Inducible Liver Cancer Models in Transgenic Zebrafish to Investigate Cancer Biology
Abstract
:Simple Summary
Abstract
1. Introduction
2. Beginning of Our Liver Cancer Journey
3. Established Transgenic Liver Cancer Models
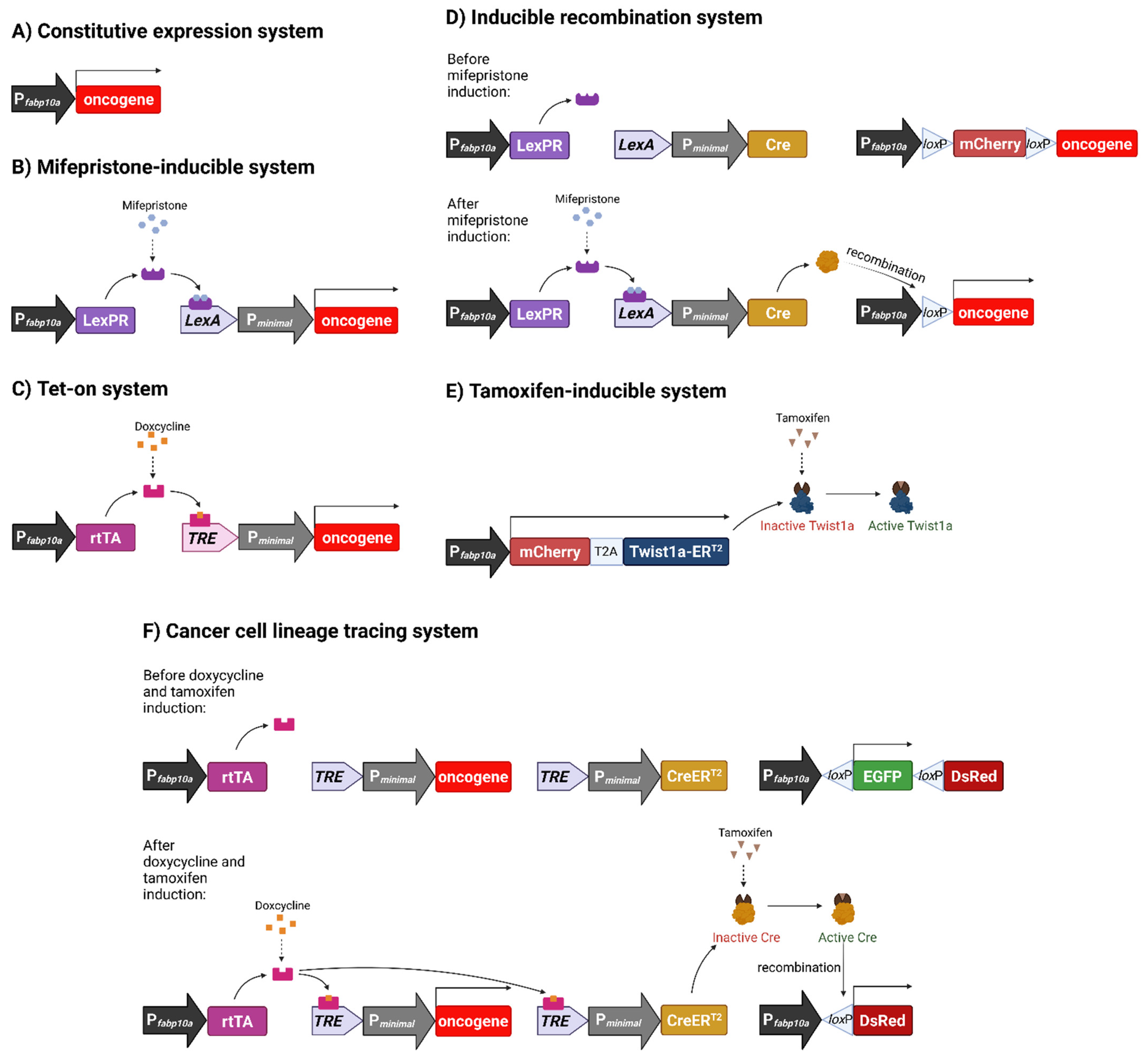
3.1. Constitutive Oncogene Expression Model
3.2. Chemically Inducible Oncogene and Cytokine Overexpression
3.3. Inducible Permanent Oncogene Expression
3.4. Metastatic Model of HCC
3.5. Histological, Transcriptomic and Molecular Validation
4. Important Findings from Our Models
4.1. Oncogene Addiction: Tumour Regression and Re-Induction
4.2. Immune Response and Inflammation
4.3. Sex Disparity and Hormones
4.4. Muscle-Wasting and Cancer Cachexia
4.5. Chemical and Toxicological Screening Using Zebrafish HCC Models
5. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Liu, Z.; Jiang, Y.; Yuan, H.; Fang, Q.; Cai, N.; Suo, C.; Jin, L.; Zhang, T.; Chen, X. The trends in incidence of primary liver cancer caused by specific etiologies: Results from the Global Burden of Disease Study 2016 and implications for liver cancer prevention. J. Hepatol. 2019, 70, 674–683. [Google Scholar] [CrossRef]
- Marin, J.J.G.; Briz, O.; Herraez, E.; Lozano, E.; Asensio, M.; Di Giacomo, S.; Romero, M.R.; Osorio-Padilla, L.M.; Santos-Llamas, A.I.; Serrano, M.A.; et al. Molecular bases of the poor response of liver cancer to chemotherapy. Clin. Res. Hepatol. Gastroenterol. 2018, 42, 182–192. [Google Scholar] [CrossRef]
- Ko, K.L.; Mak, L.Y.; Cheung, K.S.; Yuen, M.F. Hepatocellular carcinoma: Recent advances and emerging medical therapies. F1000Research 2020, 9. [Google Scholar] [CrossRef] [PubMed]
- Recio-Boiles, A.; Babiker, H.M. Liver Cancer; StatPearls Publishing LLC: Treasure Island, FL, USA, 2021. [Google Scholar]
- Eisen, J.S. History of Zebrafish Research. In The Zebrafish in Biomedical Research; Cartner, S.C., Eisen, J.S., Farmer, S.C., Guillemin, K.J., Kent, M.L., Sanders, G.E., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 3–14. [Google Scholar]
- Thisse, C.; Thisse, B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 2008, 3, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Rafferty, S.A.; Quinn, T.A. A beginner’s guide to understanding and implementing the genetic modification of zebrafish. Prog. Biophys. Mol. Biol. 2018, 138, 3–19. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Leach, S.D. Zebrafish models for cancer. Annu. Rev. Pathol. 2011, 6, 71–93. [Google Scholar] [CrossRef]
- McConnell, A.M.; Noonan, H.R.; Zon, L.I. Reeling in the Zebrafish Cancer Models. Annu. Rev. Cancer Biol. 2021, 5, 331–350. [Google Scholar] [CrossRef]
- Barbazuk, W.B.; Korf, I.; Kadavi, C.; Heyen, J.; Tate, S.; Wun, E.; Bedell, J.A.; McPherson, J.D.; Johnson, S.L. The syntenic relationship of the zebrafish and human genomes. Genome Res. 2000, 10, 1351–1358. [Google Scholar] [CrossRef] [Green Version]
- Howe, K.; Clark, M.D.; Torroja, C.F.; Torrance, J.; Berthelot, C.; Muffato, M.; Collins, J.E.; Humphray, S.; McLaren, K.; Matthews, L.; et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature 2013, 496, 498–503. [Google Scholar] [CrossRef] [Green Version]
- Xu, C.; Zon, L.I. The zebrafish as a model for human disease. In Fish Physiology; Perry, S.F., Ekker, M., Farrell, A.P., Brauner, C.J., Eds.; Academic Press: Cambridge, MA, USA, 2010; Volume 29, pp. 345–365. [Google Scholar]
- Goldsmith, J.R.; Jobin, C. Think Small: Zebrafish as a Model System of Human Pathology. J. Biomed. Biotechnol. 2012, 2012, 1–12. [Google Scholar] [CrossRef]
- Santoriello, C.; Zon, L.I. Hooked! Modeling human disease in zebrafish. J. Clin. Investig. 2012, 122, 2337–2343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bradford, Y.M.; Toro, S.; Ramachandran, S.; Ruzicka, L.; Howe, D.G.; Eagle, A.; Kalita, P.; Martin, R.; Taylor Moxon, S.A.; Schaper, K.; et al. Zebrafish Models of Human Disease: Gaining Insight into Human Disease at ZFIN. ILAR J. 2017, 58, 4–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evason, K.J.; Francisco, M.T.; Juric, V.; Balakrishnan, S.; Lopez Pazmino Mdel, P.; Gordan, J.D.; Kakar, S.; Spitsbergen, J.; Goga, A.; Stainier, D.Y. Identification of Chemical Inhibitors of beta-Catenin-Driven Liver Tumorigenesis in Zebrafish. PLoS Genet. 2015, 11, e1005305. [Google Scholar] [CrossRef]
- Lu, J.W.; Liao, C.Y.; Yang, W.Y.; Lin, Y.M.; Jin, S.L.; Wang, H.D.; Yuh, C.H. Overexpression of endothelin 1 triggers hepatocarcinogenesis in zebrafish and promotes cell proliferation and migration through the AKT pathway. PLoS ONE 2014, 9, e85318. [Google Scholar] [CrossRef] [Green Version]
- Mudbhary, R.; Hoshida, Y.; Chernyavskaya, Y.; Jacob, V.; Villanueva, A.; Fiel, M.I.; Chen, X.; Kojima, K.; Thung, S.; Bronson, R.T.; et al. UHRF1 overexpression drives DNA hypomethylation and hepatocellular carcinoma. Cancer Cell 2014, 25, 196–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, J.W.; Yang, W.Y.; Tsai, S.M.; Lin, Y.M.; Chang, P.H.; Chen, J.R.; Wang, H.D.; Wu, J.L.; Jin, S.L.; Yuh, C.H. Liver-specific expressions of HBx and src in the p53 mutant trigger hepatocarcinogenesis in zebrafish. PLoS ONE 2013, 8, e76951. [Google Scholar] [CrossRef]
- Rekha, R.D.; Amali, A.A.; Her, G.M.; Yeh, Y.H.; Gong, H.Y.; Hu, S.Y.; Lin, G.H.; Wu, J.L. Thioacetamide accelerates steatohepatitis, cirrhosis and HCC by expressing HCV core protein in transgenic zebrafish Danio rerio. Toxicology 2008, 243, 11–22. [Google Scholar] [CrossRef]
- Mizgirev, I.; Revskoy, S. Generation of clonal zebrafish lines and transplantable hepatic tumors. Nat. Protoc. 2010, 5, 383–394. [Google Scholar] [CrossRef]
- Avci, M.E.; Keskus, A.G.; Targen, S.; Isilak, M.E.; Ozturk, M.; Atalay, R.C.; Adams, M.M.; Konu, O. Development of a novel zebrafish xenograft model in ache mutants using liver cancer cell lines. Sci. Rep. 2018, 8, 1570. [Google Scholar] [CrossRef] [Green Version]
- Khan, N.; Mahajan, N.K.; Sinha, P.; Jayandharan, G.R. An efficient method to generate xenograft tumor models of acute myeloid leukemia and hepatocellular carcinoma in adult zebrafish. Blood Cells Mol. Dis. 2019, 75, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Wrighton, P.J.; Oderberg, I.M.; Goessling, W. There Is Something Fishy About Liver Cancer: Zebrafish Models of Hepatocellular Carcinoma. Cell. Mol. Gastroenterol. Hepatol. 2019, 8, 347–363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakayama, J.; Gong, Z. Transgenic zebrafish for modeling hepatocellular carcinoma. MedComm 2020, 1, 140–156. [Google Scholar] [CrossRef]
- Li, X.; Benjamin, I.S.; Alexander, B. Reproducible production of thioacetamide-induced macronodular cirrhosis in the rat with no mortality. J. Hepatol. 2002, 36, 488–493. [Google Scholar] [CrossRef]
- Groos, J.; Bannasch, P.; Schwarz, M.; Kopp-Schneider, A. Comparison of mode of action of four hepatocarcinogens: A model-based approach. Toxicol. Sci. 2007, 99, 446–454. [Google Scholar] [CrossRef] [Green Version]
- Spitsbergen, J.M.; Tsai, H.W.; Reddy, A.; Miller, T.; Arbogast, D.; Hendricks, J.D.; Bailey, G.S. Neoplasia in zebrafish (Danio rerio) treated with N-methyl-N′-nitro-N-nitrosoguanidine by three exposure routes at different developmental stages. Toxicol. Pathol. 2000, 28, 716–725. [Google Scholar] [CrossRef]
- Spitsbergen, J.M.; Tsai, H.W.; Reddy, A.; Miller, T.; Arbogast, D.; Hendricks, J.D.; Bailey, G.S. Neoplasia in zebrafish (Danio rerio) treated with 7,12-dimethylbenz[a]anthracene by two exposure routes at different developmental stages. Toxicol. Pathol. 2000, 28, 705–715. [Google Scholar] [CrossRef]
- Mizgireuv, I.V.; Majorova, I.G.; Gorodinskaya, V.M.; Khudoley, V.V.; Revskoy, S.Y. Carcinogenic effect of N-nitrosodimethylamine on diploid and triploid zebrafish (Danio rerio). Toxicol. Pathol. 2004, 32, 514–518. [Google Scholar] [CrossRef] [Green Version]
- Lam, S.H.; Wu, Y.L.; Vega, V.B.; Miller, L.D.; Spitsbergen, J.; Tong, Y.; Zhan, H.; Govindarajan, K.R.; Lee, S.; Mathavan, S.; et al. Conservation of gene expression signatures between zebrafish and human liver tumors and tumor progression. Nat. Biotechnol. 2006, 24, 73–75. [Google Scholar] [CrossRef]
- Lam, S.H.; Gong, Z. Modeling Liver Cancer Using Zebrafish: A Comparative Oncogenomics Approach. Cell Cycle 2006, 5, 573–577. [Google Scholar] [CrossRef]
- Nguyen, A.T.; Emelyanov, A.; Koh, C.H.; Spitsbergen, J.M.; Lam, S.H.; Mathavan, S.; Parinov, S.; Gong, Z. A high level of liver-specific expression of oncogenic Kras(V12) drives robust liver tumorigenesis in transgenic zebrafish. Dis. Model. Mech. 2011, 4, 801–813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arrington, A.K.; Heinrich, E.L.; Lee, W.; Duldulao, M.; Patel, S.; Sanchez, J.; Garcia-Aguilar, J.; Kim, J. Prognostic and predictive roles of KRAS mutation in colorectal cancer. Int. J. Mol. Sci. 2012, 13, 12153–12168. [Google Scholar] [CrossRef] [Green Version]
- Valtorta, E.; Misale, S.; Sartore-Bianchi, A.; Nagtegaal, I.D.; Paraf, F.; Lauricella, C.; Dimartino, V.; Hobor, S.; Jacobs, B.; Ercolani, C.; et al. KRAS gene amplification in colorectal cancer and impact on response to EGFR-targeted therapy. Int. J. Cancer 2013, 133, 1259–1265. [Google Scholar] [CrossRef] [PubMed]
- Favazza, L.A.; Parseghian, C.M.; Kaya, C.; Nikiforova, M.N.; Roy, S.; Wald, A.I.; Landau, M.S.; Proksell, S.S.; Dueker, J.M.; Johnston, E.R.; et al. KRAS amplification in metastatic colon cancer is associated with a history of inflammatory bowel disease and may confer resistance to anti-EGFR therapy. Mod. Pathol. 2020, 33, 1832–1843. [Google Scholar] [CrossRef] [PubMed]
- Karachaliou, N.; Mayo, C.; Costa, C.; Magri, I.; Gimenez-Capitan, A.; Molina-Vila, M.A.; Rosell, R. KRAS mutations in lung cancer. Clin. Lung Cancer 2013, 14, 205–214. [Google Scholar] [CrossRef]
- Wagner, P.L.; Perner, S.; Rickman, D.S.; LaFargue, C.J.; Kitabayashi, N.; Johnstone, S.F.; Weir, B.A.; Meyerson, M.; Altorki, N.K.; Rubin, M.A. In situ evidence of KRAS amplification and association with increased p21 levels in non-small cell lung carcinoma. Am. J. Clin. Pathol. 2009, 132, 500–505. [Google Scholar] [CrossRef] [Green Version]
- Wagner, P.L.; Stiedl, A.C.; Wilbertz, T.; Petersen, K.; Scheble, V.; Menon, R.; Reischl, M.; Mikut, R.; Rubin, M.A.; Fend, F.; et al. Frequency and clinicopathologic correlates of KRAS amplification in non-small cell lung carcinoma. Lung Cancer 2011, 74, 118–123. [Google Scholar] [CrossRef]
- Delire, B.; Starkel, P. The Ras/MAPK pathway and hepatocarcinoma: Pathogenesis and therapeutic implications. Eur. J. Clin. Investig. 2015, 45, 609–623. [Google Scholar] [CrossRef]
- Emelyanov, A.; Parinov, S. Mifepristone-inducible LexPR system to drive and control gene expression in transgenic zebrafish. Dev. Biol. 2008, 320, 113–121. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, A.T.; Emelyanov, A.; Koh, C.H.; Spitsbergen, J.M.; Parinov, S.; Gong, Z. An inducible kras(V12) transgenic zebrafish model for liver tumorigenesis and chemical drug screening. Dis. Models Mech. 2012, 5, 63–72. [Google Scholar] [CrossRef] [Green Version]
- Loew, R.; Heinz, N.; Hampf, M.; Bujard, H.; Gossen, M. Improved Tet-responsive promoters with minimized background expression. BMC Biotechnol. 2010, 10, 81. [Google Scholar] [CrossRef] [Green Version]
- Chew, T.W.; Liu, X.J.; Liu, L.; Spitsbergen, J.M.; Gong, Z.; Low, B.C. Crosstalk of Ras and Rho: Activation of RhoA abates Kras-induced liver tumorigenesis in transgenic zebrafish models. Oncogene 2014, 33, 2717–2727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Huang, X.; Zhan, H.; Zeng, Z.; Li, C.; Spitsbergen, J.M.; Meierjohann, S.; Schartl, M.; Gong, Z. Inducible and repressable oncogene-addicted hepatocellular carcinoma in Tet-on xmrk transgenic zebrafish. J. Hepatol. 2012, 56, 419–425. [Google Scholar] [CrossRef]
- Li, Z.; Zheng, W.; Wang, Z.; Zeng, Z.; Zhan, H.; Li, C.; Zhou, L.; Yan, C.; Spitsbergen, J.M.; Gong, Z. A transgenic zebrafish liver tumor model with inducible Myc expression reveals conserved Myc signatures with mammalian liver tumors. Dis. Models Mech. 2013, 6, 414–423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monroe, J.D.; Basheer, F.; Gibert, Y. Xmrks the Spot: Fish Models for Investigating Epidermal Growth Factor Receptor Signaling in Cancer Research. Cells 2021, 10, 1132. [Google Scholar] [CrossRef]
- Berasain, C.; Ujue Latasa, M.; Urtasun, R.; Goni, S.; Elizalde, M.; Garcia-Irigoyen, O.; Azcona, M.; Prieto, J.; Avila, M.A. Epidermal Growth Factor Receptor (EGFR) Crosstalks in Liver Cancer. Cancers 2011, 3, 2444–2461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Wu, M.C.; Sham, J.S.; Zhang, W.; Wu, W.Q.; Guan, X.Y. Prognostic significance of c-myc and AIB1 amplification in hepatocellular carcinoma. A broad survey using high-throughput tissue microarray. Cancer 2002, 95, 2346–2352. [Google Scholar] [CrossRef] [PubMed]
- Thorgeirsson, S.S.; Santoni-Rugiu, E. Transgenic mouse models in carcinogenesis: Interaction of c-myc with transforming growth factor alpha and hepatocyte growth factor in hepatocarcinogenesis. Br. J. Clin. Pharmacol. 1996, 42, 43–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conner, E.A.; Lemmer, E.R.; Sanchez, A.; Factor, V.M.; Thorgeirsson, S.S. E2F1 blocks and c-Myc accelerates hepatic ploidy in transgenic mouse models. Biochem. Biophys. Res. Commun. 2003, 302, 114–120. [Google Scholar] [CrossRef]
- Sun, L.; Nguyen, A.T.; Spitsbergen, J.M.; Gong, Z. Myc-induced liver tumors in transgenic zebrafish can regress in tp53 null mutation. PLoS ONE 2015, 10, e0117249. [Google Scholar] [CrossRef]
- Yang, Q.; Yan, C.; Gong, Z. Activation of liver stromal cells is associated with male-biased liver tumor initiation in xmrk and Myc transgenic zebrafish. Sci. Rep. 2017, 7, 10315. [Google Scholar] [CrossRef] [Green Version]
- Yan, C.; Yang, Q.; Shen, H.M.; Spitsbergen, J.M.; Gong, Z. Chronically high level of tgfb1a induction causes both hepatocellular carcinoma and cholangiocarcinoma via a dominant Erk pathway in zebrafish. Oncotarget 2017, 8, 77096–77109. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, A.T.; Koh, V.; Spitsbergen, J.M.; Gong, Z. Development of a conditional liver tumor model by mifepristone-inducible Cre recombination to control oncogenic kras V12 expression in transgenic zebrafish. Sci. Rep. 2016, 6, 19559. [Google Scholar] [CrossRef] [Green Version]
- Nakayama, J.; Lu, J.W.; Makinoshima, H.; Gong, Z. A Novel Zebrafish Model of Metastasis Identifies the HSD11beta1 Inhibitor Adrenosterone as a Suppressor of Epithelial-Mesenchymal Transition and Metastatic Dissemination. Mol. Cancer Res. 2020, 18, 477–487. [Google Scholar] [CrossRef]
- Lu, J.-W.; Sun, Y.; Lin, L.-I.; Liu, D.; Gong, Z. Exacerbation of Liver Tumor Metastasis in twist1a+/xmrk+ Double Transgenic Zebrafish Following Lipopolysaccharide or Dextran Sulphate Sodium Exposure. Pharmaceuticals 2021, 14, 867. [Google Scholar] [CrossRef]
- Yan, C.; Yang, Q.; Gong, Z. Tumor-Associated Neutrophils and Macrophages Promote Gender Disparity in Hepatocellular Carcinoma in Zebrafish. Cancer Res. 2017, 77, 1395–1407. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Li, H.; Spitsbergen, J.M.; Gong, Z. Males develop faster and more severe hepatocellular carcinoma than females in kras(V12) transgenic zebrafish. Sci. Rep. 2017, 7, 41280. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Yan, C.; Yin, C.; Gong, Z. Serotonin Activated Hepatic Stellate Cells Contribute to Sex Disparity in Hepatocellular Carcinoma. Cell. Mol. Gastroenterol. Hepatol. 2017, 3, 484–499. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Zheng, W.; Li, H.; Li, C.; Gong, Z. Synergistic Induction of Potential Warburg Effect in Zebrafish Hepatocellular Carcinoma by Co-Transgenic Expression of Myc and xmrk Oncogenes. PLoS ONE 2015, 10, e0132319. [Google Scholar] [CrossRef] [PubMed]
- Hoshida, Y.; Nijman, S.M.; Kobayashi, M.; Chan, J.A.; Brunet, J.P.; Chiang, D.Y.; Villanueva, A.; Newell, P.; Ikeda, K.; Hashimoto, M.; et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 2009, 69, 7385–7392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Luo, H.; Li, C.; Huo, X.; Yan, C.; Huang, X.; Al-Haddawi, M.; Mathavan, S.; Gong, Z. Transcriptomic analysis of a transgenic zebrafish hepatocellular carcinoma model reveals a prominent role of immune responses in tumour progression and regression. Int. J. Cancer 2014, 135, 1564–1573. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, W.; Li, Z.; Nguyen, A.T.; Li, C.; Emelyanov, A.; Gong, Z. Xmrk, kras and myc transgenic zebrafish liver cancer models share molecular signatures with subsets of human hepatocellular carcinoma. PLoS ONE 2014, 9, e91179. [Google Scholar] [CrossRef] [Green Version]
- Yan, C.; Yang, Q.; Huo, X.; Li, H.; Zhou, L.; Gong, Z. Chemical inhibition reveals differential requirements of signaling pathways in kras(V12)- and Myc-induced liver tumors in transgenic zebrafish. Sci. Rep. 2017, 7, 45796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Min, L.; He, B.; Hui, L. Mitogen-activated protein kinases in hepatocellular carcinoma development. Semin. Cancer Biol. 2011, 21, 10–20. [Google Scholar] [CrossRef]
- Wang, W.; Smits, R.; Hao, H.; He, C. Wnt/beta-Catenin Signaling in Liver Cancers. Cancers 2019, 11, 926. [Google Scholar] [CrossRef] [Green Version]
- He, S.; Tang, S. WNT/beta-catenin signaling in the development of liver cancers. Biomed. Pharmacother. 2020, 132, 110851. [Google Scholar] [CrossRef]
- Kunst, C.; Haderer, M.; Heckel, S.; Schlosser, S.; Müller, M. The p53 family in hepatocellular carcinoma. Transl. Cancer Res. 2016, 5, 632–638. [Google Scholar] [CrossRef]
- Link, T.; Iwakuma, T. Roles of p53 in extrinsic factor-induced liver carcinogenesis. Hepatoma Res. 2017, 3, 95–104. [Google Scholar] [CrossRef] [Green Version]
- Zhan, P.; Ji, Y.N.; Yu, L.K. TP53 mutation is associated with a poor outcome for patients with hepatocellular carcinoma: Evidence from a meta-analysis. Hepatobiliary Surg. Nutr. 2013, 2, 260–265. [Google Scholar] [PubMed]
- Morse, M.A.; Sun, W.; Kim, R.; He, A.R.; Abada, P.B.; Mynderse, M.; Finn, R.S. The Role of Angiogenesis in Hepatocellular Carcinoma. Clin. Cancer Res. 2019, 25, 912–920. [Google Scholar] [CrossRef] [Green Version]
- Mousa, A.B. Sorafenib in the treatment of advanced hepatocellular carcinoma. Saudi J. Gastroenterol. 2008, 14, 40–42. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Li, J.; Wu, L.; Yu, Q.; Ji, J.; Wu, J.; Dai, W.; Guo, C. Emerging roles and the regulation of aerobic glycolysis in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2020, 39, 126. [Google Scholar] [CrossRef]
- Yang, Y.; Li, C.; Nie, X.; Feng, X.; Chen, W.; Yue, Y.; Tang, H.; Deng, F. Metabonomic studies of human hepatocellular carcinoma using high-resolution magic-angle spinning 1H NMR spectroscopy in conjunction with multivariate data analysis. J. Proteome Res. 2007, 6, 2605–2614. [Google Scholar] [CrossRef]
- Beyoglu, D.; Imbeaud, S.; Maurhofer, O.; Bioulac-Sage, P.; Zucman-Rossi, J.; Dufour, J.F.; Idle, J.R. Tissue metabolomics of hepatocellular carcinoma: Tumor energy metabolism and the role of transcriptomic classification. Hepatology 2013, 58, 229–238. [Google Scholar] [CrossRef] [Green Version]
- Desai, S.; Ding, M.; Wang, B.; Lu, Z.; Zhao, Q.; Shaw, K.; Yung, W.K.; Weinstein, J.N.; Tan, M.; Yao, J. Tissue-specific isoform switch and DNA hypomethylation of the pyruvate kinase PKM gene in human cancers. Oncotarget 2014, 5, 8202–8210. [Google Scholar] [CrossRef] [Green Version]
- Jessy, T. Immunity over inability: The spontaneous regression of cancer. J. Nat. Sci. Biol. Med. 2011, 2, 43–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weinstein, I.B.; Joe, A. Oncogene addiction. Cancer Res. 2008, 68, 3077–3080; discussion 3080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Agrawal, I.; Gong, Z. Reversion of tumor hepatocytes to normal hepatocytes during liver tumor regression in an oncogene-expressing transgenic zebrafish model. Dis. Models Mech. 2019, 12, dmm039578. [Google Scholar] [CrossRef] [Green Version]
- Hou, J.; Zhang, H.; Sun, B.; Karin, M. The immunobiology of hepatocellular carcinoma in humans and mice: Basic concepts and therapeutic implications. J. Hepatol. 2020, 72, 167–182. [Google Scholar] [CrossRef]
- Roderburg, C.; Wree, A.; Demir, M.; Schmelzle, M.; Tacke, F. The role of the innate immune system in the development and treatment of hepatocellular carcinoma. Hepat. Oncol. 2020, 7, HEP17. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.Z.; Lee, S.D.; Sarkar, D.; Lee, H.M.; Khan, A.; Bhati, C.; Sharma, A.; Kumaran, V.; Bruno, D.; Cotterell, A.; et al. Immunological characterization of hepatocellular carcinoma. Hepatoma Res. 2021, 7, 6. [Google Scholar]
- Hall, C.; Flores, M.V.; Storm, T.; Crosier, K.; Crosier, P. The zebrafish lysozyme C promoter drives myeloid-specific expression in transgenic fish. BMC Dev. Biol 2007, 7, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ellett, F.; Pase, L.; Hayman, J.W.; Andrianopoulos, A.; Lieschke, G.J. mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood 2011, 117, e49–e56. [Google Scholar] [CrossRef] [Green Version]
- Yan, C.; Huo, X.; Wang, S.; Feng, Y.; Gong, Z. Stimulation of hepatocarcinogenesis by neutrophils upon induction of oncogenic kras expression in transgenic zebrafish. J. Hepatol. 2015, 63, 420–428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Q.; Yan, C.; Gong, Z. Interaction of hepatic stellate cells with neutrophils and macrophages in the liver following oncogenic kras activation in transgenic zebrafish. Sci. Rep. 2018, 8, 8495. [Google Scholar] [CrossRef]
- Zhao, Y.; Huang, X.; Ding, T.W.; Gong, Z. Enhanced angiogenesis, hypoxia and neutrophil recruitment during Myc-induced liver tumorigenesis in zebrafish. Sci. Rep. 2016, 6, 31952. [Google Scholar] [CrossRef] [Green Version]
- Huo, X.; Li, H.; Li, Z.; Yan, C.; Agrawal, I.; Mathavan, S.; Liu, J.; Gong, Z. Transcriptomic profiles of tumor-associated neutrophils reveal prominent roles in enhancing angiogenesis in liver tumorigenesis in zebrafish. Sci. Rep. 2019, 9, 1509. [Google Scholar] [CrossRef] [Green Version]
- Yan, C.; Yang, Q.; Gong, Z. Transgenic expression of tgfb1a induces hepatic inflammation, fibrosis and metastasis in zebrafish. Biochem. Biophys. Res. Commun. 2019, 509, 175–181. [Google Scholar] [CrossRef]
- Helal, M.; Yan, C.; Gong, Z. Stimulation of hepatocarcinogenesis by activated cholangiocytes via Il17a/f1 pathway in kras transgenic zebrafish model. Sci. Rep. 2021, 11, 1372. [Google Scholar] [CrossRef]
- Hefaiedh, R.; Ennaifer, R.; Romdhane, H.; Ben Nejma, H.; Arfa, N.; Belhadj, N.; Gharbi, L.; Khalfallah, T. Gender difference in patients with hepatocellular carcinoma. Tunis Med. 2013, 91, 505–508. [Google Scholar]
- Wu, E.M.; Wong, L.L.; Hernandez, B.Y.; Ji, J.F.; Jia, W.; Kwee, S.A.; Kalathil, S. Gender differences in hepatocellular cancer: Disparities in nonalcoholic fatty liver disease/steatohepatitis and liver transplantation. Hepatoma Res. 2018, 4, 66. [Google Scholar] [CrossRef]
- Natri, H.M.; Wilson, M.A.; Buetow, K.H. Distinct molecular etiologies of male and female hepatocellular carcinoma. BMC Cancer 2019, 19, 951. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Lu, J.W.; Huo, X.; Li, Y.; Li, Z.; Gong, Z. Effects of sex hormones on liver tumor progression and regression in Myc/xmrk double oncogene transgenic zebrafish. Gen. Comp. Endocrinol. 2019, 277, 112–121. [Google Scholar] [CrossRef]
- Li, H.; Li, Y.; Lu, J.W.; Huo, X.; Gong, Z. Liver-specific androgen receptor knockout attenuates early liver tumor development in zebrafish. Sci. Rep. 2019, 9, 10645. [Google Scholar] [CrossRef]
- Dhanapal, R.; Saraswathi, T.; Govind, R.N. Cancer cachexia. J. Oral Maxillofac. Pathol. 2011, 15, 257–260. [Google Scholar] [CrossRef] [PubMed]
- Baracos, V.E.; Martin, L.; Korc, M.; Guttridge, D.C.; Fearon, K.C.H. Cancer-associated cachexia. Nat. Rev. Dis. Primers 2018, 4, 17105. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, C.; Spiro, A.; Ahern, R.; Emery, P.W. Oral nutritional interventions in malnourished patients with cancer: A systematic review and meta-analysis. J. Natl. Cancer Inst. 2012, 104, 371–385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Safdie, F.M.; Dorff, T.; Quinn, D.; Fontana, L.; Wei, M.; Lee, C.; Cohen, P.; Longo, V.D. Fasting and cancer treatment in humans: A case series report. Aging 2009, 1, 988–1007. [Google Scholar] [CrossRef] [Green Version]
- Zhong, Z.; Sanchez-Lopez, E.; Karin, M. Autophagy, Inflammation, and Immunity: A Troika Governing Cancer and Its Treatment. Cell 2016, 166, 288–298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Q.; Yan, C.; Wang, X.; Gong, Z. Leptin induces muscle wasting in a zebrafish kras-driven hepatocellular carcinoma (HCC) model. Dis. Models Mech. 2019, 12, dmm038240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelesidis, T.; Kelesidis, I.; Chou, S.; Mantzoros, C.S. Narrative review: The role of leptin in human physiology: Emerging clinical applications. Ann. Intern. Med. 2010, 152, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Taylor, K.L.; Grant, N.J.; Temperley, N.D.; Patton, E.E. Small molecule screening in zebrafish: An in vivo approach to identifying new chemical tools and drug leads. Cell Commun. Signal. 2010, 8, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, T.; Peterson, R.T. Zebrafish as a Platform for Drug Screening. In The Zebrafish in Biomedical Research; Cartner, S.C., Eisen, J.S., Farmer, S.C., Guillemin, K.J., Kent, M.L., Sanders, G.E., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 659–675. [Google Scholar]
- Huang, X.; Nguyen, A.T.; Li, Z.; Emelyanov, A.; Parinov, S.; Gong, Z. One Step Forward: The Use of Transgenic Zebrafish Tumor Model in Drug Screens. Birth Defects Res. C 2011, 93, 173–181. [Google Scholar] [CrossRef]
- Cully, M. Zebrafish earn their drug discovery stripes. Nat. Rev. Drug Discov. 2019, 18, 811–813. [Google Scholar] [CrossRef]
- Patton, E.E.; Zon, L.I.; Langenau, D.M. Zebrafish disease models in drug discovery: From preclinical modelling to clinical trials. Nat. Rev. Drug Discov. 2021, 20, 611–628. [Google Scholar] [CrossRef] [PubMed]
- North, T.E.; Goessling, W.; Walkley, C.R.; Lengerke, C.; Kopani, K.R.; Lord, A.M.; Weber, G.J.; Bowman, T.V.; Jang, I.-H.; Grosser, T.; et al. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature 2007, 447, 1007–1011. [Google Scholar] [CrossRef]
- Goessling, W.; Allen, R.S.; Guan, X.; Jin, P.; Uchida, N.; Dovey, M.; Harris, J.M.; Metzger, M.E.; Bonifacino, A.C.; Stroncek, D.; et al. Prostaglandin E2 enhances engraftment of human cord blood stem cells and shows long-term safety in preclinical non-human primate transplant models. Cell Stem Cell 2011, 8, 445–458. [Google Scholar] [CrossRef] [Green Version]
- Cutler, C.; Multani, P.; Robbins, D.; Kim, H.T.; Le, T.; Hoggatt, J.; Pelus, L.M.; Desponts, C.; Chen, Y.-B.; Rezner, B.; et al. Prostaglandin-modulated umbilical cord blood hematopoietic stem cell transplantation. Blood 2013, 122, 3074–3081. [Google Scholar] [CrossRef]
- Mandelbaum, J.; Shestopalov, I.A.; Henderson, R.E.; Chau, N.G.; Knoechel, B.; Wick, M.J.; Zon, L.I. Zebrafish blastomere screen identifies retinoic acid suppression of MYB in adenoid cystic carcinoma. J. Exp. Med. 2018, 215, 2673–2685. [Google Scholar] [CrossRef]
- Yang, Q.; Salim, L.; Yan, C.; Gong, Z. Rapid Analysis of Effects of Environmental Toxicants on Tumorigenesis and Inflammation Using a Transgenic Zebrafish Model for Liver Cancer. Mar. Biotechnol. 2019, 21, 396–405. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Dang, Y.; Gong, Z.; Letcher, R.J.; Liu, C. Progression of liver tumor was promoted by tris(1,3-dichloro-2-propyl) phosphate through the induction of inflammatory responses in kras transgenic zebrafish. Environ. Pollut. 2019, 255, 113315. [Google Scholar] [CrossRef]
- Chen, S.; Gong, Z.; Letcher, R.J.; Liu, C. Promotion effect of liver tumor progression in male kras transgenic zebrafish induced by tris (1, 3-dichloro-2-propyl) phosphate. Ecotoxicol. Environ. Saf. 2020, 191, 110220. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Yang, D.; Duan, X.; Zhang, Y.; Chen, D.; Gong, Z.; Liu, C. Perfluorooctane sulfonate promotes doxycycline-induced liver tumor progression in male Kras(v12) transgenic zebrafish. Environ. Res. 2021, 196, 110962. [Google Scholar] [CrossRef] [PubMed]
- Argiles, J.M.; Busquets, S.; Stemmler, B.; Lopez-Soriano, F.J. Cancer cachexia: Understanding the molecular basis. Nat. Rev. Cancer 2014, 14, 754–762. [Google Scholar] [CrossRef]
- Springer, J.; Tschirner, A.; Haghikia, A.; von Haehling, S.; Lal, H.; Grzesiak, A.; Kaschina, E.; Palus, S.; Potsch, M.; von Websky, K.; et al. Prevention of liver cancer cachexia-induced cardiac wasting and heart failure. Eur. Heart J. 2014, 35, 932–941. [Google Scholar] [CrossRef]

| Transgene | Transposon System | Inducible System | Permanent Recombination | Tumour Phenotypes * | References |
|---|---|---|---|---|---|
| krasV12 | Ac/Ds | - | - | 3 mpf: 36% HP 6 mpf: 22% HA 9 mpf: 26% HCC | [34] |
| Ac/Ds | LexPR | No | 1 wpi: 100% HP 4 wpi: 100% HCC | [43] | |
| None | Tet-on | No | 7 dpi: 40% HCC and 50–60% HA in males. 30–40% HA and 60–70% HP in females. 10 dpi: 100% HCC in males. 70% HCC, 20% HA and 10% HP in females. 5mpi: 100% multinodular HCC in males, 100% homogenous HCC in females. | [45,59,60,61] | |
| Ac/Ds | LexPR | Yes | Severity dependent on age of induction. Most fish developed mosaic livers with more than 1 tumour type. | [56] | |
| xmrk | None | Tet-on | No | 7 dpi: 50% HCC, 30% HA and 20% HP in males. 20% HA and 80% HP in females. 6 wpi: 100% HCC | [46,54,62] |
| Mouse Myc | None | Tet-on | No | 3 wpi: 30% HA, 70% HP 16 wpi: 7 out of 8 showed HA, 1 showed HCC | [47] |
| Zebrafish myca and mycb | Ac/Ds | LexPR | No | Liver tumour progression from HP, HA to multinodular HCC | [53] |
| tgfβ1a | Ac/Ds | LexPR | No | 6 wpi: 30% mixed HCC + CCA, 20% HCC + BHP, 10% HCC, 20% HA, 20% HP + BHP | [55] |
| Twist1a-ERT2/xmrk | Ac/Ds | Tet-on | No | 8 dpi: 46% showed distant dissemination of tumour cells, 39% showed abdominal dissemination and the rest showed no dissemination | [57] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, A.Q.; Li, Y.; Gong, Z. Inducible Liver Cancer Models in Transgenic Zebrafish to Investigate Cancer Biology. Cancers 2021, 13, 5148. https://doi.org/10.3390/cancers13205148
Lee AQ, Li Y, Gong Z. Inducible Liver Cancer Models in Transgenic Zebrafish to Investigate Cancer Biology. Cancers. 2021; 13(20):5148. https://doi.org/10.3390/cancers13205148
Chicago/Turabian StyleLee, Ai Qi, Yan Li, and Zhiyuan Gong. 2021. "Inducible Liver Cancer Models in Transgenic Zebrafish to Investigate Cancer Biology" Cancers 13, no. 20: 5148. https://doi.org/10.3390/cancers13205148
APA StyleLee, A. Q., Li, Y., & Gong, Z. (2021). Inducible Liver Cancer Models in Transgenic Zebrafish to Investigate Cancer Biology. Cancers, 13(20), 5148. https://doi.org/10.3390/cancers13205148





