafish. J. Vis. Exp. 2009, e1645. [Google Scholar] [CrossRef] [PubMed]
- Gutzman, J.H.; Sive, H. Zebrafish Brain Ventricle Injection. J. Vis. Exp. 2009, e1218. [Google Scholar] [CrossRef]
- Tanimoto, M.; Ota, Y.; Inoue, M.; Oda, Y. Origin of Inner Ear Hair Cells: Morphological and Functional Differentiation from Ciliary Cells into Hair Cells in Zebrafish Inner Ear. J. Neurosci. 2011, 31, 3784–3794. [Google Scholar] [CrossRef]
- Li, X.-Y.; Huang, L.-T.; Wu, J.-Q.; He, M.-F.; Zhu, S.-H.; Zhan, P.; Lv, T.-F.; Song, Y. Zebrafish Xenograft Model of Human Lung Cancer for Evaluating Osimertinib Resistance. BioMed Res. Int. 2019, 2019, 3129748-10. [Google Scholar] [CrossRef] [PubMed]
- Westerfield, M. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Danio Rerio), 4th ed.; University of Oregon Press: Eugene, OR, USA, 2000. [Google Scholar]
- Montgomery, D.C. Blocks, Latin squares and related designs of Experiments. In Design and Analysis of Experiments, 8th ed.; Ratts, L.M.A., Ed.; John Wiley & Sons: Hoboken, NJ, USA, 2012; p. 168. [Google Scholar]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Chem. Biol. 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Dice, L.R. Measures of the Amount of Ecologic Association Between Species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]
- Guo, Y.; Veneman, W.J.; Spaink, H.P.; Verbeek, F.J. Three-dimensional reconstruction and measurements of zebrafish larvae from high-throughput axial-view in vivo imaging. Biomed. Opt. Express 2017, 8, 2611–2634. [Google Scholar] [CrossRef]
- Molenberghs, G.; Bijnens, L.; Shaw, D. Linear mixed models and missing data. In Dependence in Probability and Statistics; Springer Science and Business Media LLC.: Berlin/Heidelberg, Germany, 1997; pp. 191–274. [Google Scholar]
- Ibrahim, J.G.; Molenberghs, G. Missing data methods in longitudinal studies: A review. TEST 2009, 18, 1–43. [Google Scholar] [CrossRef]
- Amidon, G.L.; Lennernäs, H.; Shah, V.P.; Crison, J.R. A Theoretical Basis for a Biopharmaceutic Drug Classification: The Correlation of in Vitro Drug Product Dissolution and in Vivo Bioavailability. Pharm. Res. 1995, 12, 413–420. [Google Scholar] [CrossRef]
- Yu, L.X.; Amidon, G.L.; Polli, J.E.; Zhao, H.; Mehta, M.; Conner, D.P.; Shah, V.P.; Lesko, L.J.; Chen, M.; Lee, V.H.L.; et al. Biopharmaceutics Classification System: The Scientific Basis for Biowaiver Extensions. Pharm. Res. 2002, 19, 921–925. [Google Scholar] [CrossRef]
- Fda. Center for Drug Evaluation Application Number: 212306Orig1s000 Product Quality Review (S); U.S. Food and Drug Administraiton: Silver Spring, MD, USA, 2019.
- DrugBank. Available online: https://go.drugbank.com/ (accessed on 9 March 2021).
- Rainero, A.; Angaroni, F.; D’Avila, F.; Conti, A.; Pirrone, C.; Micheloni, G.; Tararà, L.; Millefanti, G.; Maserati, E.; Valli, R.; et al. gDNA qPCR is statistically more reliable than mRNA analysis in detecting leukemic cells to monitor CML. Cell Death Dis. 2018, 9, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Gauert, A.; Olk, N.; Pimentel-Gutiérrez, H.; Astrahantseff, K.; Jensen, L.D.; Cao, Y.; Eggert, A.; Eckert, C.; Hagemann, A.I. Fast, In Vivo Model for Drug-Response Prediction in Patients with B-Cell Precursor Acute Lymphoblastic Leukemia. Cancers 2020, 12, 1883. [Google Scholar] [CrossRef] [PubMed]
- Teng, Y.; Xie, X.; Walker, S.; White, D.T.; Mumm, J.S.; Cowell, J.K. Evaluating human cancer cell metastasis in zebrafish. BMC Cancer 2013, 13, 453. [Google Scholar] [CrossRef]
- Kimmel, C.B.; Ballard, W.W.; Kimmel, S.R.; Ullmann, B.; Schilling, T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995, 203, 253–310. [Google Scholar] [CrossRef] [PubMed]
- Pontes, K.C.D.S.; Groenewoud, A.; Cao, J.; Ataíde, L.; Snaar-Jagalska, E.; Jager, M.J. Evaluation of (fli:GFP) Casper Zebrafish Embryos as a Model for Human Conjunctival Melanoma. Investig. Opthalmology Vis. Sci. 2017, 58, 6065–6071. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kislyuk, S. Zebrafish—A Small-Animal Model for ADME Studies. Ph.D. Thesis, KU Leuven, Leuven, Belgium, 2017. [Google Scholar]
- De Almeida, C.R.; Mendes, R.V.; Pezzarossa, A.; Gago, J.; Carvalho, C.; Alves, A.; Nunes, V.; Brito, M.J.; Cardoso, M.J.; Ribeiro, J.; et al. Zebrafish xenografts as a fast screening platform for bevacizumab cancer therapy. Commun. Biol. 2020, 3, 1–13. [Google Scholar] [CrossRef]
- Farren, M.R.; Hennessey, R.; Shakya, R.; Elnaggar, O.; Young, G.; Kendra, K.; Landesman, Y.; Elloul, S.; Crochiere, M.; Klebanov, B.; et al. The Exportin-1 Inhibitor Selinexor Exerts Superior Antitumor Activity when Combined with T-Cell Checkpoint Inhibitors. Mol. Cancer Ther. 2017, 16, 417–427. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Letrado, P.; Mole, H.; Montoya, M.; Palacios, I.; Barriuso, J.; Hurlstone, A.; Díez-Martínez, R.; Oyarzabal, J. Systematic Roadmap for Cancer Drug Screening Using Zebrafish Embryo Xenograft Cancer Models: Melanoma Cell Line as a Case Study. Cancers 2021, 13, 3705. https://doi.org/10.3390/cancers13153705
Letrado P, Mole H, Montoya M, Palacios I, Barriuso J, Hurlstone A, Díez-Martínez R, Oyarzabal J. Systematic Roadmap for Cancer Drug Screening Using Zebrafish Embryo Xenograft Cancer Models: Melanoma Cell Line as a Case Study. Cancers. 2021; 13(15):3705. https://doi.org/10.3390/cancers13153705
Chicago/Turabian StyleLetrado, Patricia, Holly Mole, María Montoya, Irene Palacios, Jorge Barriuso, Adam Hurlstone, Roberto Díez-Martínez, and Julen Oyarzabal. 2021. "Systematic Roadmap for Cancer Drug Screening Using Zebrafish Embryo Xenograft Cancer Models: Melanoma Cell Line as a Case Study" Cancers 13, no. 15: 3705. https://doi.org/10.3390/cancers13153705
APA StyleLetrado, P., Mole, H., Montoya, M., Palacios, I., Barriuso, J., Hurlstone, A., Díez-Martínez, R., & Oyarzabal, J. (2021). Systematic Roadmap for Cancer Drug Screening Using Zebrafish Embryo Xenograft Cancer Models: Melanoma Cell Line as a Case Study. Cancers, 13(15), 3705. https://doi.org/10.3390/cancers13153705






