Pan-Cancer Analysis of Prognostic and Immune Infiltrates for CXCs
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Oncomine Analysis
2.2. TISCH Analysis
2.3. GEPIA Analysis
2.4. Kaplan–Meier Plotter
2.5. OncoLnc Dataset
2.6. TRRUST Dataset
2.7. KnockTF Dataset
2.8. MiRWalk Dataset
2.9. CBioPortal Dataset
2.10. STRING Dataset
2.11. GO (Gene Ontology) and KEGG (Kyoto Encyclopedia of Genes and Genomes) Analysis
2.12. TIMER Analysis
2.13. Amadis
2.14. Bacterial Culture
2.15. Mice
2.16. Western Blot
2.17. Enzyme-Linked Immunosorbent Assay (ELISA)
2.18. Histology and Immunohistochemistry (IHC)
2.19. Statistical Analysis
3. Results
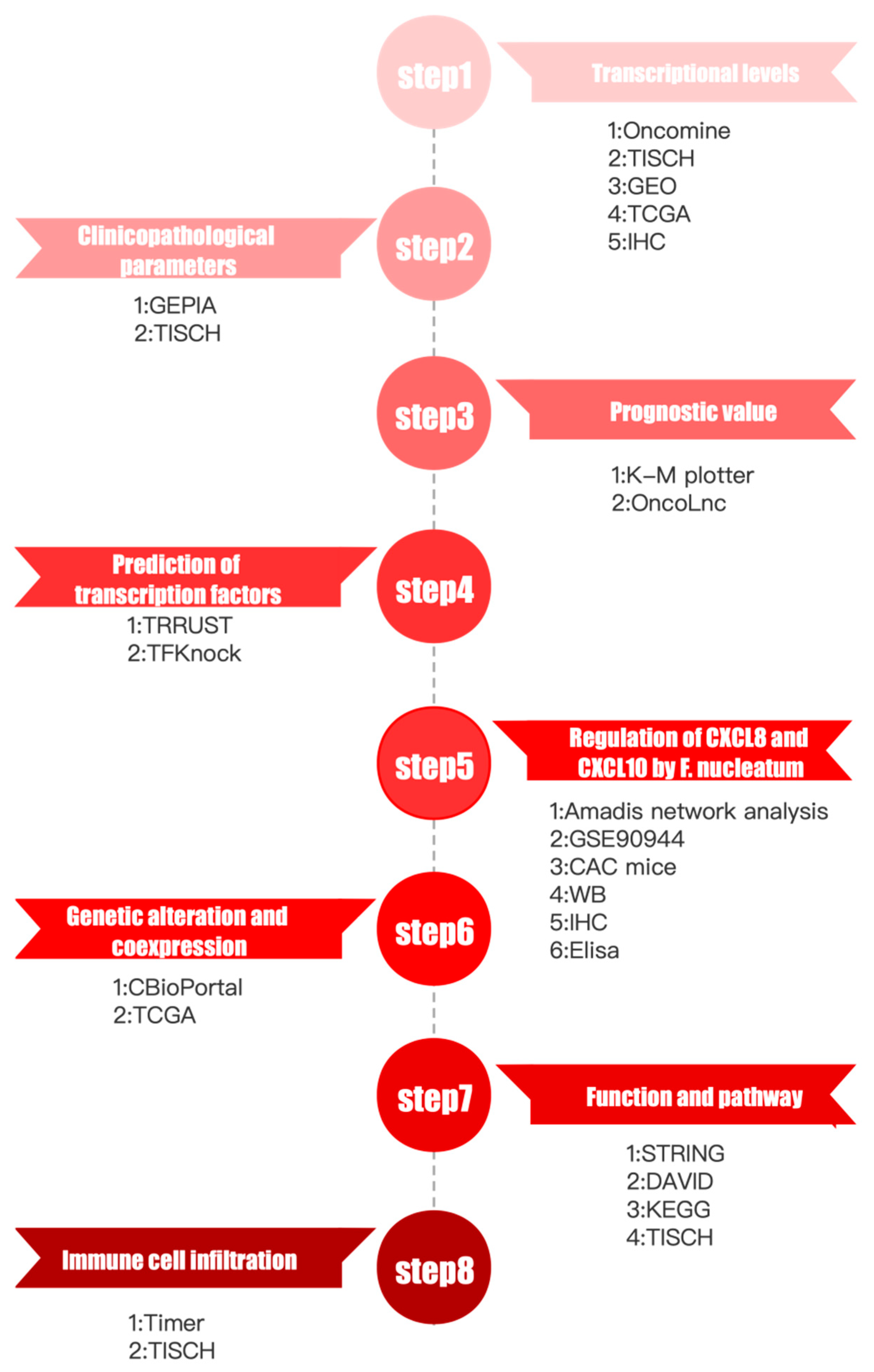
3.1. Analysis Process and Data Processing
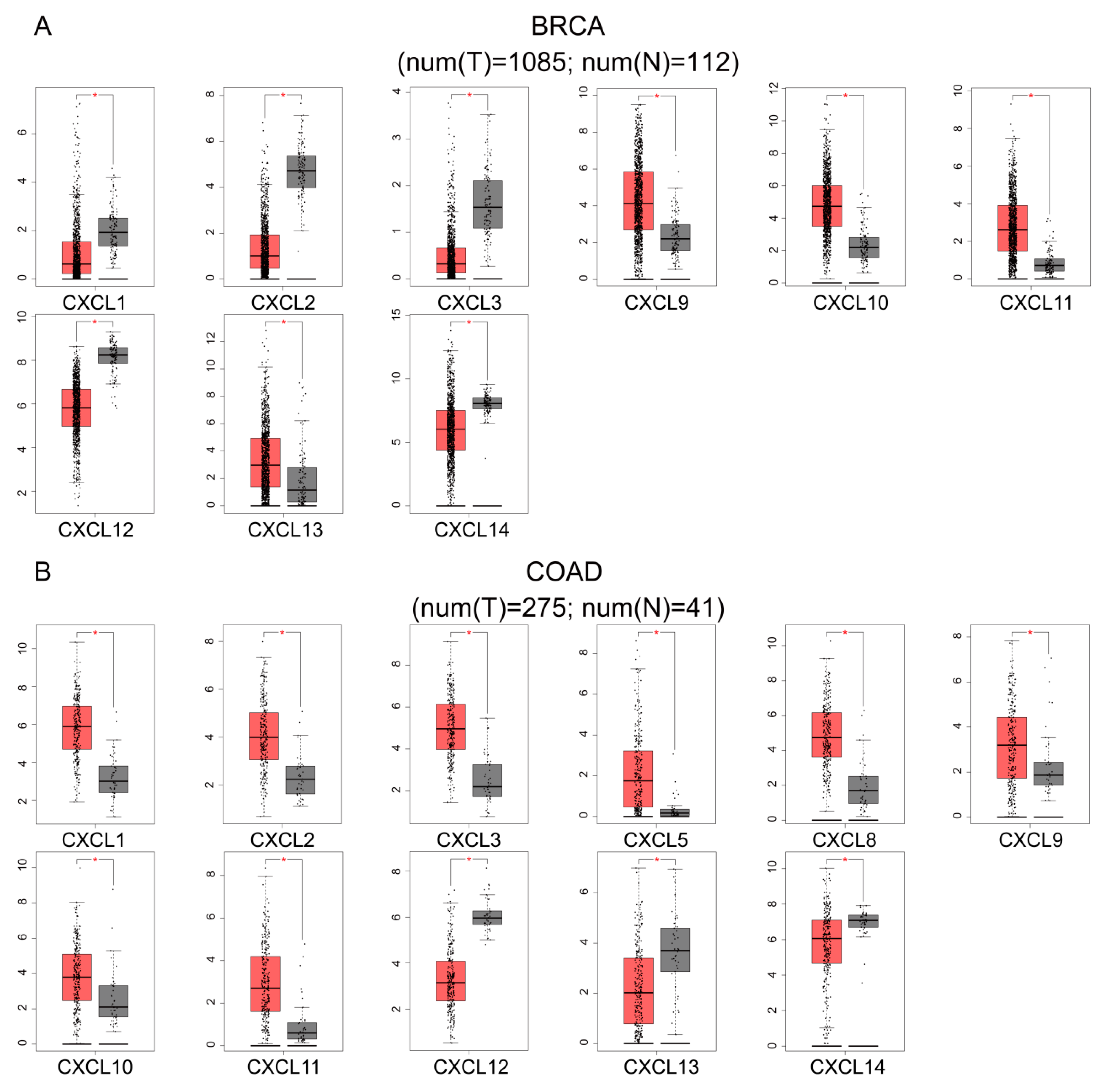
3.2. Transcriptional Levels of CXCs in Various Cancers
3.3. mRNA and Protein Expression of CXCs in Three Kinds of Cancer
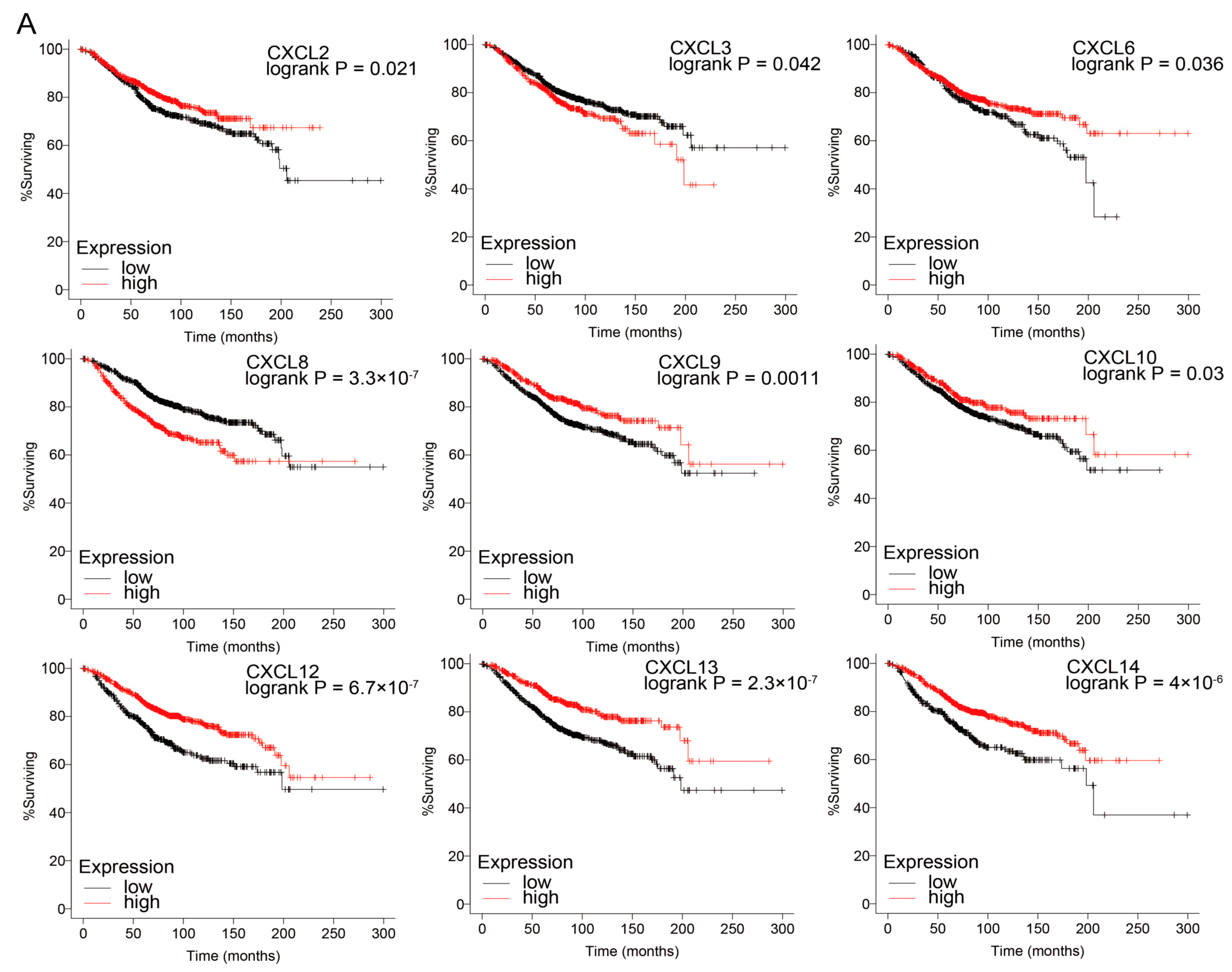
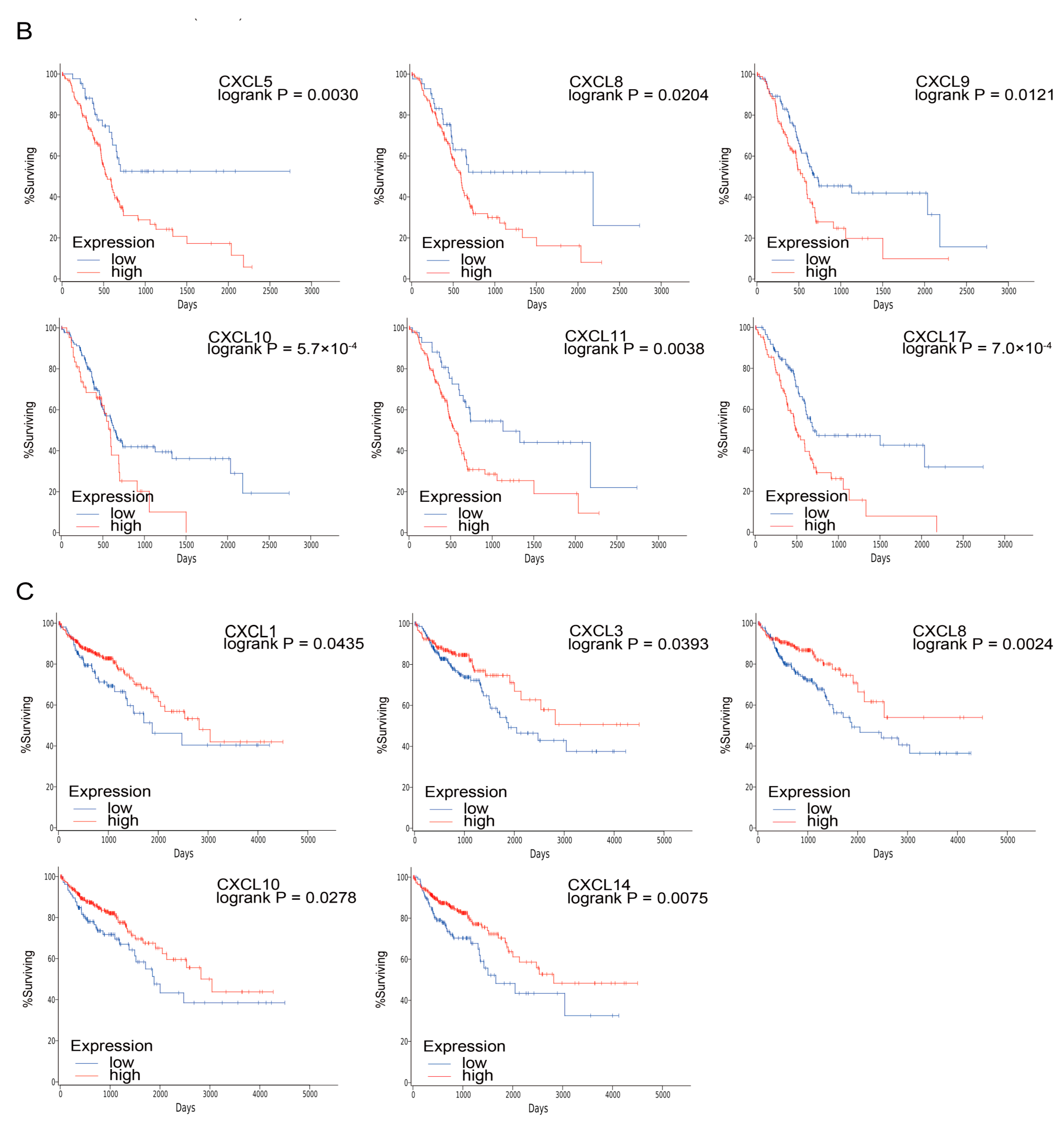
3.4. Prognostic Value of CXCs in Three Kinds of Cancer
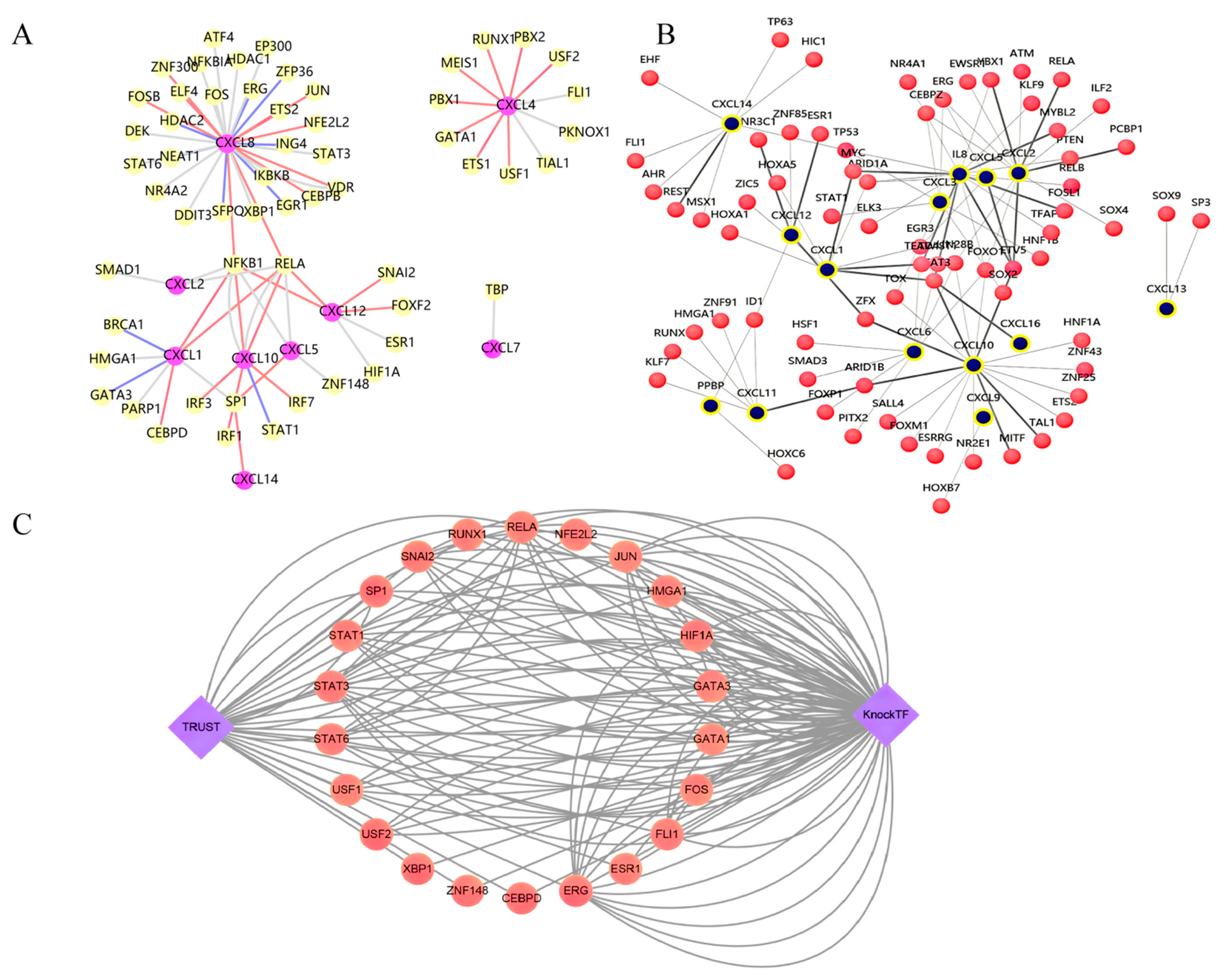
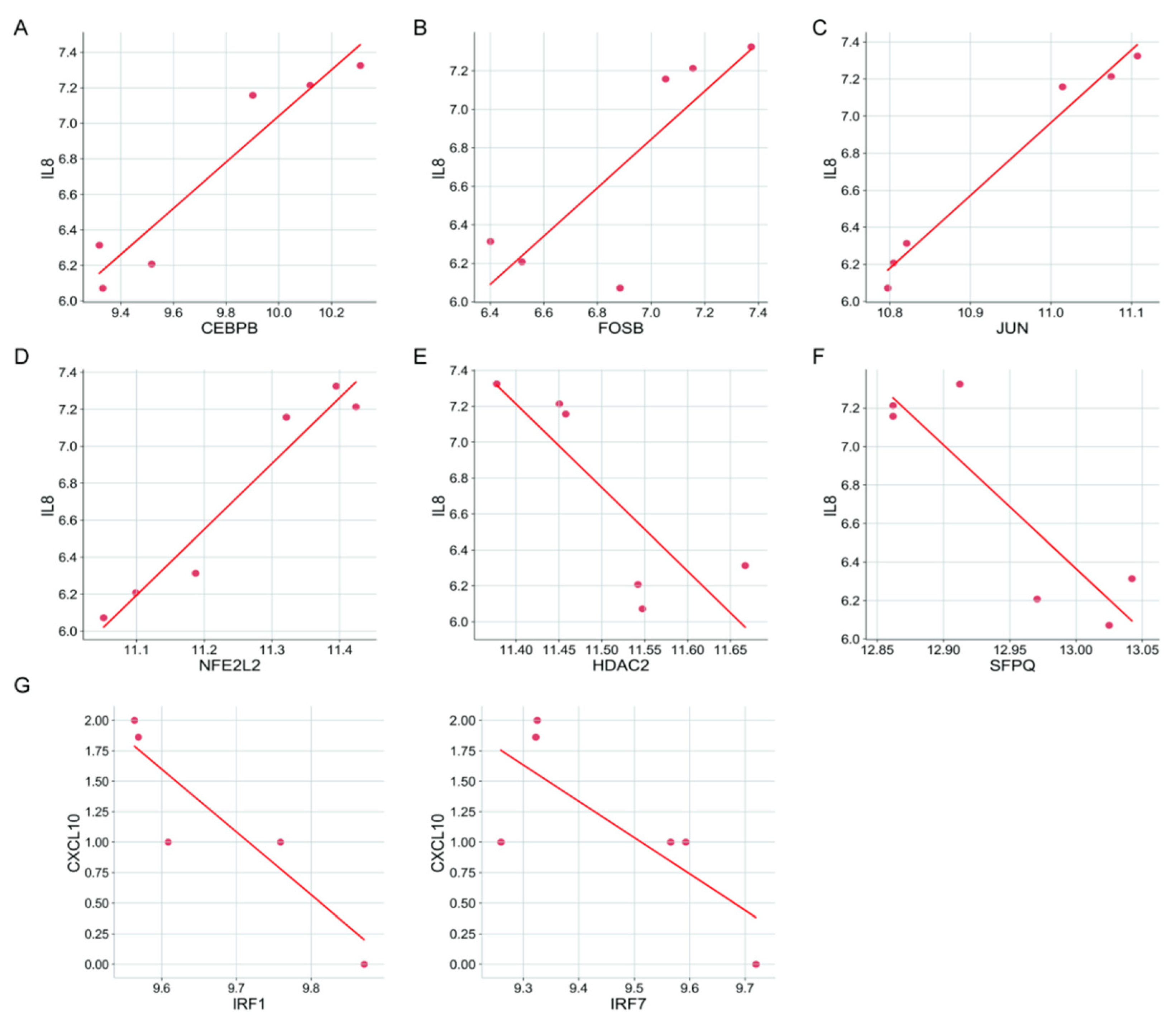
3.5. Prediction of Transcription Factors (TFs) Regulating CXCs
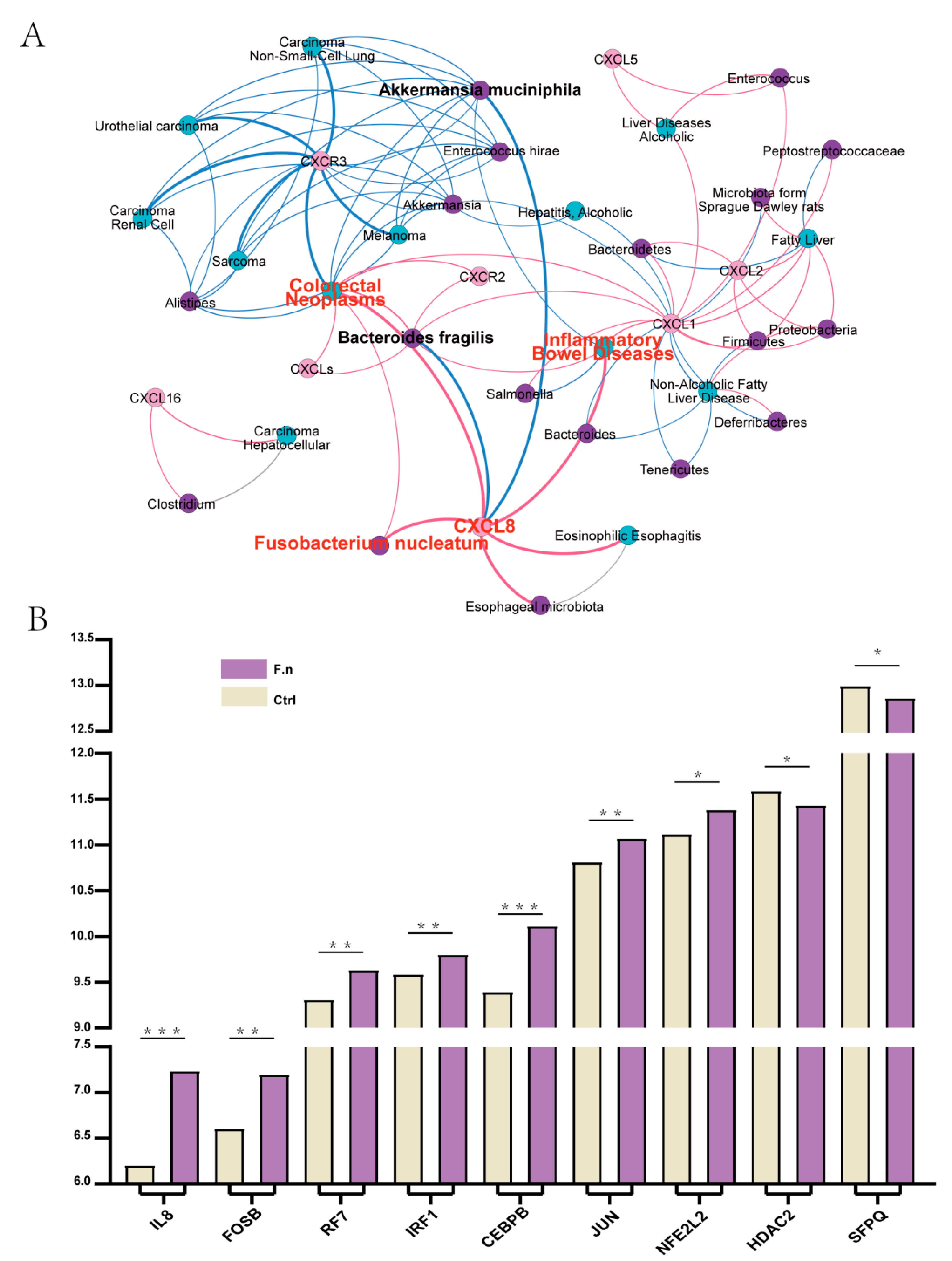
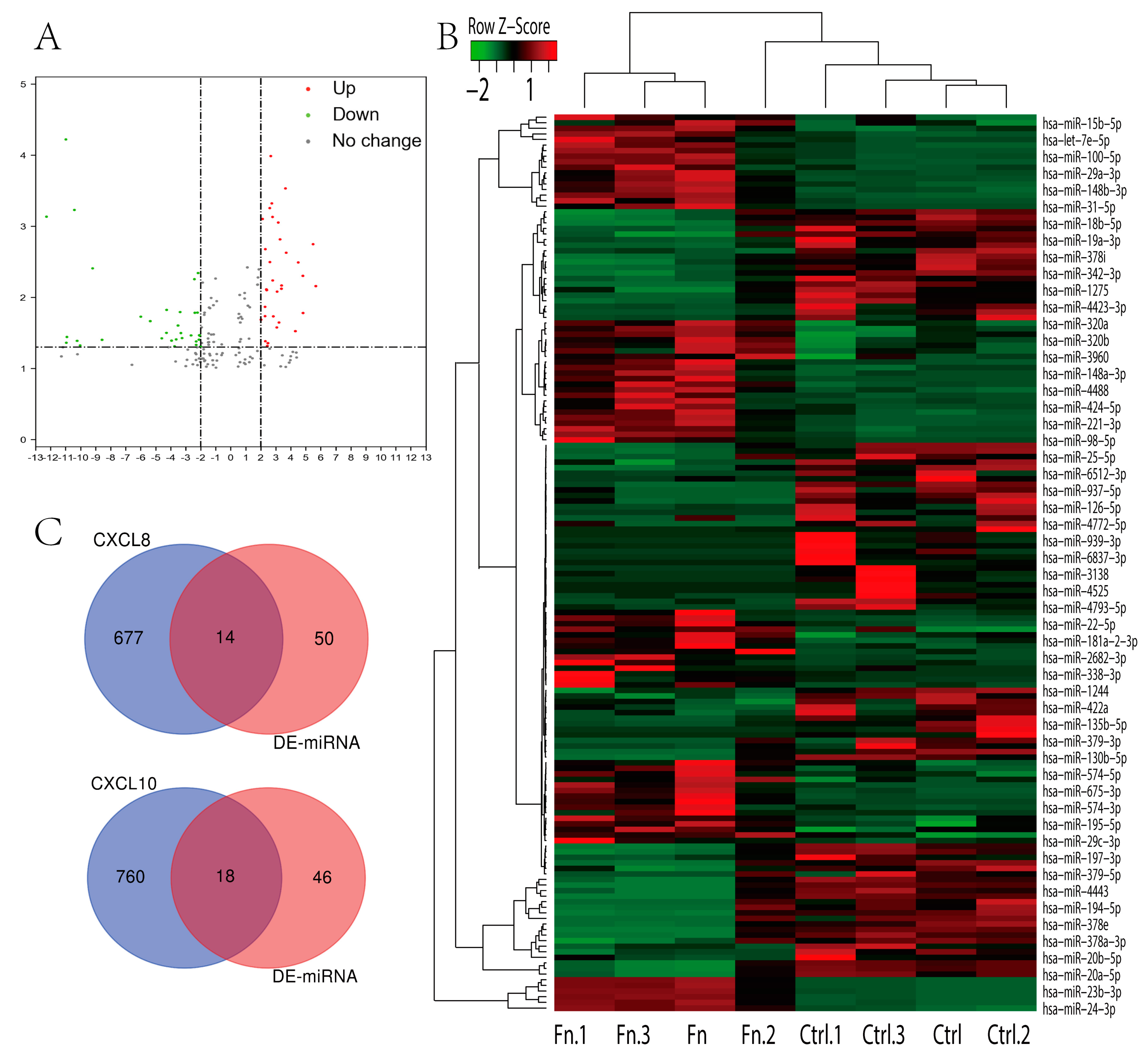
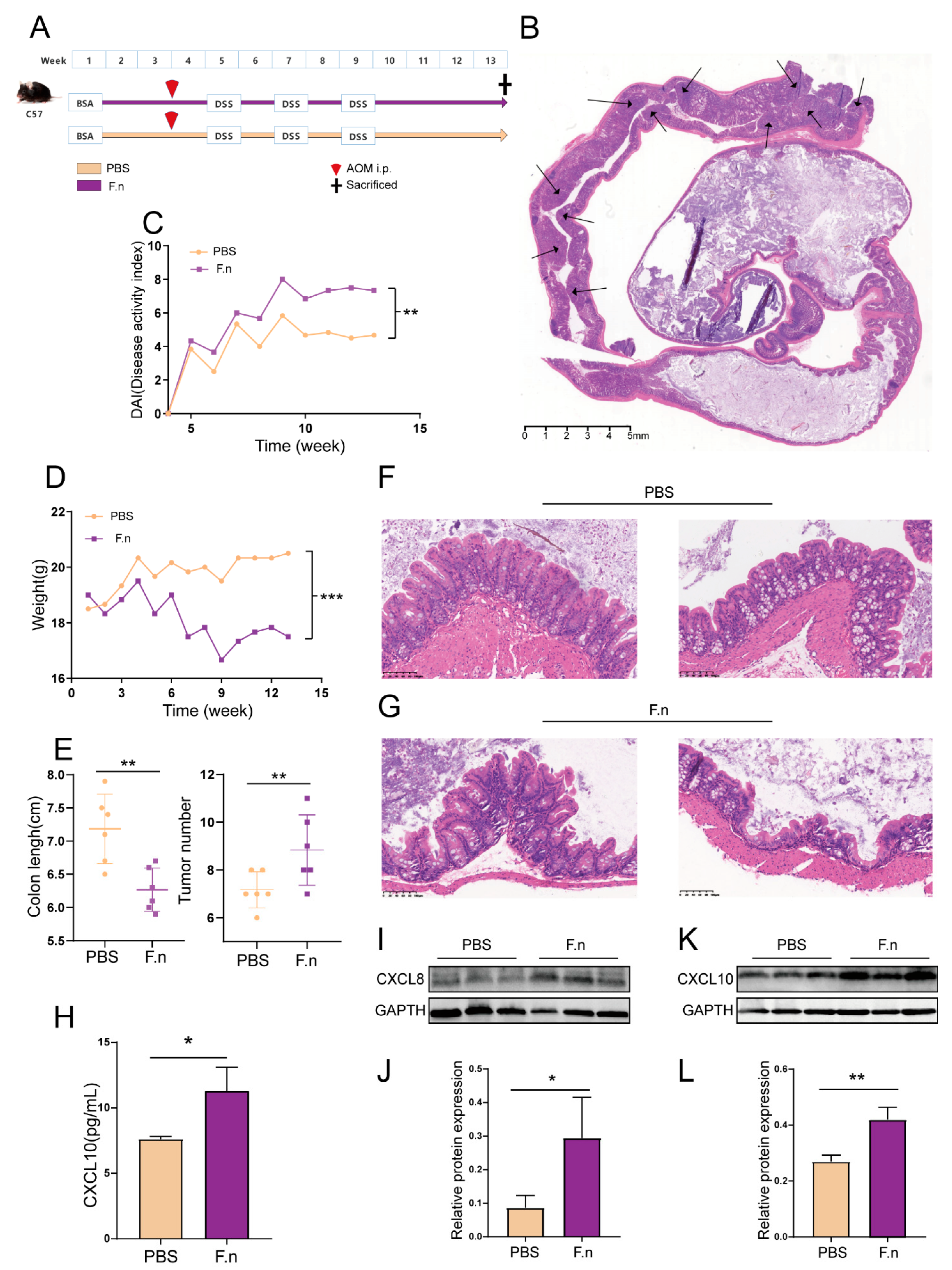
3.6. Regulation of CXCL8 and 10 by F. nucleatum in CAC
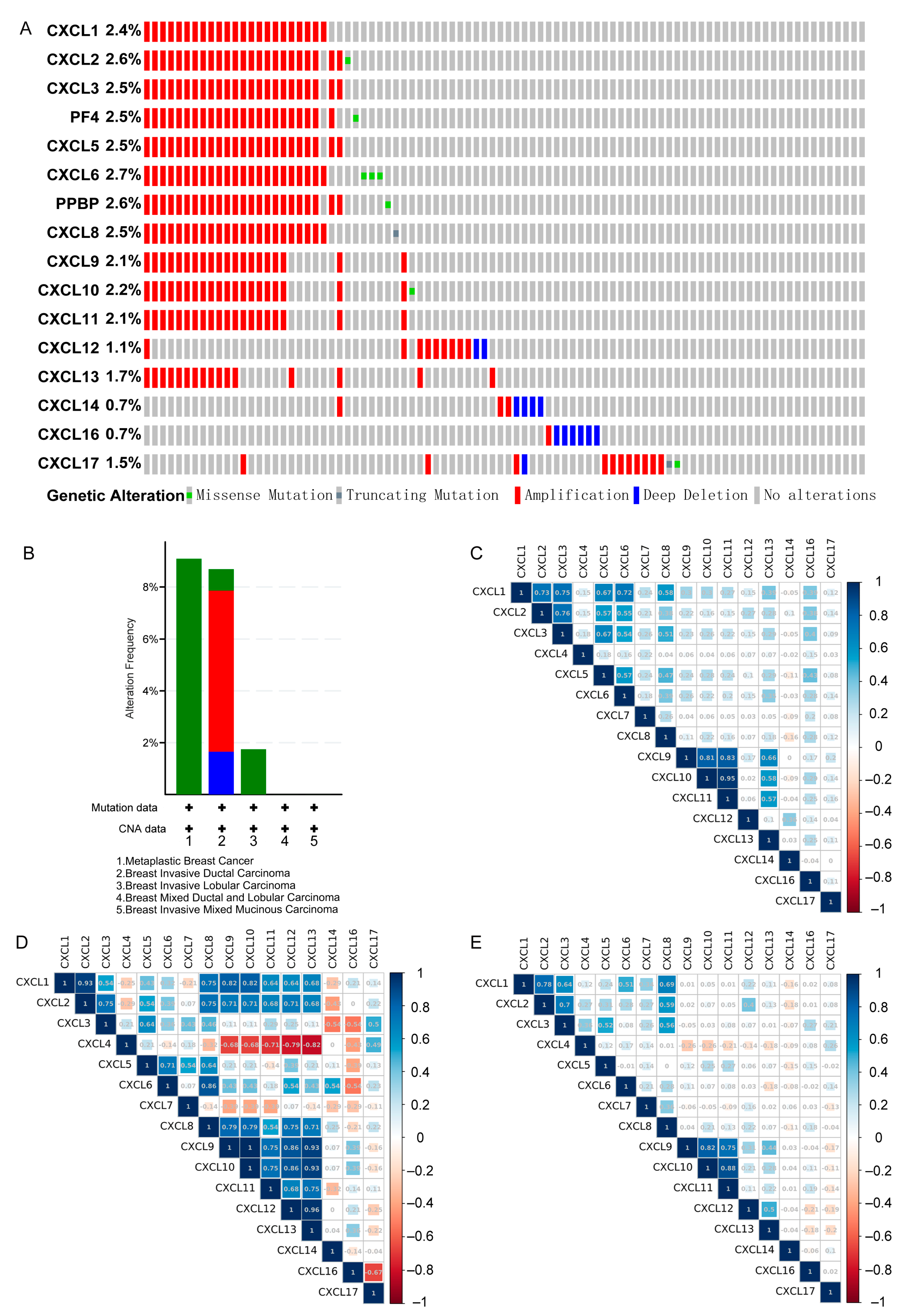
3.7. Genetic Alteration and Co-Expression of CXCs in Three Types of Cancer
3.8. Prediction of CXC-Interacting Proteins and Their Functions and Pathways
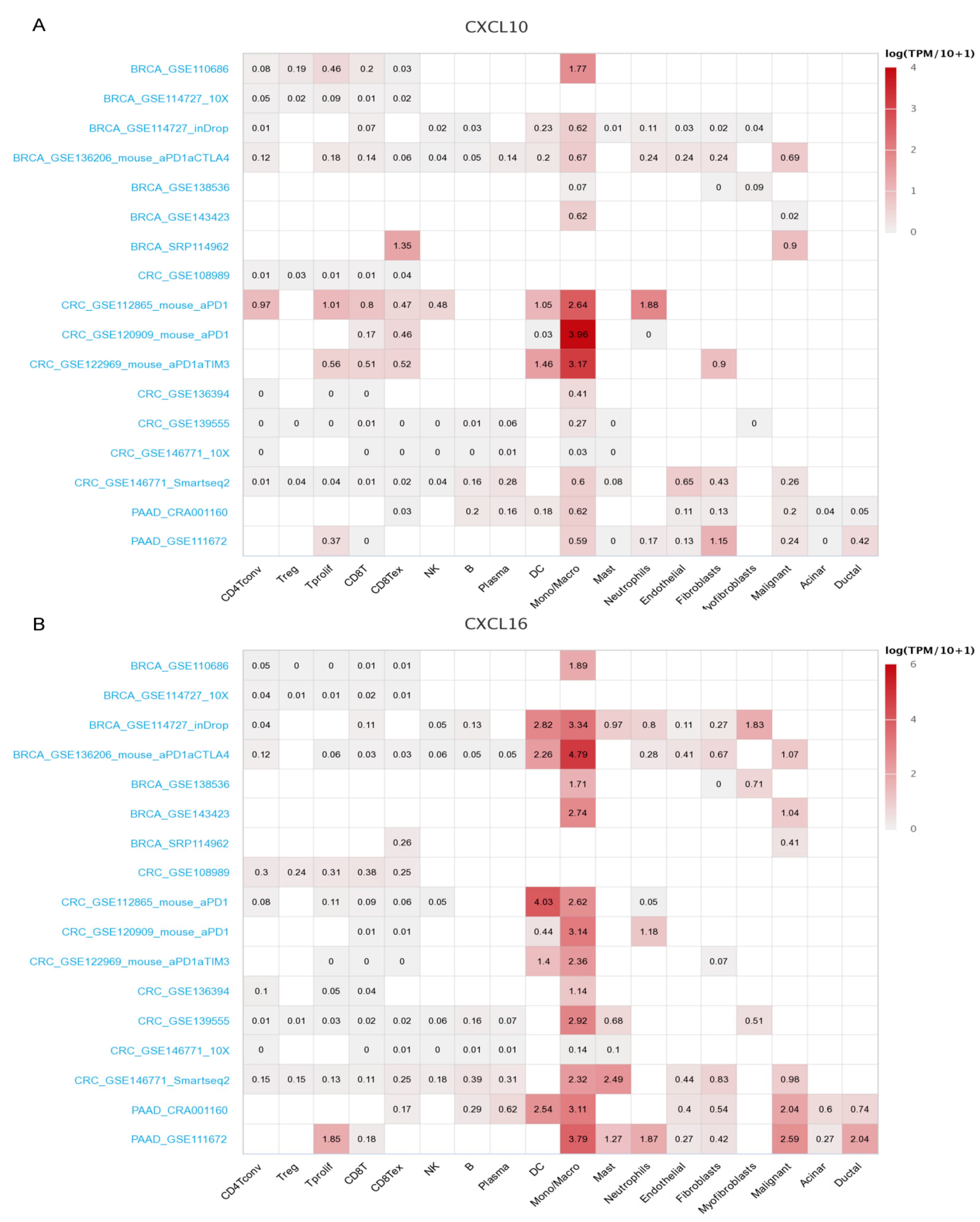
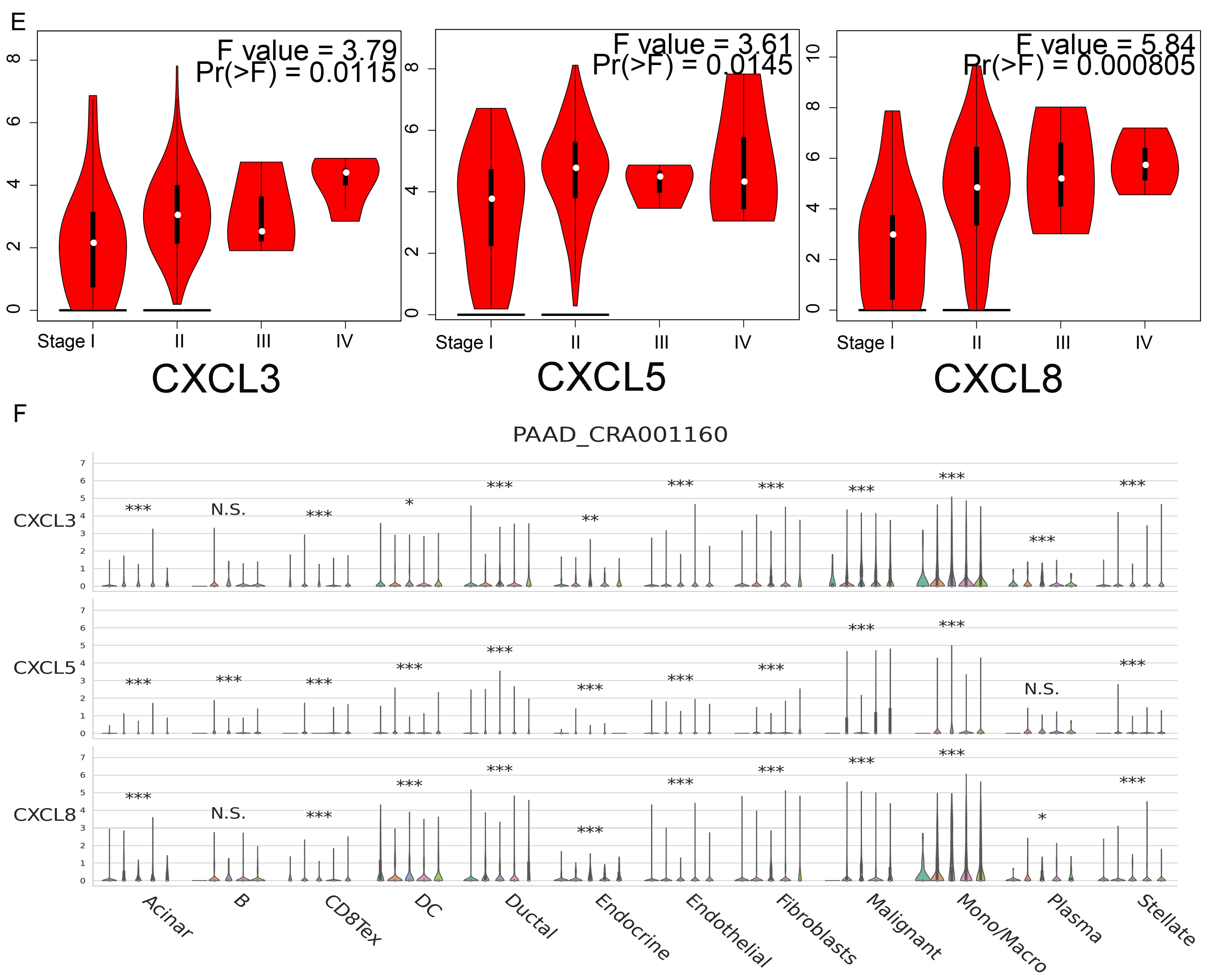
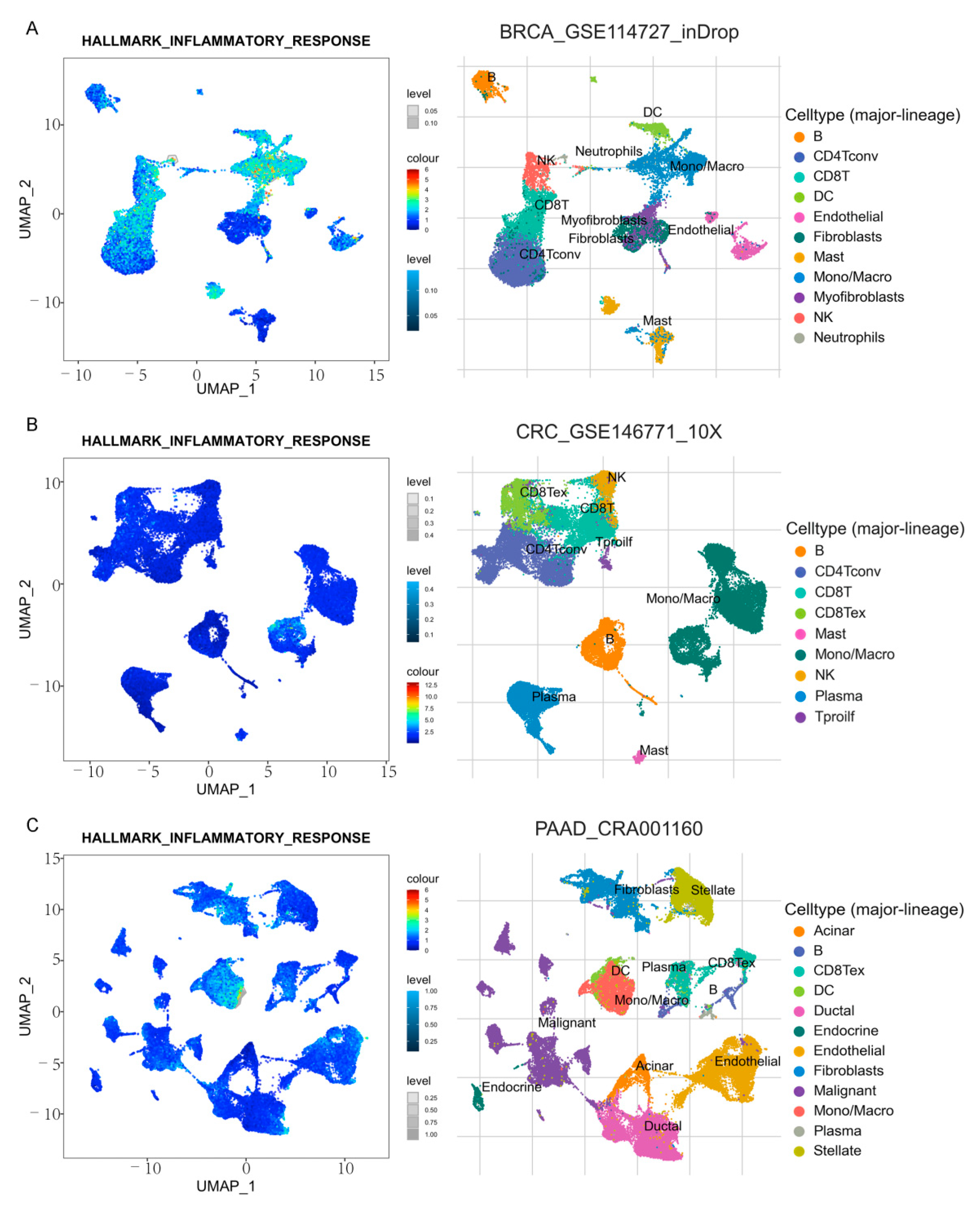
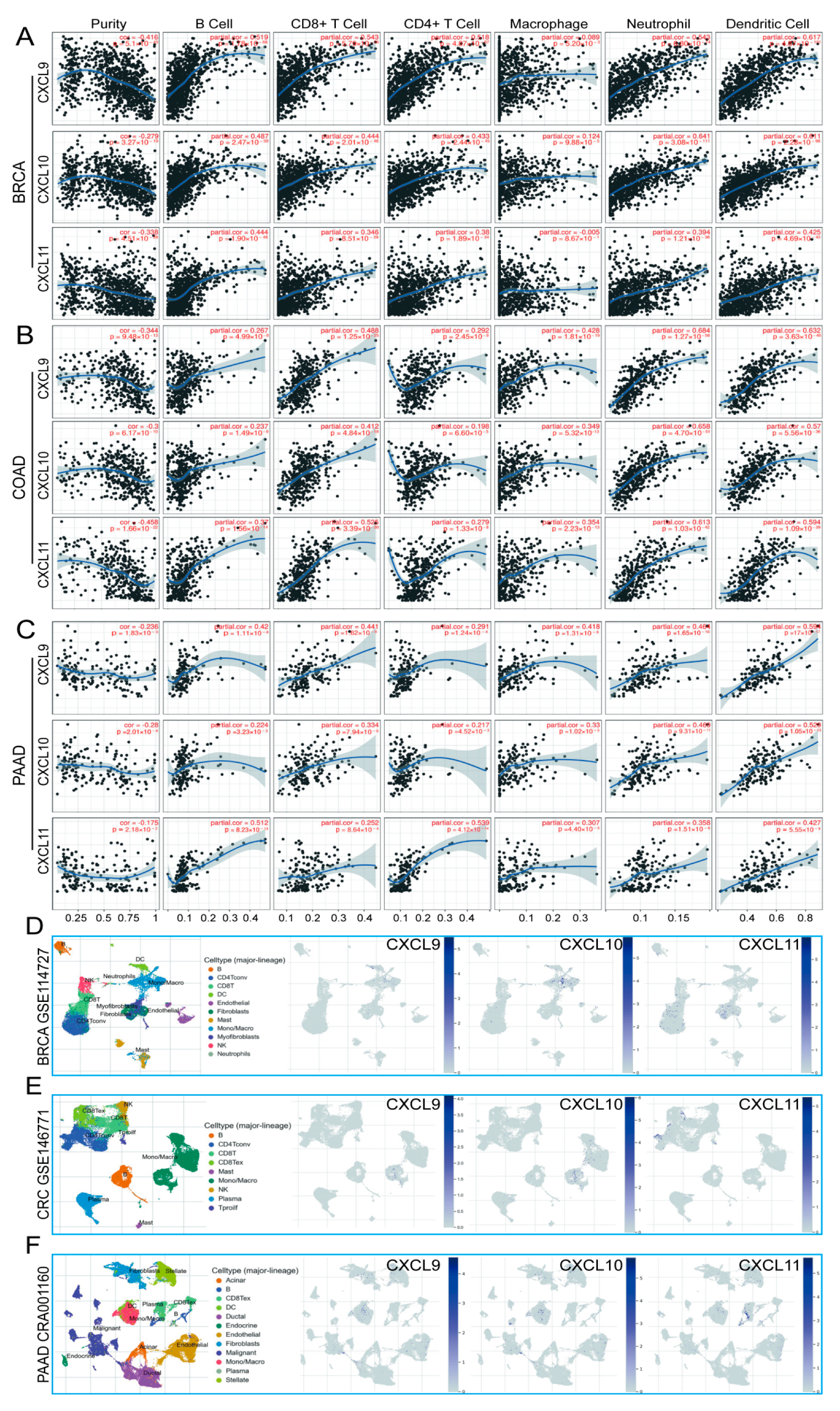
3.9. Immune Cell Infiltration and CXCs in Three Types of Cancer
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Vandercappellen, J.; Van Damme, J.; Struyf, S. The role of CXC chemokines and their receptors in cancer. Cancer Lett. 2008, 267, 226–244. [Google Scholar] [CrossRef]
- Keeley, E.C.; Mehrad, B.; Strieter, R.M. CXC Chemokines in Cancer Angiogenesis and Metastases. Adv. Cancer Res. 2010, 106, 91–111. [Google Scholar] [CrossRef] [Green Version]
- Vinader, V.; Afarinkia, K. The emerging role of CXC chemokines and their receptors in cancer. Futur. Med. Chem. 2012, 4, 853–867. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Q.; Sun, S.; Li, Y.; Li, X.; Li, Z.; Liang, H. Identification of Therapeutic Targets and Prognostic Biomarkers Among CXC Chemokines in the Renal Cell Carcinoma Microenvironment. Front. Oncol. 2020, 9, 1555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Ye, Y.-L.; Li, M.-X.; Ye, S.-B.; Huang, W.-R.; Cai, T.-T.; He, J.; Peng, J.-Y.; Duan, T.-H.; Cui, J.; et al. CXCL2/MIF-CXCR2 signaling promotes the recruitment of myeloid-derived suppressor cells and is correlated with prognosis in bladder cancer. Oncogene 2016, 36, 2095–2104. [Google Scholar] [CrossRef]
- Qi, Y.; Li, Y.; Man, X.; Sui, H.; Zhao, X.; Zhang, P.; Qu, X.; Zhang, H.; Wang, B.; Li, J.; et al. CXCL3 overexpression promotes the tumorigenic potential of uterine cervical cancer cells via the MAPK/ERK pathway. J. Cell. Physiol. 2020, 235, 4756–4765. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; He, H.; Liu, H.; Li, R.; Chen, Y.; Qi, Y.; Jiang, Q.; Chen, L.; Zhang, P.; Zhang, H.; et al. Tumour-associated macrophages-derived CXCL8 determines immune evasion through autonomous PD-L1 expression in gastric cancer. Gut 2019, 68, 1764–1773. [Google Scholar] [CrossRef]
- Yang, C.; Yu, H.; Chen, R.; Tao, K.; Jian, L.; Peng, M.; Li, X.; Liu, M.; Liu, S. CXCL1 stimulates migration and invasion in ER-negative breast cancer cells via activation of the ERK/MMP2/9 signaling axis. Int. J. Oncol. 2019, 55, 684–696. [Google Scholar] [CrossRef] [Green Version]
- Dayer, R.; Babashah, S.; Jamshidi, S.; Sadeghizadeh, M. Upregulation of CXC chemokine receptor 4-CXC chemokine ligand 12 axis ininvasive breast carcinoma: A potent biomarker predicting lymph node metastasis. J. Cancer Res. Ther. 2018, 14, 345–350. [Google Scholar] [PubMed]
- Heras, S.C.-D.L.; Martínez-Balibrea, E. CXC family of chemokines as prognostic or predictive biomarkers and possible drug targets in colorectal cancer. World J. Gastroenterol. 2018, 24, 4738–4749. [Google Scholar] [CrossRef]
- Wang, D.; Sun, H.; Wei, J.; Cen, B.; Dubois, R.N. CXCL1 Is Critical for Premetastatic Niche Formation and Metastasis in Colorectal Cancer. Cancer Res. 2017, 77, 3655–3665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.; Jiao, C.; Zhuoxin, C.; Liang, D.; Zao, M.; Meng, X.; Gao, J.; Deshen, L.; Liu, W.; Hou, J.; et al. Activation of CXCL12/CXCR4 renders colorectal cancer cells less sensitive to radiotherapy via up-regulating the expression of survivin. Exp. Biol. Med. 2017, 242, 429–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, N.-H.; Nikfarjam, M.; He, H. Functions of the CXC ligand family in the pancreatic tumor microenvironment. Pancreatology 2018, 18, 705–716. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, D.R.; Yu, J.; Shanker, K.; Deshpande, N.; Varambally, R.; Ghosh, D.; Barrette, T.; Pander, A.; Chinnaiyan, A.M. ONCOMINE: A Cancer Microarray Database and Integrated Data-Mining Platform. Neoplasia 2004, 6, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Sun, D.; Wang, J.; Han, Y.; Dong, X.; Ge, J.; Zheng, R.; Shi, X.; Wang, B.; Li, Z.; Ren, P.; et al. TISCH: A comprehensive web resource enabling interactive single-cell transcriptome visualization of tumor microenvironment. Nucleic Acids Res. 2021, 49, D1420–D1430. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [Green Version]
- Györffy, B.; Lanczky, A.; Eklund, A.C.; Denkert, C.; Budczies, J.; Li, Q.; Szallasi, Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1809 patients. Breast Cancer Res. Treat. 2009, 123, 725–731. [Google Scholar] [CrossRef] [Green Version]
- Han, H.; Cho, J.-W.; Lee, S.-Y.; Yun, A.; Kim, H.; Bae, D.; Yang, S.; Kim, C.Y.; Lee, M.; Kim, E.; et al. TRRUST v2: An expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 2018, 46, D380–D386. [Google Scholar] [CrossRef]
- Feng, C.; Song, C.; Liu, Y.; Qian, F.; Gao, Y.; Ning, Z.; Wang, Q.; Jiang, Y.; Li, Y.; Li, M.; et al. KnockTF: A comprehensive human gene expression profile database with knockdown/knockout of transcription factors. Nucleic Acids Res. 2020, 48, D93–D100. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [Green Version]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [Green Version]
- Huang, D.W.; Lempicki, R.A.; Sherman, B.T. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Li, T.; Fan, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S.; Li, B.; Liu, X.S. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Jing, Q.; Yan, S.; Liu, X.; Sun, Y.; Zhu, D.; Wang, D.; Hao, C.; Xue, D. Amadis: A Comprehensive Database for Association Between Microbiota and Disease. Front. Physiol. 2021, 12, 1074. [Google Scholar] [CrossRef]
- Curtis, C.; Shah, S.P.; Chin, S.-F.; Turashvili, G.; Rueda, O.M.; Dunning, M.J.; Speed, D.; Lynch, A.G.; Samarajiwa, S.; METABRIC Group; et al. The genomic and transcriptomic architecture of 2000 breast tumours reveals novel subgroups. Nature 2012, 486, 346–352. [Google Scholar] [CrossRef]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehár, J.; Kryukov, G.V.; Sonkin, D.; et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef] [PubMed]
- Pei, H.; Li, L.; Fridley, B.L.; Jenkins, G.D.; Kalari, K.; Lingle, W.; Petersen, G.; Lou, Z.; Wang, L. FKBP51 Affects Cancer Cell Response to Chemotherapy by Negatively Regulating Akt. Cancer Cell 2009, 16, 259–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Tan, X.; Luo, J.; Cui, B.; Lei, S.; Si, Z.; Shen, L.; Yao, H. GNA13 promotes tumor growth and angiogenesis by upregulating CXC chemokines via the NF-κB signaling pathway in colorectal cancer cells. Cancer Med. 2018, 7, 5611–5620. [Google Scholar] [CrossRef]
- Unver, N. Macrophage chemoattractants secreted by cancer cells: Sculptors of the tumor microenvironment and another crucial piece of the cancer secretome as a therapeutic target. Cytokine Growth Factor Rev. 2019, 50, 13–18. [Google Scholar] [CrossRef] [PubMed]
- Bagheri, H.; Pourhanifeh, M.H.; Derakhshan, M.; Mahjoubin-Tehran, M.; Ghasemi, F.; Mousavi, S.; Rafiei, R.; Abbaszadeh-Goudarzi, K.; Mirzaei, H.R.; Mirzaei, H. CXCL-10: A new candidate for melanoma therapy? Cell. Oncol. 2020, 43, 353–365. [Google Scholar] [CrossRef]
- Pączek, S.; Łukaszewicz-Zając, M.; Gryko, M.; Mroczko, P.; Kulczynska-Przybik, A.; Mroczko, B. CXCL-8 in Preoperative Colorectal Cancer Patients: Significance for Diagnosis and Cancer Progression. Int. J. Mol. Sci. 2020, 21, 2040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, K.; Wu, L.; Yuan, S.; Wu, M.; Xu, Y.; Sun, Q.; Li, S.; Zhao, S.; Hua, T.; Liu, Z.-J. Structural basis of CXC chemokine receptor 2 activation and signalling. Nat. Cell Biol. 2020, 585, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhang, C.; Zhang, Z.; Qian, W.; Sun, Y.; Ji, B.; Zhang, Y.; Zhu, C.; Ji, D.; Wang, Q.; et al. Transcriptome analysis in primary colorectal cancer tissues from patients with and without liver metastases using next-generation sequencing. Cancer Med. 2017, 6, 1976–1987. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.-S.; Li, Y.-F.; Tan, J.; Sun, B.; Xiao, Y.-C.; Fang, X.-B.; Zhang, X.-F.; Li, Q.; Dong, J.-H.; Li, M.; et al. CCL20 and CXCL8 synergize to promote progression and poor survival outcome in patients with colorectal cancer by collaborative induction of the epithelial–mesenchymal transition. Cancer Lett. 2014, 348, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Weng, W.; Peng, J.; Hong, L.; Yang, L.; Toiyama, Y.; Gao, R.; Liu, M.; Yin, M.; Pan, C.; et al. Fusobacterium nucleatum Increases Proliferation of Colorectal Cancer Cells and Tumor Development in Mice by Activating Toll-Like Receptor 4 Signaling to Nuclear Factor−κB, and Up-regulating Expression of MicroRNA-21. Gastroenterology 2017, 152, 851–866.e24. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Tang, L.; Feng, Y.; Zhao, S.; Han, M.; Zhang, C.; Yuan, G.; Zhu, J.; Cao, S.; Wu, Q.; et al. A purified membrane protein from Akkermansia muciniphila or the pasteurised bacterium blunts colitis associated tumourigenesis by modulation of CD8+ T cells in mice. Gut 2020, 69, 1988–1997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viennois, E.; Chassaing, B.; Tahsin, A.; Pujada, A.; Wang, L.; Gewirtz, A.T.; Merlin, D. Host-derived fecal microRNAs can indicate gut microbiota healthiness and ability to induce inflammation. Theranostics 2019, 9, 4542–4557. [Google Scholar] [CrossRef]
- Yu, T.; Guo, F.; Yu, Y.; Sun, T.; Ma, D.; Han, J.; Qian, Y.; Kryczek, I.; Sun, D.; Nagarsheth, N.; et al. Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell 2017, 170, 548–563.e16. [Google Scholar] [CrossRef] [Green Version]





| CXCs | Type | Fold Change | p Value | t-Test | Dataset |
|---|---|---|---|---|---|
| CXCL1 | Invasive ductal breast carcinoma | −6.050 | 9.30 × 10−30 | −14.245 | TCGA |
| Breast cancer | −3.805 | 9.82 × 10−75 | −20.522 | Bittner Multi-cancer Statistics | |
| Colorectal cancer | 6.308 | 7.49 × 10−121 | 29.225 | Bittner Multi-cancer Statistics | |
| Colon adenocarcinoma | 6.365 | 4.39 × 10−12 | 10.236 | TCGA | |
| CXCL2 | Invasive ductal breast carcinoma | −36.063 | 4.18 × 10−59 | −32.023 | TCGA |
| Invasive ductal breast carcinoma | −3.671 | 1.28 × 10−51 | −23.342 | Curtis Breast Statistics | |
| Colorectal cancer | 3.055 | 4.69 × 10−71 | 20.072 | Bittner Multi-cancer Statistics | |
| Pancreatic cancer | 3.904 | 2.20 × 10−7 | 5.798 | Barretina CellLine Statistics | |
| CXCL3 | Breast cancer | −3.484 | 1.51 × 10−61 | −18.125 | Bittner Multi-cancer Statistics |
| Invasive ductal breast carcinoma | −7.462 | 4.22 × 10−34 | −19.062 | TCGA | |
| Colorectal cancer | 9.737 | 7.64 × 10−152 | 36.102 | Bittner Multi-cancer Statistics | |
| Colon adenocarcinoma | 8.551 | 2.61 × 10−13 | 11.423 | TCGA | |
| Pancreatic carcinoma | 5.062 | 3.80 × 10−7 | 5.806 | Pei Pancreas Statistics | |
| Pancreatic ductal adenocarcinoma | 3.907 | 1.19 × 10−8 | 6.423 | Badea Pancreas Statistics | |
| Pancreatic cancer | 2.088 | 4.70 × 10−4 | 3.536 | Barretina CellLine Statistics | |
| CXCL4 | Colorectal cancer | 2.314 | 7.21 × 10−40 | 14.283 | Bittner Multi-cancer Statistics |
| CXCL5 | Colon adenocarcinoma | 2.739 | 3.6 × 10−22 | 11.937 | TCGA |
| Colorectal cancer | 4.493 | 1.43 × 10−42 | 15.077 | Bittner Multi-cancer Statistics | |
| Pancreatic carcinoma | 12.881 | 5.58 × 10−9 | 6.828 | Pei Pancreas Statistics | |
| Pancreatic cancer | 7.439 | 1.22 × 10−6 | 5.395 | Barretina CellLine Statistics | |
| CXCL6 | Breast cancer | −3.256 | 1.33 × 10−54 | −16.970 | Bittner Multi-cancer Statistics |
| Colon adenocarcinoma | 2.859 | 5.44 × 10−12 | 8.66 | TCGA | |
| CXCL7 | Breast cancer | −2.707 | 3.97 × 10−33 | −12.629 | Bittner Multi-cancer Statistics |
| Colorectal cancer | 3.02 | 3.35 × 10−27 | 11.438 | Bittner Multi-cancer Statistics | |
| CXCL8 | Breast cancer | −4.229 | 3.32 × 10−60 | −17.871 | Bittner Multi-cancer Statistics |
| Invasive ductal breast carcinoma | −2.746 | 2.20 × 10−2 | −2.338 | Curtis Breast Statistics | |
| Colorectal cancer | 7.457 | 2.12 × 10−85 | 23.265 | Bittner Multi-cancer Statistics | |
| Pancreatic ductal adenocarcinoma | 9.800 | 9.92 × 10−12 | 7.971 | Badea Pancreas Statistics | |
| Pancreatic carcinoma | 8.378 | 3.51 × 10−6 | 5.473 | Pei Pancreas Statistics | |
| CXCL9 | Invasive ductal breast carcinoma | 4.663 | 7.27 × 10−91 | 29.060 | Curtis Breast Statistics |
| CXCL10 | Invasive ductal breast carcinoma | 5.022 | 4.40 × 10−71 | 26.711 | Curtis Breast Statistics |
| Pancreatic carcinoma | 3.950 | 1.41 × 10−4 | 4.190 | Pei Pancreas Statistics | |
| CXCL11 | Colon adenocarcinoma | 2.304 | 7.74 × 10−5 | 3.885 | TCGA |
| CXCL12 | Invasive ductal breast carcinoma | −5.734 | 2.59 × 10−110 | −41.690 | Curtis Breast Statistics |
| Invasive ductal breast carcinoma | −6.730 | 3.41 × 10−51 | −25.411 | TCGA | |
| Colon adenocarcinoma | −10.511 | 5.53 × 10−34 | −22.305 | TCGA | |
| CXCL13 | Ductal breast carcinoma | 2.238 | 5.72 × 10−4 | 3.358 | Bittner Breast Statistics |
| CXCL14 | Invasive ductal breast carcinoma | −2.991 | 4.38 × 10−55 | −20.207 | Curtis Breast Statistics |
| Invasive ductal breast carcinoma | −3.074 | 5.92 × 10−34 | −14.493 | TCGA | |
| CXCL16 | Pancreatic ductal adenocarcinoma | 2.311 | 6.64 × 10−12 | 8.384 | Badea Pancreas Statistics |
| Pancreatic cancer | 2.395 | 2.11 × 10−6 | 5.172 | Barretina CellLine Statistics | |
| CXCL17 | Colorectal cancer | −2.896 | 4.16 × 10−41 | −13.988 | Bittner Multi-cancer Statistics |
| Key TF | Description | p Value | Overlapped Genes |
|---|---|---|---|
| RELA | A subunit of NF-kappa B that is primarily responsible for its transactivation function. It contains a C-terminal transactivation domain and an N-terminal domain with homology to PROTO-ONCOGENE PROTEINS C-REL. | 1.09 × 10−7 | CXCL10, CXCL8, CXCL12, CXCL2, CXCL5, CXCL1 |
| NFKB1 | Nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 protein, human. | 1.14 × 10−7 | CXCL12, CXCL8, CXCL10, CXCL5, CXCL1, CXCL2 |
| SP1 | Promoter-specific RNA polymerase II transcription factor that binds to the GC box, one of the upstream promoter elements, in mammalian cells. | 0.00683 | CXCL5, CXCL1, CXCL14 |
| CXCs | Type | TF | Ref (PMID) |
|---|---|---|---|
| CXCL1 | Activation | CEBPD, NFKB1, RELA | 23028973; 10530453; 15958549 |
| Repression | BRCA1, GATA3 | 22120723 | |
| Unknown | HMGA1, NFKB1, PARP1, RELA, SP1 | 11112786; 16040075; 7479086 | |
| CXCL2 | Unknown | NFKB1, RELA, SMAD1 | 17363596 |
| PF4 | Activation | ETS1, GATA1, MEIS1, PBX1, PBX2, RUNX1, USF1, USF2 | 12609849; 12732210; 21129147; 15187018 |
| Repression | RUNX1 | 17150917 | |
| Unknown | ETS1, FLI1, PKNOX1, RUNX1, TIAL1 | 23848403; 12732210; 9207209 | |
| CXCL5 | Activation | SP1 | 11559712 |
| Unknown | NFKB1, RELA, ZNF148 | 11559712 | |
| PPBP | Unknown | TBP | 7958954 |
| CXCL8 | Activation | CEBPB, ELF4, ETS2, FOSB, JUN, NFE2L2, NFKB1, RELA, VDR, ZNF300 | 15085176; 15688424; 14625302; 12438253; 20826776; 16701870; 18684922; 21925595; 11512674; 16764699; 17040605; 17041011; 17917246; 18074095; 18996370; 22340043; 10506755; 10530453; 11297551; 11564889; 12058956; 15350531; 15387324; 15958549; 22340043; 22891766; 7876168; 9431991; 21777376 |
| Repression | EGR1, ELF4, ERG, HDAC2, ING4, NEAT1, NFKB1, RELA, SFPQ, ZFP36 | 12438253; 19359602; 22235125; 19411311; 17848618; 20707719; 15517885; 8413215; 24507715; 21593445 | |
| Unknown | ATF4, CEBPB, DDIT3, DEK, EGR1, EP300, FOS, HDAC1, HDAC2, IKBKB, JUN, NFKB1, NFKBIA, NR4A2, RELA, STAT3, STAT6, VDR, XBP1 | 16931790; 11120852; 18772138; 16829531; 19837667; 19966855; 12218154; 11564889; 12296854; 12643600; 12716652; 14670967; 10477716; 11342414; 11953364; 12485925; 12707271; 14631383; 14670967; 15289496; 15950427; 16004996; 16105834; 16803583; 17045242; 18281166; 19376732; 19732956; 19786024; 8878392; 9698090; 8617886; 17726017; 24250750 | |
| CXCL10 | Activation | IRF1, IRF3, IRF7, NFKB1, RELA | 19342664; 18691624; 24257594; 16982926; 20164184; 24701034 |
| Repression | STAT1 | 22022583 | |
| Unknown | NFKB1, RELA, STAT1 | 16818736; 19479051; 24257594; 23153456 | |
| CXCL12 | Activation | FOXF2, NFKB1, RELA, SNAI2 | 19562724; 17530707; 22074556 |
| Unknown | ESR1, HIF1A | 18563714; 12384916 | |
| CXCL14 | Activation | SP1 | 22382027 |
| TF | Intergenes | Numbers | p Value | FDR |
|---|---|---|---|---|
| AHR | CXCL16; CXCL11; CXCL14; CXCL9; CXCL6; CXCL3; CXCL5 | 7 | 1.13 × 10−6 | 0.000347 |
| ARID1A | IL8; CXCL1; CXCL5; CXCL12; CXCL2; CXCL6 | 6 | 0.00161 | 0.0216 |
| CREB1 | CXCL13; CXCL1; CXCL12; CXCL9; PF4; CXCL5; CXCL17; CXCL14; CXCL11; CXCL6; CXCL10; CXCL16; CXCL2 | 13 | 0.0039 | 0.0428 |
| EGR3 | CXCL1; CXCL6; CXCL3; CXCL5; CXCL13; CXCL10; CXCL2; CXCL11; CXCL9; CXCL16 | 10 | 1.59 × 10−5 | 0.000976 |
| ERG | CXCL3; CXCL5; CXCL2; CXCL1; CXCL6; PPBP; PF4; CXCL10; CXCL12; CXCL9; CXCL16; CXCL17; CXCL14; CXCL11 | 14 | 3.21 × 10−5 | 0.00164 |
| ESRRG | CXCL5; CXCL1; IL8; CXCL6; CXCL10 | 5 | 0.000465 | 0.00892 |
| ETV5 | CXCL10; IL8; CXCL5; CXCL2; CXCL1; CXCL6 | 6 | 0.000321 | 0.00704 |
| FOSL1 | IL8; CXCL6; CXCL10; CXCL5 | 4 | 0.000703 | 0.0121 |
| FOXO1 | CXCL14; CXCL12; CXCL6; CXCL3; IL8; CXCL2; CXCL10; CXCL1; CXCL5 | 9 | 7.42 × 10−5 | 0.00228 |
| FOXP1 | PPBP; CXCL3; PF4; CXCL5; CXCL13; CXCL6; IL8; CXCL1; CXCL2; CXCL11; CXCL17; CXCL14; CXCL10; CXCL9; CXCL16; CXCL12 | 16 | 8.60 × 10−6 | 0.000976 |
| HNF1A | CXCL12; CXCL10; CXCL11; CXCL9; CXCL3; CXCL1; CXCL2; CXCL5; PF4; IL8; CXCL17; CXCL14; CXCL6; CXCL16 | 14 | 1.31 × 10−5 | 0.000976 |
| HNF1B | CXCL3; CXCL1; CXCL5; CXCL16; PPBP; CXCL14 | 6 | 0.00329 | 0.0404 |
| HOXA5 | CXCL1; IL8; CXCL2; CXCL16 | 4 | 1.23 × 10−5 | 0.000976 |
| HOXD9 | CXCL6; CXCL16; CXCL5; CXCL10; PPBP; CXCL17; CXCL1; PF4; CXCL13; CXCL3; CXCL2; CXCL11 | 12 | 0.000157 | 0.00402 |
| KLF4 | CXCL5; CXCL17; CXCL2; CXCL14; CXCL10; PPBP | 6 | 0.000429 | 0.00878 |
| LIN28B | CXCL1; CXCL6; CXCL10; CXCL2; CXCL3 | 5 | 0.00162 | 0.0216 |
| MYBL2 | IL8; PPBP | 2 | 0.00405 | 0.0429 |
| NR2F2 | CXCL13; CXCL9; CXCL14; PF4; CXCL3; CXCL10; CXCL11; CXCL6; CXCL1; PPBP; CXCL5; CXCL2; CXCL16 | 13 | 0.00358 | 0.0423 |
| PITX2 | CXCL10; CXCL6; CXCL1 | 3 | 0.00111 | 0.017 |
| POLR3A | PPBP; CXCL14; CXCL5; CXCL3; CXCL13; CXCL9; CXCL11; CXCL10; CXCL6 | 9 | 0.00378 | 0.0428 |
| Prox1 | CXCL3; CXCL13; CXCL11; CXCL5; CXCL2; CXCL16; CXCL1; CXCL17; PPBP; CXCL6 | 10 | 0.00122 | 0.0178 |
| PTEN | CXCL14; CXCL13; CXCL16; PF4; PPBP; CXCL9; CXCL17; CXCL12; CXCL10; IL8; CXCL1; CXCL3; CXCL11; CXCL2; CXCL5 | 15 | 6.24 × 10−5 | 0.00228 |
| RARA | PPBP; CXCL9; CXCL13; CXCL17; CXCL11; CXCL3; CXCL1; CXCL2; CXCL14 | 9 | 0.00021 | 0.00496 |
| SOX17 | PPBP; IL8; CXCL6; CXCL10; CXCL1; CXCL11; PF4; CXCL3; CXCL2; CXCL12; CXCL5 | 11 | 8.18 × 10−5 | 0.00228 |
| SOX4 | CXCL2; CXCL3; CXCL14; CXCL1; CXCL13; CXCL16; IL8; CXCL10 | 8 | 0.00101 | 0.0163 |
| SOX9 | CXCL13; CXCL5; CXCL6; CXCL3; PF4; CXCL17; PPBP; CXCL1; CXCL10; CXCL2 | 10 | 5.16 × 10−5 | 0.00226 |
| SPDEF | CXCL2; PF4; CXCL14; CXCL13; PPBP; CXCL11; CXCL6; CXCL1; CXCL9 | 9 | 0.00071 | 0.0121 |
| YY1 | CXCL11; CXCL6; CXCL12; PF4; CXCL9; CXCL17; CXCL13; CXCL5; CXCL16; CXCL3 | 10 | 6.72 × 10−5 | 0.00228 |
| ZNF25 | CXCL10; CXCL5; CXCL6 | 3 | 0.0029 | 0.0371 |
| Binding Gene | Mirnaid | Binding p Value | Accessibility | Number of Pairings | Binding Region Length | Longest Consecutive Pairings |
|---|---|---|---|---|---|---|
| CXCL8 | hsa-miR-19a-3p | 0.923077 | 0.016853 | 17 | 22 | 10 |
| hsa-miR-3138 | 0.846154 | 0.009999 | 19 | 23 | 9 | |
| hsa-miR-4306 | 0.846154 | 0.019455 | 13 | 16 | 7 | |
| hsa-miR-4793-3p | 1 | 0.006677 | 17 | 24 | 9 | |
| hsa-miR-6793-5p | 1 | 2.62 × 10−5 | 19 | 29 | 12 | |
| hsa-miR-6837-5p | 1 | 0.000791 | 17 | 20 | 12 | |
| hsa-miR-6837-3p | 0.846154 | 0.000961 | 18 | 26 | 8 | |
| CXCL10 | hsa-let-7b-5p | 0.923077 | 0.000462 | 18 | 21 | 10 |
| hsa-miR-23a-3p | 0.846154 | 0.079181 | 12 | 18 | 8 | |
| hsa-miR-196a-5p | 0.923077 | 0.000483 | 17 | 19 | 17 | |
| hsa-miR-34a-5p | 0.846154 | 0.000568 | 15 | 19 | 6 | |
| hsa-let-7i-5p | 0.923077 | 0.000462 | 17 | 22 | 8 | |
| hsa-miR-23b-3p | 0.846154 | 0.079181 | 12 | 18 | 8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, L.; Yao, W.; Yan, S.; Dong, X.; Lv, Z.; Jing, Q.; Wang, Q.; Ma, B.; Hao, C.; Xue, D.; et al. Pan-Cancer Analysis of Prognostic and Immune Infiltrates for CXCs. Cancers 2021, 13, 4153. https://doi.org/10.3390/cancers13164153
Li L, Yao W, Yan S, Dong X, Lv Z, Jing Q, Wang Q, Ma B, Hao C, Xue D, et al. Pan-Cancer Analysis of Prognostic and Immune Infiltrates for CXCs. Cancers. 2021; 13(16):4153. https://doi.org/10.3390/cancers13164153
Chicago/Turabian StyleLi, Long, Wenchao Yao, Sen Yan, Xianghui Dong, Zhenyi Lv, Qingxu Jing, Qiang Wang, Biao Ma, Chenjun Hao, Dongbo Xue, and et al. 2021. "Pan-Cancer Analysis of Prognostic and Immune Infiltrates for CXCs" Cancers 13, no. 16: 4153. https://doi.org/10.3390/cancers13164153





