Clinical Categorization Algorithm (CLICAL) and Machine Learning Approach (SRF-CLICAL) to Predict Clinical Benefit to Immunotherapy in Metastatic Melanoma Patients: Real-World Evidence from the Istituto Nazionale Tumori IRCCS Fondazione Pascale, Napoli, Italy
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Statistics
2.2.1. Survival Analysis
2.2.2. The Clinical Categorization Algorithm (CLICAL)
2.2.3. Application of the Machine Learning Survival Random Forest Clinical Categorization Algorithm (SRF-CLICAL) and Definition of Prognostic Signatures
2.2.4. Cox Proportional Hazards Analysis
2.2.5. Survival Random Forest Model
2.2.6. Kaplan–Meier Survival Curves
3. Results
3.1. Clinicopathological Features of Melanoma Patients
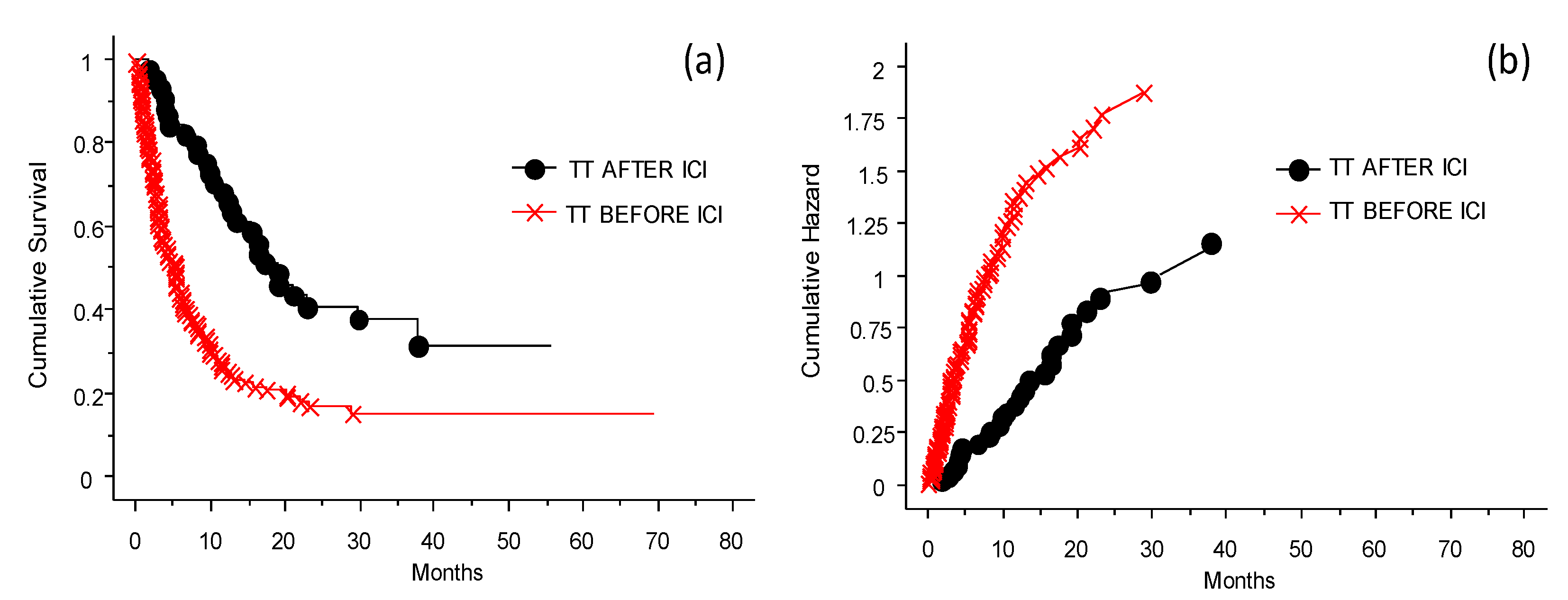
3.2. The Efficacy of ICI Depending on the Previous Treatment
3.3. The Response to Immunotherapy
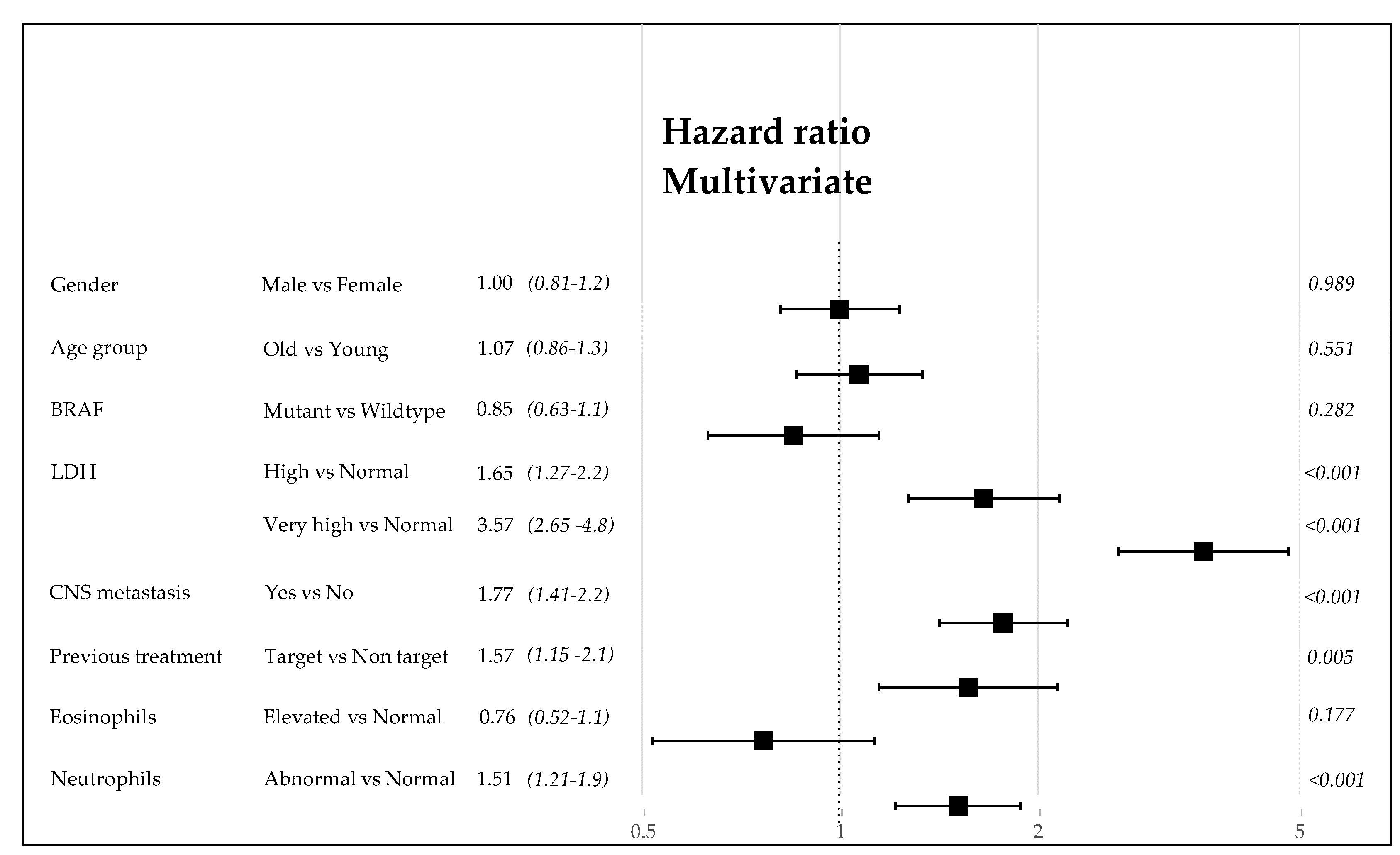
3.4. Analysis of the Factors Related to the Efficacy of Immunotherapy (ICI Program)
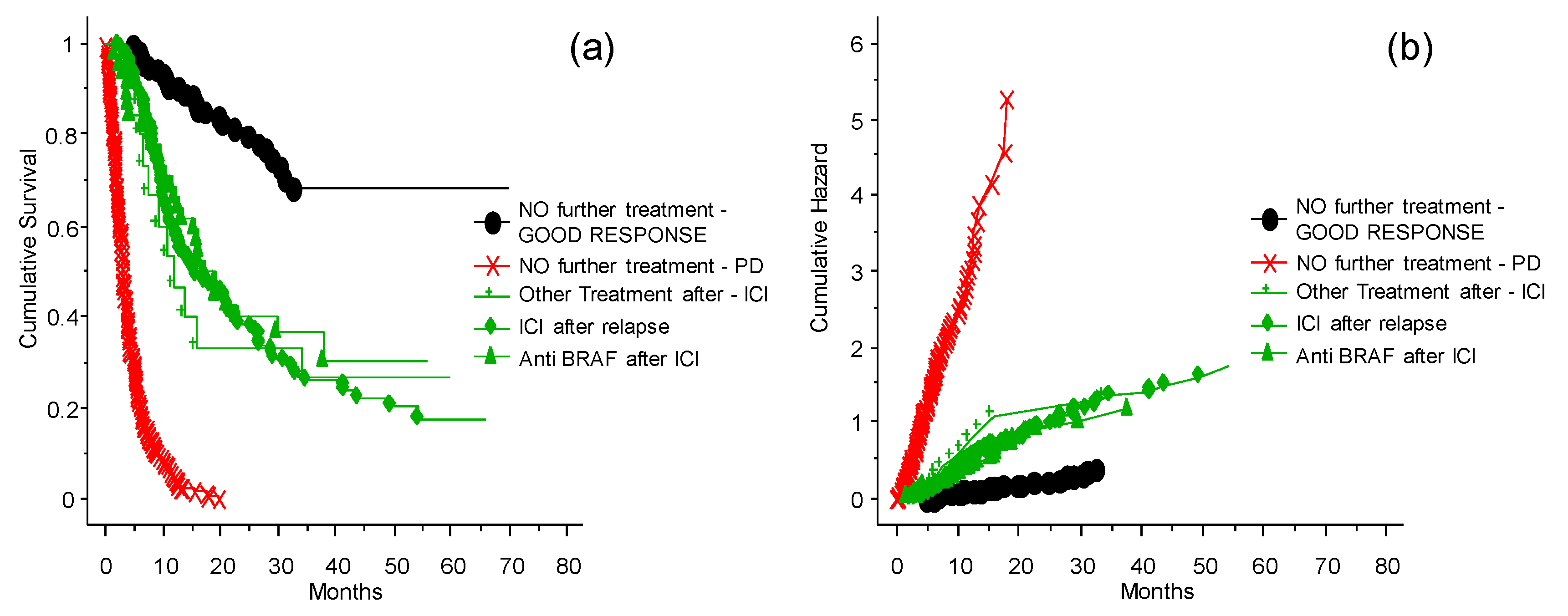
Role of the Different Treatments Given before Inclusion to the INT-NA
3.5. Treatment of Patients Who Relapsed after the ICI Program at the INT-NA
3.6. Analysis of the Predictive Power of Clinical Variables at Inclusion
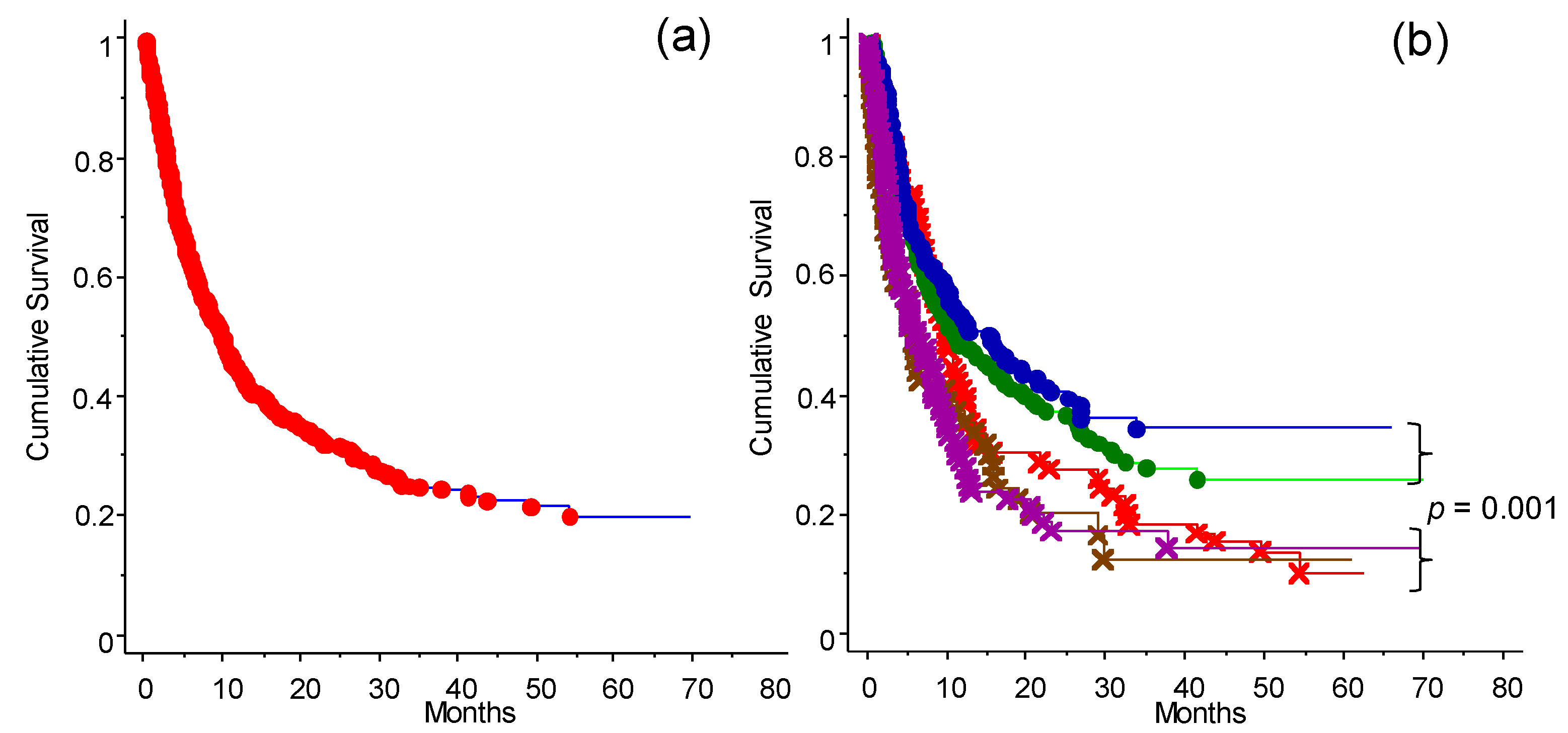
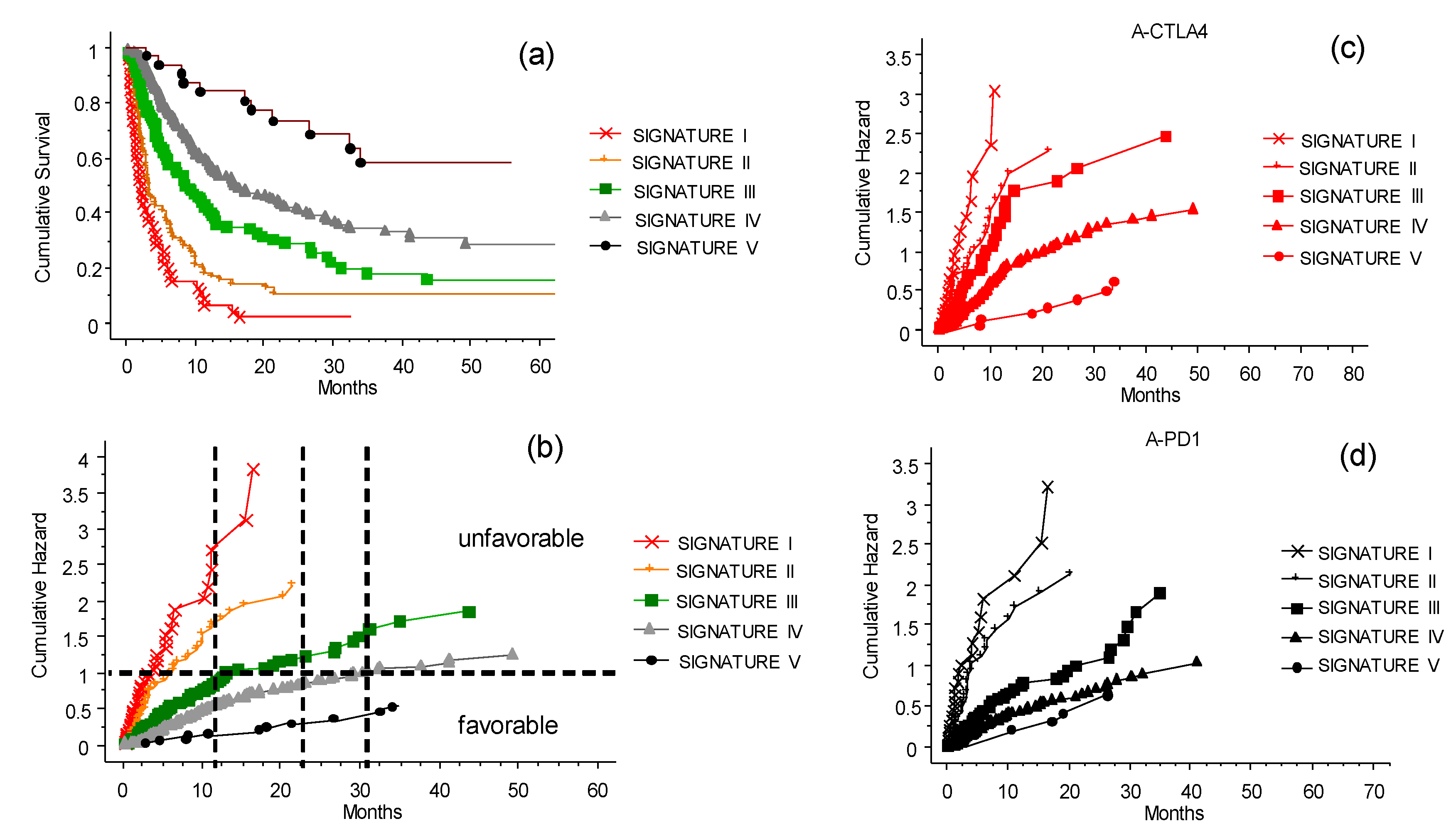
3.7. The CLICAL Signature and Prediction of Survival Rates
3.8. The CLICAL Signature and Prediction of Response to ICIs
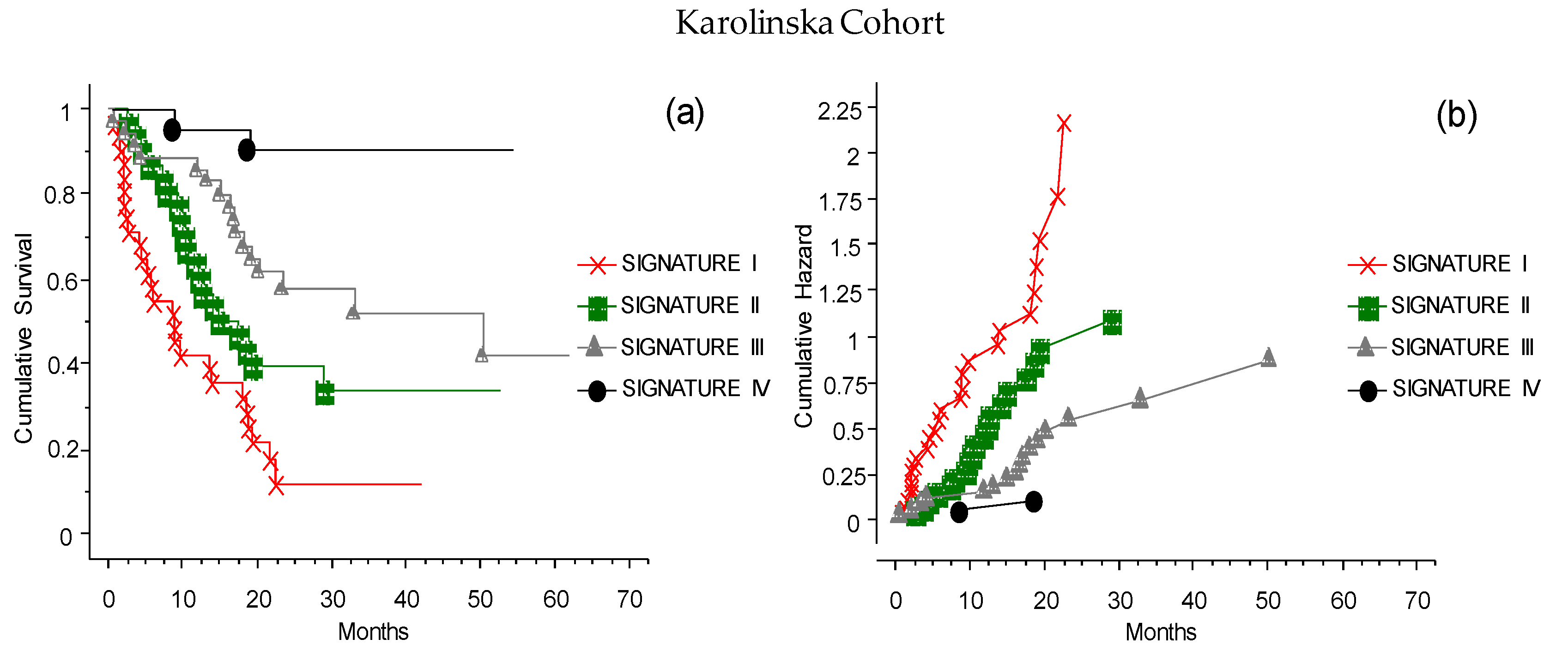
3.9. The CLICAL Signature Applied to an External Cohort
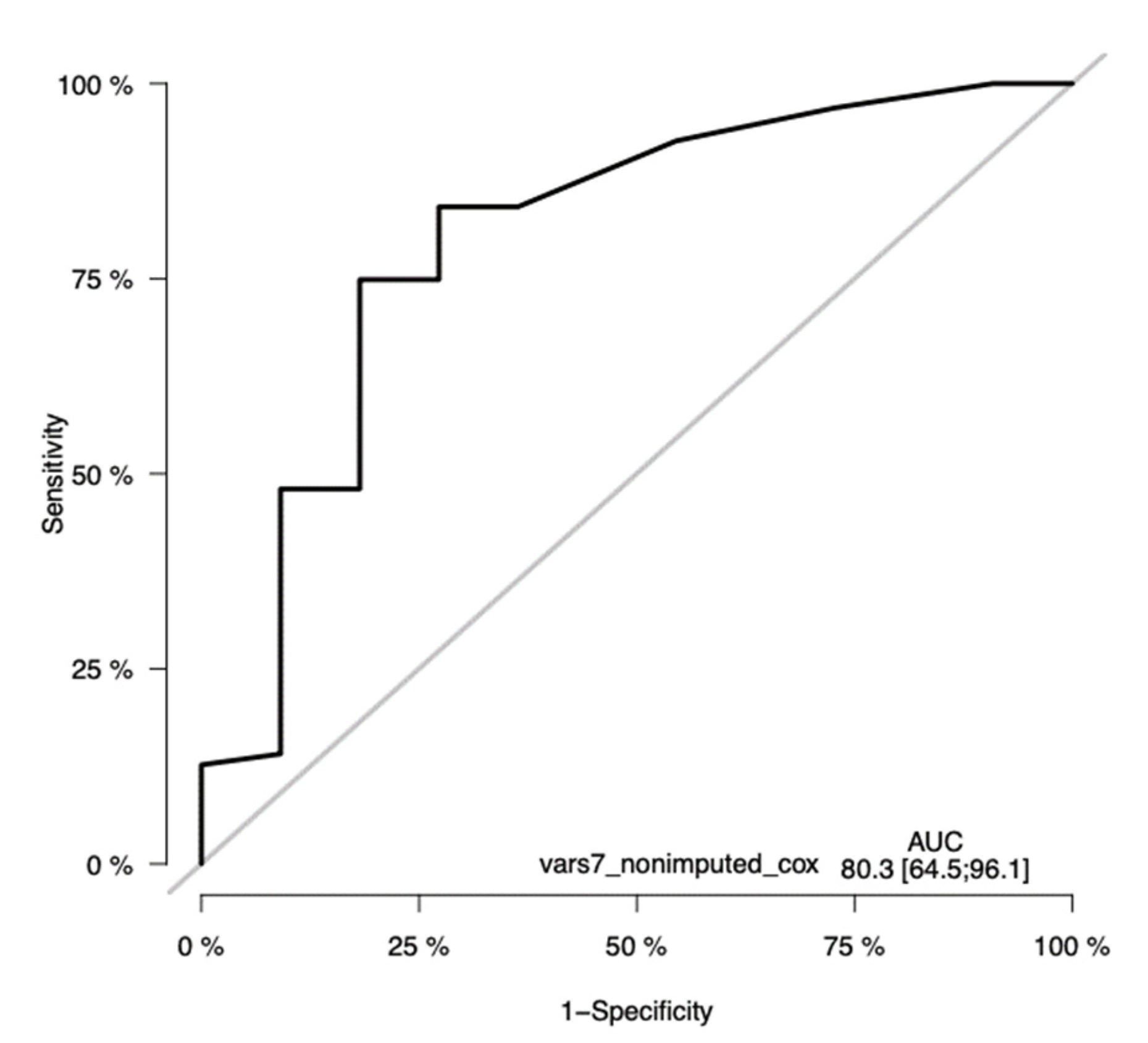
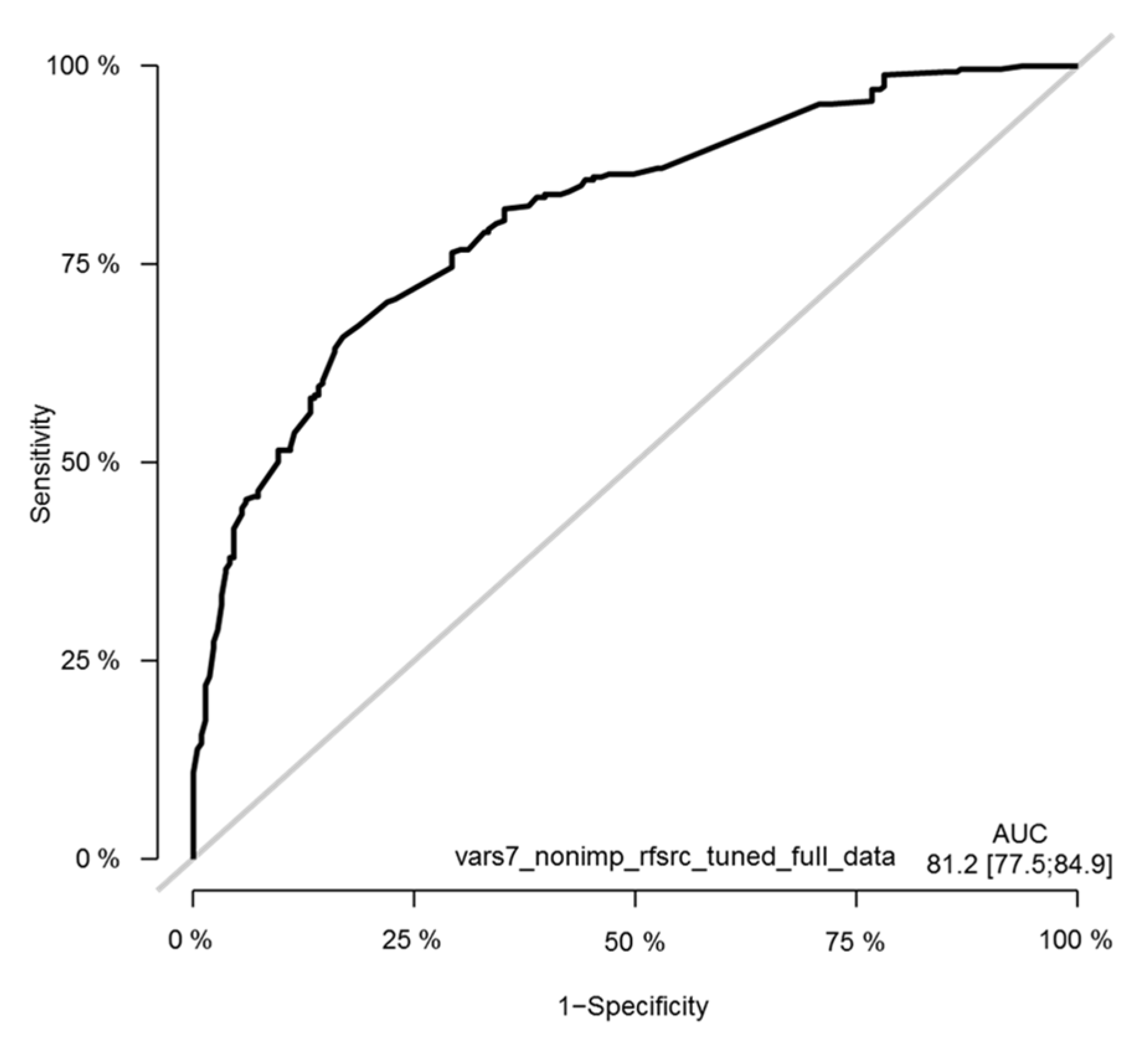
3.10. The Validation of the CLICAL Algorithm’s Efficiency by Machine Learning Survival Random Forest Analysis (SRF-CLICAL)
3.11. Survival Random Forest Model
3.12. The SRF-CLICAL Signature
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ascierto, P.A.; Marincola, F.M. What have we learned from cancer immunotherapy in the last 3 years? J. Transl. Med. 2014, 12, 141. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Schadendorf, D.; Dummer, R.; Hauschild, A.; Robert, C.; Hamid, O.; Daud, A.; Eertwegh, A.V.D.; Cranmer, L.; O’Day, S.; Puzanov, I.; et al. Health-related quality of life in the randomised KEYNOTE-002 study of pembrolizumab versus chemotherapy in patients with ipilimumab-refractory melanoma. Eur. J. Cancer 2016, 67, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Schadendorf, D.; Larkin, J.; Wolchok, J.; Hodi, F.S.; Sileni, V.C.; Gonzalez, R.; Rutkowski, P.; Grob, J.-J.; Cowey, C.L.; Lao, C.; et al. Health-related quality of life results from the phase III CheckMate 067 study. Eur. J. Cancer 2017, 82, 80–91. [Google Scholar] [CrossRef]
- Petrella, T.M.; Robert, C.; Richtig, E.; Miller, W.H.; Masucci, G.V.; Walpole, E.; Lebbe, C.; Steven, N.; Middleton, M.R.; Hille, D.; et al. Patient-reported outcomes in KEYNOTE-006, a randomised study of pembrolizumab versus ipilimumab in patients with advanced melanoma. Eur. J. Cancer 2017, 86, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Mamoor, M.; Postow, M.A.; A Lavery, J.; Baxi, S.S.; Khan, N.; Mao, J.J.; Rogak, L.J.; Sidlow, R.; Thom, B.; Wolchok, J.A.; et al. Quality of life in long-term survivors of advanced melanoma treated with checkpoint inhibitors. J. Immunother. Cancer 2019, 8, e000260. [Google Scholar] [CrossRef]
- Rogiers, A.; Boekhout, A.; Schwarze, J.K.; Awada, G.; Blank, C.U.; Neyns, B. Long-Term Survival, Quality of Life, and Psychosocial Outcomes in Advanced Melanoma Patients Treated with Immune Checkpoint Inhibitors. J. Oncol. 2019, 2019, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Madonna, G.; Ballesteros-Merino, C.; Feng, Z.; Bifulco, C.; Capone, M.; Giannarelli, D.; Mallardo, D.; Simeone, E.; Grimaldi, A.M.; Caracò, C.; et al. PD-L1 expression with immune-infiltrate evaluation and outcome prediction in melanoma patients treated with ipilimumab. OncoImmunology 2018, 7, e1405206. [Google Scholar] [CrossRef] [PubMed]
- Pardoll, D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 2012, 12, 252–264. [Google Scholar] [CrossRef] [PubMed]
- Hodi, F.S.; O’Day, S.J.; McDermott, D.F.; Weber, R.W.; Sosman, J.A.; Haanen, J.B.; Gonzalez, R.; Robert, C.; Schadendorf, D.; Hassel, J.C.; et al. Improved Survival with Ipilimumab in Patients with Metastatic Melanoma. N. Engl. J. Med. 2010, 363, 711–723. [Google Scholar] [CrossRef] [PubMed]
- Larkin, J.; Sileni, V.C.; Gonzalez, R.; Grob, J.-J.; Cowey, C.L.; Lao, C.D.; Schadendorf, D.; Dummer, R.; Smylie, M.; Rutkowski, P.; et al. Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. N. Engl. J. Med. 2015, 373, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Balar, A.; Weber, J.S. PD-1 and PD-L1 antibodies in cancer: Current status and future directions. Cancer Immunol. Immunother. 2017, 66, 551–564. [Google Scholar] [CrossRef]
- Larkin, J.; Minor, D.; D’Angelo, S.; Neyns, B.; Smylie, M., Jr.; Gutzmer, R.; Linette, G.; Chmielowski, B.; Lao, C.D. Overall Survival in Patients with Advanced Melanoma Who Received Nivolumab Versus Investigator’s Choice Chemotherapy in CheckMate 037: A Randomized, Controlled, Open-Label Phase III Trial. J. Clin. Oncol. 2018, 36, 383–390. [Google Scholar] [CrossRef] [PubMed]
- Schachter, J.; Ribas, A.; Long, G.; Arance, A.; Grob, J.-J.; Mortier, L.; Daud, A.; Carlino, M.S.; McNeil, C.; Lotem, M.; et al. Pembrolizumab versus ipilimumab for advanced melanoma: Final overall survival results of a multicentre, randomised, open-label phase 3 study (KEYNOTE-006). Lancet 2017, 390, 1853–1862. [Google Scholar] [CrossRef]
- Harder, N.; Schönmeyer, R.; Nekolla, K.; Meier, A.; Brieu, N.; Vanegas, C.; Madonna, G.; Capone, M.; Botti, G.; Ascierto, P.A.; et al. Automatic discovery of image-based signatures for ipilimumab response prediction in malignant melanoma. Sci. Rep. 2019, 9, 1–19. [Google Scholar] [CrossRef]
- Capone, M.; Fratangelo, F.; Giannarelli, D.; Sorrentino, C.; Turiello, R.; Zanotta, S.; Galati, D.; Madonna, G.; Tuffanelli, M.; Scarpato, L.; et al. Frequency of circulating CD8+CD73+T cells is associated with survival in nivolumab-treated melanoma patients. J. Transl. Med. 2020, 18, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Schilling, B.; Martens, A.; Foppen, M.H.G.; Gebhardt, C.; Hassel, J.C.; Rozeman, E.A.; Gesierich, A.; Gutzmer, R.; Kähler, K.C.; Livingstone, E.; et al. First-line therapy-stratified survival in BRAF-mutant melanoma: A retrospective multicenter analysis. Cancer Immunol. Immunother. 2019, 68, 765–772. [Google Scholar] [CrossRef]
- Ayers, M.; Lunceford, J.; Nebozhyn, M.; Murphy, E.; Loboda, A.; Kaufman, D.R.; Albright, A.; Cheng, J.D.; Kang, S.P.; Shankaran, V.; et al. IFN-γ–related mRNA profile predicts clinical response to PD-1 blockade. J. Clin. Investig. 2017, 127, 2930–2940. [Google Scholar] [CrossRef] [PubMed]
- Tucci, M.; Passarelli, A.; Mannavola, F.; Stucci, L.S.; Ascierto, P.A.; Capone, M.; Madonna, G.; Lopalco, P.; Silvestris, F. Serum exosomes as predictors of clinical response to ipilimumab in metastatic melanoma. OncoImmunology 2017, 7, e1387706. [Google Scholar] [CrossRef]
- Jacquelot, N.; Yamazaki, T.; Roberti, M.P.; Duong, C.P.M.; Andrews, M.C.; Verlingue, L.; Ferrere, G.; Becharef, S.; Vétizou, M.; Daillère, R.; et al. Sustained Type I interferon signaling as a mechanism of resistance to PD-1 blockade. Cell Res. 2019, 29, 846–861. [Google Scholar] [CrossRef] [PubMed]
- Madonna, G.; Capone, M.; Tuffanelli, M.; Curvietto, M.; Paone, M.; Esposito, A.; Sorrentino, A.; Palla, M.; Scarpato, L.; Mallardo, D.; et al. Real World data analysis related to metastatic melanoma patients treated with immunotherapy from 2012 to 2018 at Istituto Nazionale Tumori IRCCS Fondazione “G. Pascale” of Napoli, Italy. 34th Annual Meeting & Pre-Conference Programs of the Society for Immunotherapy of Cancer (SITC 2019): Part 1. J. Immunother. Cancer 2019, 7, 272–282. [Google Scholar] [CrossRef]
- Wolchok, J.D.; Hoos, A.; O’Day, S.; Weber, J.; Hamid, O.; Lebbé, C.; Maio, M.; Binder, M.; Bohnsack, O.; Nichol, G.; et al. Guidelines for the Evaluation of Immune Therapy Activity in Solid Tumors: Immune-Related Response Criteria. Clin. Cancer Res. 2009, 15, 7412–7420. [Google Scholar] [CrossRef]
- Krakowski, I.; Bottai, M.; Häbel, H.; Masucci, G.; Girnita, A.; Smedby, K.; Eriksson, H. Impact of modern systemic therapies and clinical markers on treatment outcome for metastatic melanoma in a real-world setting. J. Eur. Acad. Dermatol. Venereol. 2020, 35, 105–115. [Google Scholar] [CrossRef]
- Simao, F.A.; Hieta, R.; Pulman, J.A.; Madonna, G.; Ascierto, P.A.; Lewensohn, R.; Masucci, G.V. SRF-CLICAL: An approach for patient risk stratification using random forest models. bioRxiv 2021. [Google Scholar] [CrossRef]
- Therneau, T. A Package for Survival Analysis in R, Version 3.2-3. 2020. Available online: https://CRAN.R-project.org/package=survival (accessed on 13 March 2021).
- Kassambara, A.; Kosinski, M. Survminer: Drawing Survival Curves Using ‘ggplot2’ R Package Version 0.4.8. 2018. Available online: https://CRAN.R-project.org/package=survminer (accessed on 9 March 2021).
- Mogensen, U.B.; Ishwaran, H.; Gerds, T.A. Evaluating Random Forests for Survival Analysis Using Prediction Error Curves. J. Stat. Softw. 2012, 50, 1–23. Available online: http://www.jstatsoft.org/v50/i11/ (accessed on 18 September 2020). [CrossRef]
- Gerds, T.A.; Ozenne, B. Risk Regression: Risk Regression Models and Prediction Scores for Survival Analysis with Competing Risks. R Package Version 2020.02.05. 2020. Available online: https://CRAN.R-project.org/package=riskRegression (accessed on 9 December 2020).
- Ishwaran, H.; Kogalur, U.B. Fast Unified Random Forests for Survival, Regression, and Classification (RF-SRC), R Package Version 2.9.3. 2021. Available online: https://cran.r-project.org/package=randomForestSRC (accessed on 9 March 2021).
- Ruggiero, C.F.; Malpicci, D.; Fattore, L.; Madonna, G.; Vanella, V.; Mallardo, D.; Liguoro, D.; Salvati, V.; Capone, M.; Bedogni, B.; et al. ErbB3 Phosphorylation as Central Event in Adaptive Resistance to Targeted Therapy in Metastatic Melanoma: Early Detection in CTCs during Therapy and Insights into Regulation by Autocrine Neuregulin. Cancers 2019, 11, 1425. [Google Scholar] [CrossRef]
- Villani, A.; Scalvenzi, M.; Fabbrocini, G.; Ocampo-Candiani, J.; Ocampo-Garza, S.S. Looking into a Better Future: Novel Therapies for Metastatic Melanoma. Dermatol. Ther. 2021, 11, 751–767. [Google Scholar] [CrossRef]
- Luke, J.J.; Flaherty, K.T.; Ribas, A.; Long, G. Targeted agents and immunotherapies: Optimizing outcomes in melanoma. Nat. Rev. Clin. Oncol. 2017, 14, 463–482. [Google Scholar] [CrossRef] [PubMed]
- Mason, R.; Au, L.; Garces, A.I.R.; Larkin, J. Current and emerging systemic therapies for cutaneous metastatic melanoma. Expert Opin. Pharmacother. 2019, 20, 1135–1152. [Google Scholar] [CrossRef]
- Guida, M.; Bartolomeo, N.; Quaglino, P.; Madonna, G.; Pigozzo, J.; Di Giacomo, A.; Minisini, A.; Tucci, M.; Spagnolo, F.; Occelli, M.; et al. No Impact of NRAS Mutation on Features of Primary and Metastatic Melanoma or on Outcomes of Checkpoint Inhibitor Immunotherapy: An Italian Melanoma Intergroup (IMI) Study. Cancers 2021, 13, 475. [Google Scholar] [CrossRef] [PubMed]
- Ottaviano, M.; De Placido, S.; Ascierto, P.A. Recent success and limitations of immune checkpoint inhibitors for cancer: A lesson from melanoma. Virchows Arch. 2019, 474, 421–432. [Google Scholar] [CrossRef] [PubMed]
- Bedognetti, D.; Ceccarelli, M.; Galluzzi, L.; Lu, R.; Palucka, K.; Samayoa, J.; Spranger, S.; Warren, S.; Wong, K.K. Toward a comprehensive view of cancer immune responsiveness: A synopsis from the SITC workshop. J. Immunother. Cancer 2019, 7, 131. [Google Scholar] [CrossRef] [PubMed]
- Donia, M.; Ellebaek, E.; Øllegaard, T.H.; Duval, L.; Aaby, J.B.; Hoejberg, L.; Køhler, U.H.; Schmidt, H.; Bastholt, L.; Svane, I.M. The real-world impact of modern treatments on the survival of patients with metastatic melanoma. Eur. J. Cancer 2019, 108, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Moser, J.C.; Chen, D.; Hu-Lieskovan, S.; Grossmann, K.F.; Patel, S.; Colonna, S.V.; Ying, J.; Hyngstrom, J.R. Real-world survival of patients with advanced BRAF V600 mutated melanoma treated with front-line BRAF/MEK inhibitors, anti-PD-1 antibodies, or nivolumab/ipilimumab. Cancer Med. 2019, 8, 7637–7643. [Google Scholar] [CrossRef]
- Pavlick, A.C.; Fecher, L.; Ascierto, P.A.; Sullivan, R.J. Frontline Therapy for BRAF-Mutated Metastatic Melanoma: How Do You Choose, and Is There One Correct Answer? Am. Soc. Clin. Oncol. Educ. Book 2019, 39, 564–571. [Google Scholar] [CrossRef]
- Johnson, D.B.; Pectasides, E.; Feld, E.; Ye, F.; Zhao, S.; Johnpulle, R.; Merritt, R.; McDermott, D.F.; Puzanov, I.; Lawrence, D.; et al. Sequencing Treatment in BRAF V600 Mutant Melanoma: Anti-PD-1 Before and After BRAF Inhibition. J. Immunother. 2017, 40, 31–35. [Google Scholar] [CrossRef] [PubMed]
- Rozeman, E.A.; Dekker, T.; Haanen, J.B.A.G.; Blank, C.U. Advanced Melanoma: Current Treatment Options, Biomarkers, and Future Perspectives. Am. J. Clin. Dermatol. 2017, 19, 303–317. [Google Scholar] [CrossRef]
- Weide, B.; Martens, A.; Hassel, J.C.; Berking, C.; Postow, M.A.; Bisschop, K.; Simeone, E.; Mangana, J.; Schilling, B.; Di Giacomo, A.M.; et al. Baseline Biomarkers for Outcome of Melanoma Patients Treated with Pembrolizumab. Clin. Cancer Res. 2016, 22, 5487–5496. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Zhao, H.; Zhao, J. Impact of clinicopathological characteristics on survival in patients treated with immune checkpoint inhibitors for metastatic melanoma. Int. J. Cancer 2018, 144, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Nie, R.-C.; Yuan, S.-Q.; Wang, Y.; Chen, Y.-B.; Cai, Y.-Y.; Chen, S.; Li, S.-M.; Zhou, J.; Chen, G.-M.; Luo, T.-Q.; et al. Robust immunoscore model to predict the response to anti-PD1 therapy in melanoma. Aging 2019, 11, 11576–11590. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Schilling, B.; Liu, D.; Sucker, A.; Livingstone, E.; Jerby-Arnon, L.; Zimmer, L.; Gutzmer, R.; Satzger, I.; Loquai, C.; et al. Author Correction: Integrative molecular and clinical modeling of clinical outcomes to PD1 blockade in patients with metastatic melanoma. Nat. Med. 2020, 26, 1147. [Google Scholar] [CrossRef]
- Danaher, P.; Warren, S.; Lu, R.; Samayoa, J.; Sullivan, A.; Pekker, I.; Wallden, B.; Marincola, F.M.; Cesano, A. Pan-cancer adaptive immune resistance as defined by the Tumor Inflammation Signature (TIS): Results from The Cancer Genome Atlas (TCGA). J. Immunother. Cancer 2018, 6, 63. [Google Scholar] [CrossRef] [PubMed]





| Variables | All | Females | Males | |||
|---|---|---|---|---|---|---|
| n | % | n | % | n | % | |
| Clinical anamnestic variable | ||||||
| Sex | 578 | 100 | 258 | 45 | 320 | 55 |
| Median age (range) | 61.4 (23–89) | 59.9 (23–89) | 62.5 (22–87) | |||
| Age 65–100 | 317 | 55 | 132 | 51 | 185 | 58 |
| Age 18–64 | 261 | 45 | 126 | 49 | 135 | 42 |
| BRAF mut 600E tested | 548 a | 100 | 241 | 100 | 307 | 100 |
| BRAF 600E mutated | 234 | 43 | 108 | 45 | 126 | 41 |
| CNS met | 572 | 100 | 257 | 100 | 315 | 100 |
| Yes | 162 | 28 | 71 | 28 | 91 | 29 |
| No | 410 | 72 | 186 | 72 | 224 | 71 |
| LDH | 535 | 100 | 241 | 100 | 294 | 100 |
| Very high (>2 × LLR) | 77 | 14 | 39 | 16 | 38 | 13 |
| High (≥1 × ≤2 × LLR) | 107 | 20 | 43 | 18 | 64 | 22 |
| Normal (<1× LLR) | 351 | 66 | 159 | 66 | 192 | 65 |
| Eosinophil counts | 543 | 100 | 242 | 100 | 301 | 100 |
| Elevated | 47 | 9 | 18 | 7 | 29 | 10 |
| Normal | 496 | 91 | 224 | 93 | 272 | 90 |
| NLR | 551 | 100 | 248 | 100 | 303 | 100 |
| Abnormal | 247 | 45 | 104 | 42 | 143 | 47 |
| Normal | 304 | 55 | 144 | 58 | 160 | 53 |
| Treatment groups | ||||||
| None (Naïve) | 199 | 34 | 83 | 32 | 116 | 36 |
| Immunotherapy | 142 | 25 | 61 | 24 | 81 | 25 |
| Target and immunotherapy | 59 | 10 | 29 | 11 | 30 | 9 |
| Target | 102 | 18 | 48 | 19 | 54 | 18 |
| Cytostatic | 76 | 13 | 37 | 14 | 39 | 12 |
| Grouped in Pre-target /No target | ||||||
| No target | 417 | 72 | 181 | 70 | 236 | 74 |
| Pre-target | 161 | 28 | 77 | 30 | 84 | 26 |
| Treatment at inclusion to the INT-NA ICI program | ||||||
| Anti-CTLA-4 | 292 | 51 | 129 | 50 | 163 | 51 |
| Anti-PD-1 (nivolumab) | 151 | 26 | 68 | 26 | 83 | 26 |
| Anti-PD-1 (pembrolizumab) | 135 | 23 | 61 | 24 | 74 | 23 |
| RFS | OS | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| n (%) | Probability of RFS (%) at 24 Months | CI 95% Min–Max | Cumulative Hazard at 24 Months | CI 95% Min–Max | Probability of Survival (%) at 50 Months | CI 95% Min–Max | Cumulative Hazard at 50 Months | CI 95% Min–Max | |
| ALL | 578 (100) | 16 | 13–19 | 1.83 | 1.63–2.03 | 8 | 5–11 | 2.46 | 2.12.80 |
| CR | 32 (5.5) | 67 | 48–85 | 0.39 | 0.12–0.66 | 81 | 61–100 | 0.19 | 0–0.43 |
| PR | 74 (12.8) | 64 | 52–76 | 0.48 | 0.28–0.67 | 58 | 42–75 | 0.52 | 0.24–0.79 |
| SD | 103 (17.8) | 24 | 15–33 | 1.48 | 1.09–1.86 | 39 | 25–52 | 0.94 | 0.60–1.28 |
| PD | 369 (63.8) | 0 | 0 | 6.42 | 3.91–8.93 | 5 | 2–7 | 2.9 | 2.4–3.4 |
| RFS | OS | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| n (%) | Probability of RFS (%) at 24 Months | CI 95% Min–Max | Cumulative Hazard at 20 Months | CI 95% Min–Max | Probability of Survival (%) at 50 Months | CI 95% Min–Max | Cumulative Hazard at 50 Months | CI 95% Min–Max | |
| ALL | 199 (100) | 22 | 16–28 | 1.49 | 1.22–1.76 | 36 | 27–42 | 1.06 | 0.83–1.28 |
| CR | 17 (8.5) | 81 | 62–100 | 0.32 | 0–0.66 | 100 | 100 | 0 | 0 |
| PR | 29 (14.5) | 73 | 56–90 | 0.31 | 0.08–0.53 | 76 | 52–99 | 0.27 | 0–0.56 |
| SD | 36 (17.6) | 31 | 14–43 | 1.13 | 0.61–1.64 | 55 | 34–76 | 0.58 | 0.21–0.96 |
| PD | 118 (59.3) | 0 | 0 | 4.28 | 2.73–5.83 | 11 | 6–17 | 2.16 | 1.61–2.71 |
| CLICAL SCORE | n | Cumulative Hazard at 32 Months (95% CI) | Still Alive (at Risk) at 32 Months | CLICAL SIGNATURES | n (%) | Cumulative Hazard at 32 Months (95% CI) |
|---|---|---|---|---|---|---|
| 1.143 | 8 | 2.7 (0.29–5.14) # | 0 | Signature I | 46 (9.1) | 3.42 (1.87–4.96) |
| 1.286 | 38 | 3.23 (1.69–4.77) | 1 | |||
| 1.429 | 71 | 2.41 (1.59–3.24) | 2 | Signature II | 71 (14.1) | 2.41 (1.59–3.24) |
| 1.571 | 111 | 1.70 (1.24–2.16) | 10 | Signature III | 111 (22.1) | 1.70 (1.24–2.16) |
| 1.714 | 127 | 1.14 (0.86–1.43) | 10 | Signature IV | 243 (48.3) | 1.08 (0.89–1.27) |
| 1.857 | 116 | 1.00 (0.75–1.20) | 30 | |||
| 2.000 | 31 | 0.50 (0.19–0.81) | 12 | Signature V | 32 (6.4) | 0.48 (0.19–0.77) |
| 2.143 | 1 | 0.50(0.19–0.81) | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Madonna, G.; Masucci, G.V.; Capone, M.; Mallardo, D.; Grimaldi, A.M.; Simeone, E.; Vanella, V.; Festino, L.; Palla, M.; Scarpato, L.; et al. Clinical Categorization Algorithm (CLICAL) and Machine Learning Approach (SRF-CLICAL) to Predict Clinical Benefit to Immunotherapy in Metastatic Melanoma Patients: Real-World Evidence from the Istituto Nazionale Tumori IRCCS Fondazione Pascale, Napoli, Italy. Cancers 2021, 13, 4164. https://doi.org/10.3390/cancers13164164
Madonna G, Masucci GV, Capone M, Mallardo D, Grimaldi AM, Simeone E, Vanella V, Festino L, Palla M, Scarpato L, et al. Clinical Categorization Algorithm (CLICAL) and Machine Learning Approach (SRF-CLICAL) to Predict Clinical Benefit to Immunotherapy in Metastatic Melanoma Patients: Real-World Evidence from the Istituto Nazionale Tumori IRCCS Fondazione Pascale, Napoli, Italy. Cancers. 2021; 13(16):4164. https://doi.org/10.3390/cancers13164164
Chicago/Turabian StyleMadonna, Gabriele, Giuseppe V. Masucci, Mariaelena Capone, Domenico Mallardo, Antonio Maria Grimaldi, Ester Simeone, Vito Vanella, Lucia Festino, Marco Palla, Luigi Scarpato, and et al. 2021. "Clinical Categorization Algorithm (CLICAL) and Machine Learning Approach (SRF-CLICAL) to Predict Clinical Benefit to Immunotherapy in Metastatic Melanoma Patients: Real-World Evidence from the Istituto Nazionale Tumori IRCCS Fondazione Pascale, Napoli, Italy" Cancers 13, no. 16: 4164. https://doi.org/10.3390/cancers13164164
APA StyleMadonna, G., Masucci, G. V., Capone, M., Mallardo, D., Grimaldi, A. M., Simeone, E., Vanella, V., Festino, L., Palla, M., Scarpato, L., Tuffanelli, M., D'angelo, G., Villabona, L., Krakowski, I., Eriksson, H., Simao, F., Lewensohn, R., & Ascierto, P. A. (2021). Clinical Categorization Algorithm (CLICAL) and Machine Learning Approach (SRF-CLICAL) to Predict Clinical Benefit to Immunotherapy in Metastatic Melanoma Patients: Real-World Evidence from the Istituto Nazionale Tumori IRCCS Fondazione Pascale, Napoli, Italy. Cancers, 13(16), 4164. https://doi.org/10.3390/cancers13164164






