The Molecular Networks of microRNAs and Their Targets in the Drug Resistance of Colon Carcinoma
Abstract
:Simple Summary
Abstract
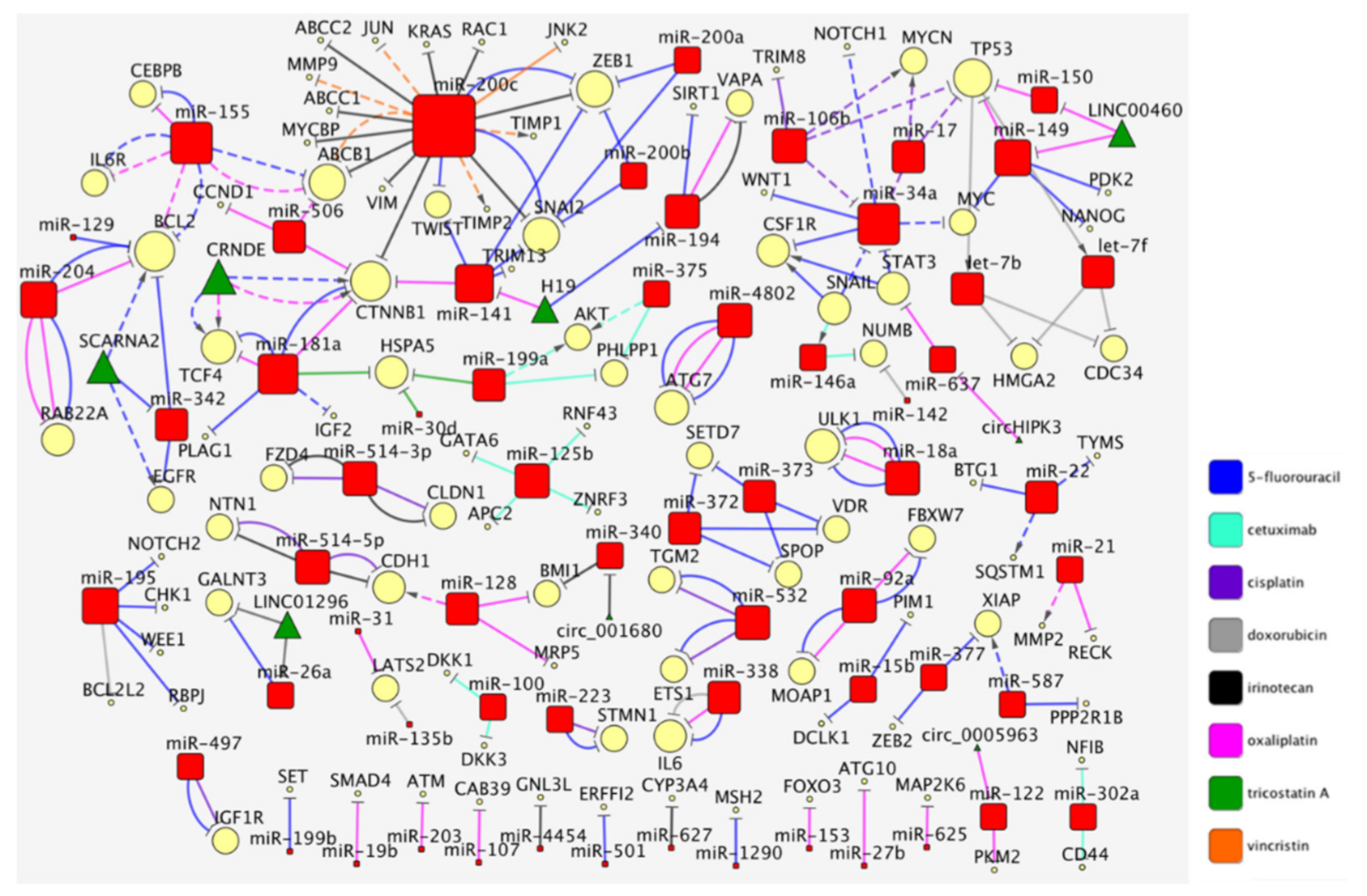
1. The Curated Networks of MiRNAs and Their Targets in Colon Cancer Drug Resistance
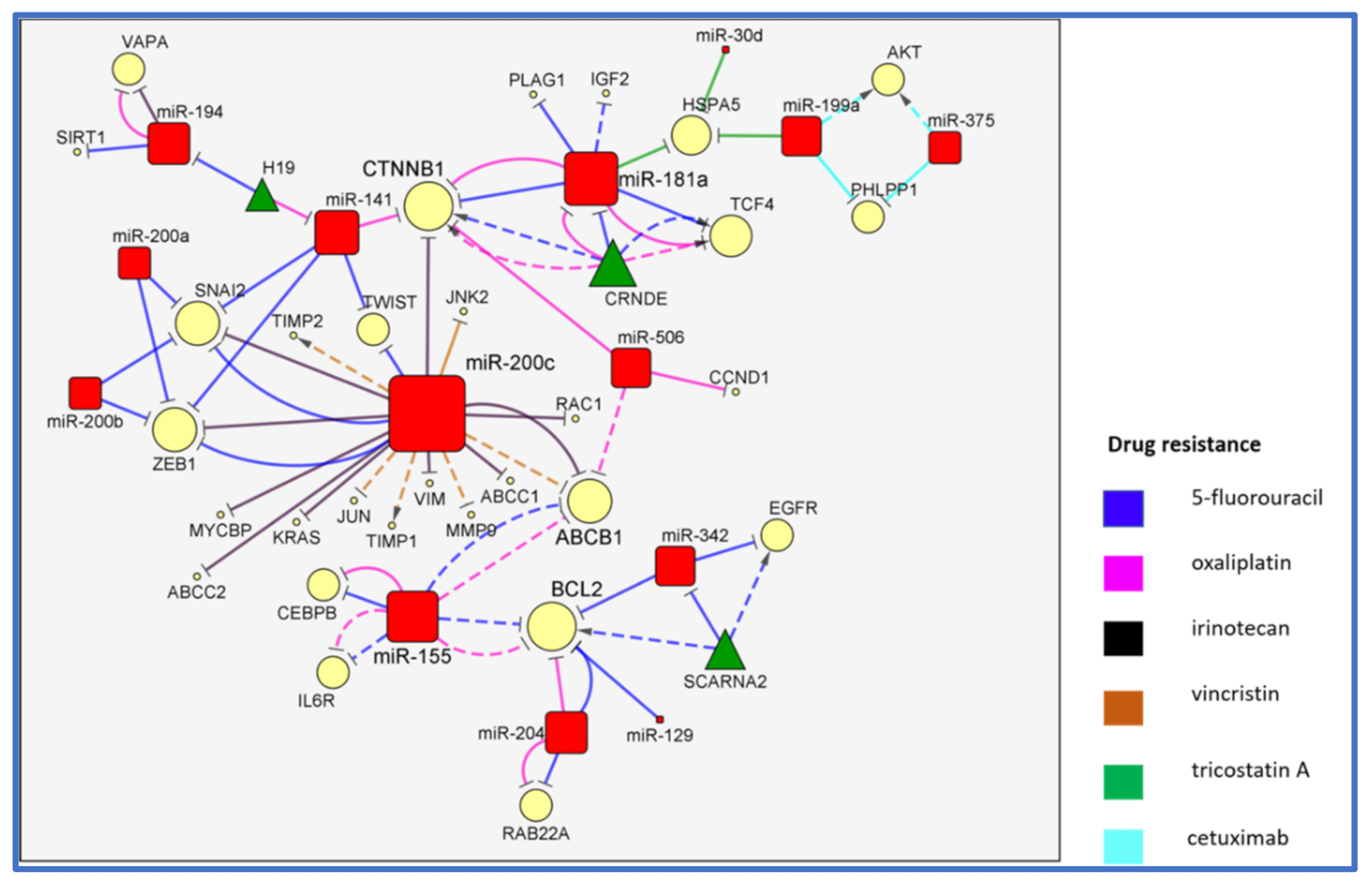
2. The MiR-200/MiR-181/MiR-155 CTNNB1 BCL2 Network
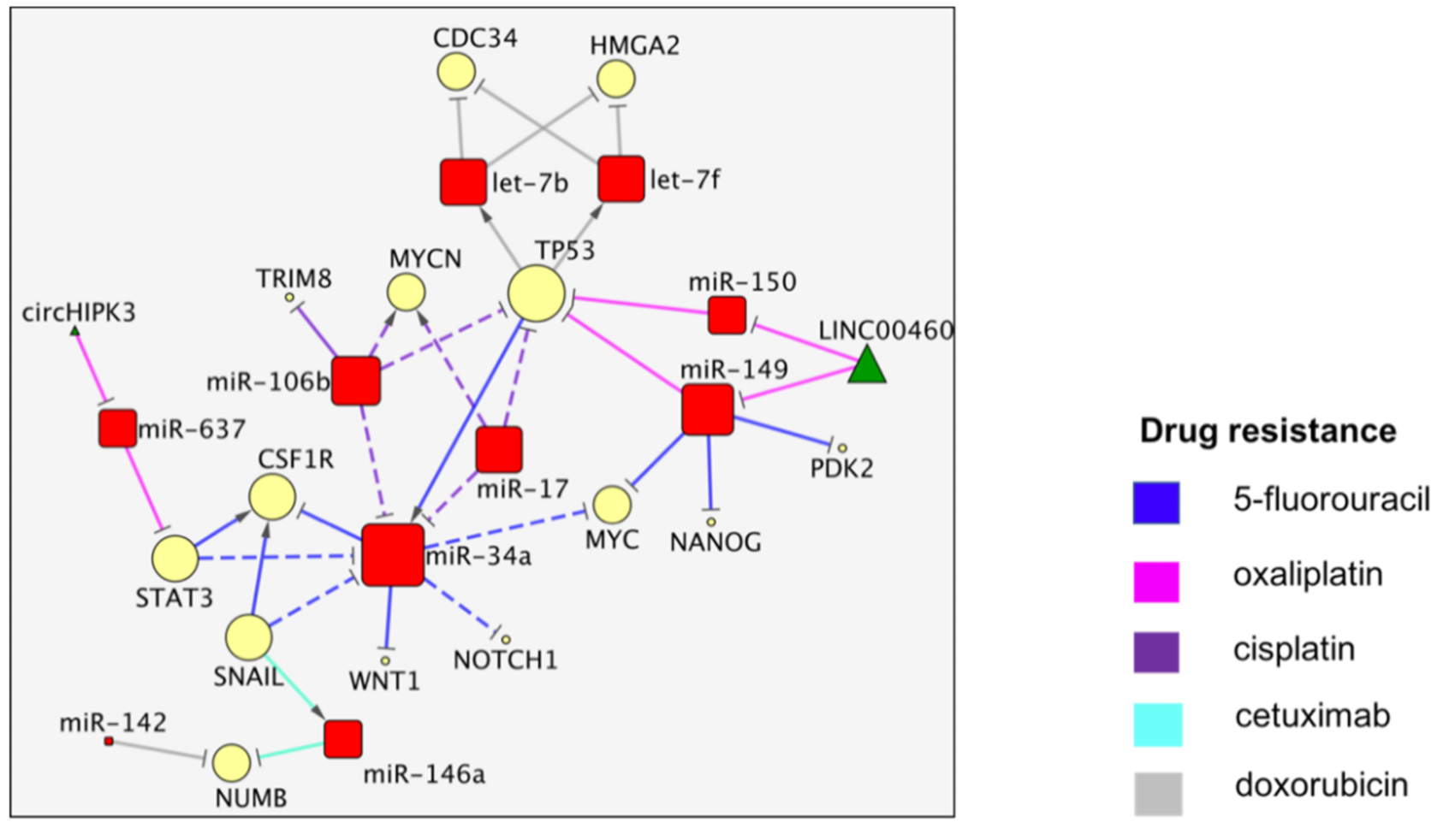
3. The TP53/miR-34a Network
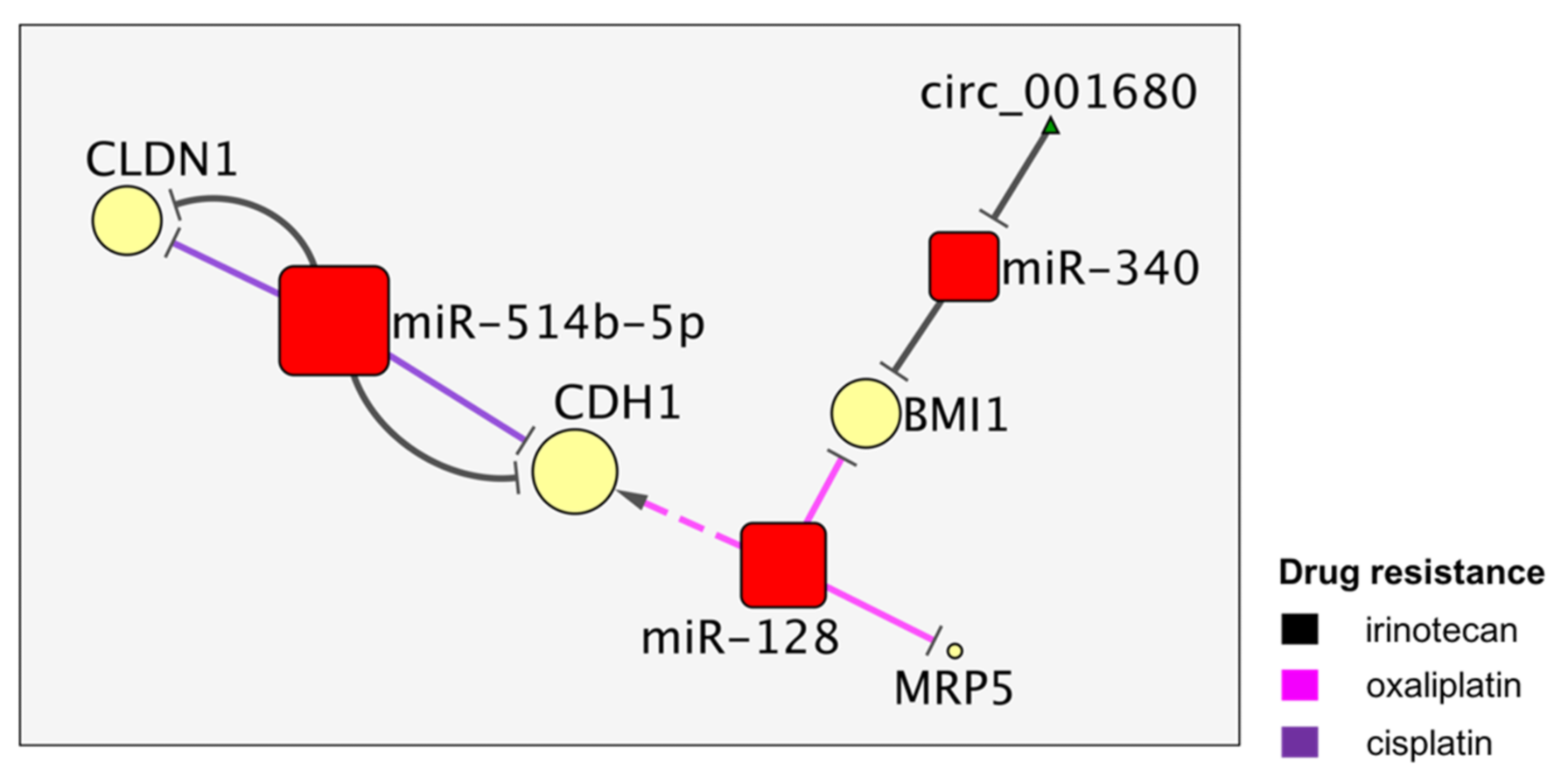
4. The MiR-514b and MiR128 Activities Converge on CDH1
5. Smaller MiRNA Networks Involved in CRC Drug Resistance
5.1. MiR-195
5.2. MiR-194
5.3. MiR-15b
6. Unconfirmed Associations of MiRNAs with Drug Resistance in CRC
6.1. 5-Fluorouracil Resistance
6.2. Irinotecan Resistance
6.3. Cetuximab Resistance
6.4. Doxorubicin Resistance
6.5. Oxaliplatin Resistance
6.6. 5-FU and Cisplatin Resistance (Multidrug)
6.7. 5-FU and L-OHP Resistance (Multidrug)
7. Drug-Centric Network and Clusters of MiRNA/Targets Interactions in CRC
8. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
References
- Corrà, F.; Agnoletto, C.; Minotti, L.; Baldassari, F.; Volinia, S. The Network of Non-coding RNAs in Cancer Drug Resistance. Front. Oncol. 2018, 8, 327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crudele, F.; Bianchi, N.; Reali, E.; Galasso, M.; Agnoletto, C.; Volinia, S. The network of non-coding RNAs and their molecular targets in breast cancer. Mol. Cancer 2020, 19, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sui, H.; Cai, G.-X.; Pan, S.-F.; Deng, W.-L.; Wang, Y.-W.; Chen, Z.-S.; Cai, S.-J.; Zhu, H.-R.; Li, Q. miR200c attenuates P-gp-mediated MDR and metastasis by targeting JNK2/c-Jun signaling pathway in colorectal cancer. Mol. Cancer Ther. 2014, 13, 3137–3151. [Google Scholar] [CrossRef] [Green Version]
- Zhou, H.; Lin, C.; Zhang, Y.; Zhang, X.; Zhang, C.; Zhang, P.; Xie, X.; Ren, Z. miR-506 enhances the sensitivity of human colorectal cancer cells to oxaliplatin by suppressing MDR1/P-gp expression. Cell Prolif. 2017, 50, e12341. [Google Scholar] [CrossRef] [PubMed]
- Juang, V.; Chang, C.-H.; Wang, C.-S.; Wang, H.-E.; Lo, Y.-L. pH-Responsive PEG-Shedding and Targeting Peptide-Modified Nanoparticles for Dual-Delivery of Irinotecan and microRNA to Enhance Tumor-Specific Therapy. Small 2019, 15, 1903296. [Google Scholar] [CrossRef]
- Senfter, D.; Holzner, S.; Kalipciyan, M.; Staribacher, A.; Walzl, A.; Huttary, N.; Krieger, S.; Brenner, S.; Jäger, W.; Krupitza, G.; et al. Loss of miR-200 family in 5-fluorouracil resistant colon cancer drives lymphendothelial invasiveness in vitro. Hum. Mol. Genet. 2015, 24, 3689–3698. [Google Scholar] [CrossRef] [Green Version]
- Moon, S.U.; Park, Y.; Park, M.G.; Song, S.K.; Jeong, S.H.; Lee, Y.S.; Heo, H.J.; Jung, W.Y.; Kim, S. Theragnosis by a miR-141-3p molecular beacon: Simultaneous detection and sensitization of 5-fluorouracil resistant colorectal cancer cells through the activation of the TRIM13-associated apoptotic pathway. Chem. Commun. 2019, 55, 7466–7469. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Ding, L.; Zhang, D.; Shi, G.; Xu, Q.; Shen, S.; Wang, Y.; Wang, T.; Hou, Y. Carcinoma-associated fibroblasts promote the stemness and chemoresistance of colorectal cancer by transferring exosomal lncRNA H19. Theranostics 2018, 8, 3932–3948. [Google Scholar] [CrossRef]
- Wang, M.; Han, D.; Yuan, Z.; Hu, H.; Zhao, Z.; Yang, R.; Jin, Y.; Zou, C.; Chen, Y.; Wang, G.; et al. Long non-coding RNA H19 confers 5-Fu resistance in colorectal cancer by promoting SIRT1-mediated autophagy. Cell Death Dis. 2018, 9, 1149. [Google Scholar] [CrossRef] [PubMed]
- Han, P.; Li, J.-W.; Zhang, B.-M.; Lv, J.-C.; Li, Y.-M.; Gu, X.-Y.; Yu, Z.-W.; Jia, Y.-H.; Bai, X.-F.; Li, L.; et al. The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/β-catenin signaling. Mol. Cancer 2017, 16, 9. [Google Scholar] [CrossRef] [Green Version]
- Shi, L.; Li, X.; Wu, Z.; Li, X.; Nie, J.; Guo, M.; Mei, Q.; Han, W. DNA methylation-mediated repression of miR-181a/135a/302c expression promotes the microsatellite-unstable colorectal cancer development and 5-FU resistance via targeting PLAG1. J. Genet. Genom. 2018, 45, 205–214. [Google Scholar] [CrossRef]
- Su, S.-F.; Chang, Y.-W.; Andreu-Vieyra, C.; Fang, J.Y.; Yang, Z.; Han, B.; Lee, A.S.; Liang, G. miR-30d, miR-181a and miR-199a-5p cooperatively suppress the endoplasmic reticulum chaperone and signaling regulator GRP78 in cancer. Oncogene 2013, 32, 4694–4701. [Google Scholar] [CrossRef] [Green Version]
- Mussnich, P.; Rosa, R.; Bianco, R.; Fusco, A.; D’Angelo, D. MiR-199a-5p and miR-375 affect colon cancer cell sensitivity to cetuximab by targeting PHLPP1. Expert Opin. Ther. Targets 2015, 19, 1017–1026. [Google Scholar] [CrossRef] [Green Version]
- Yin, Y.; Yao, S.; Hu, Y.; Feng, Y.; Li, M.; Bian, Z.; Zhang, J.; Qin, Y.; Qi, X.; Zhou, L.; et al. The Immune-microenvironment Confers Chemoresistance of Colorectal Cancer through Macrophage-Derived IL6. Clin. Cancer Res. 2017, 23, 7375–7387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, Y.; Zhang, B.; Wang, W.; Fei, B.; Quan, C.; Zhang, J.; Song, M.; Bian, Z.; Wang, Q.; Ni, S.; et al. miR-204-5p inhibits proliferation and invasion and enhances chemotherapeutic sensitivity of colorectal cancer cells by downregulating RAB22A. Clin. Cancer Res. 2014, 20, 6187–6199. [Google Scholar] [CrossRef] [Green Version]
- Karaayvaz, M.; Zhai, H.; Ju, J. miR-129 promotes apoptosis and enhances chemosensitivity to 5-fluorouracil in colorectal cancer. Cell Death Dis 2013, 4, e659. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.-F.; Wu, J.; Wu, Y.; Huang, W.; Liu, M.; Dong, Z.-R.; Xu, B.-Y.; Jin, Y.; Wang, F.; Zhang, X.-M. The lncRNA SCARNA2 mediates colorectal cancer chemoresistance through a conserved microRNA-342-3p target sequence. J. Cell Physiol. 2019, 234, 10157–10165. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Muruganujan, A.; Huang, X.; Ebert, D.; Mills, C.; Guo, X.; Thomas, P.D. Protocol Update for Large-Scale Genome and Gene Function Analysis with PANTHER Classification System (v.14.0). Nat. Protoc. 2019, 14, 703–721. [Google Scholar] [CrossRef]
- Dufresne, M.; Seva, C.; Fourmy, D. Cholecystokinin and Gastrin Receptors. Physiol. Rev. 2006, 86, 805–847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, X.; Kaller, M.; Rokavec, M.; Kirchner, T.; Horst, D.; Hermeking, H. Characterization of a p53/miR-34a/CSF1R/STAT3 Feedback Loop in Colorectal Cancer. Cell Mol. Gastroenterol. Hepatol. 2020, 10, 391–418. [Google Scholar] [CrossRef]
- Cai, M.-H.; Xu, X.-G.; Yan, S.-L.; Sun, Z.; Ying, Y.; Wang, B.-K.; Tu, Y.-X. Regorafenib suppresses colon tumorigenesis and the generation of drug resistant cancer stem-like cells via modulation of miR-34a associated signaling. J. Exp. Clin. Cancer Res. 2018, 37, 151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mastropasqua, F.; Marzano, F.; Valletti, A.; Aiello, I.; Di Tullio, G.; Morgano, A.; Liuni, S.; Ranieri, E.; Guerrini, L.; Gasparre, G.; et al. TRIM8 restores p53 tumour suppressor function by blunting N-MYC activity in chemo-resistant tumours. Mol. Cancer 2017, 16, 67. [Google Scholar] [CrossRef] [Green Version]
- Findlay, V.J.; Wang, C.; Nogueira, L.M.; Hurst, K.; Quirk, D.; Ethier, S.P.; Staveley O’Carroll, K.F.; Watson, D.K.; Camp, E.R. SNAI2 modulates colorectal cancer 5-fluorouracil sensitivity through miR145 repression. Mol. Cancer Ther. 2014, 13, 2713–2726. [Google Scholar] [CrossRef] [Green Version]
- Liang, Y.; Hou, L.; Li, L.; Li, L.; Zhu, L.; Wang, Y.; Huang, X.; Hou, Y.; Zhu, D.; Zou, H.; et al. Dichloroacetate restores colorectal cancer chemosensitivity through the p53/miR-149-3p/PDK2-mediated glucose metabolic pathway. Oncogene 2020, 39, 469–485. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Sun, W.; Yu, J.; Zhou, Y.; Gu, Y.; Han, J.; Zhou, L.; Jiang, X.; Wang, C. LINC00460-miR-149-5p/miR-150-5p-Mutant p53 Feedback Loop Promotes Oxaliplatin Resistance in Colorectal Cancer. Mol. Ther. Nucleic Acids 2020, 22, 1004–1015. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Kim, H.J.; Yoon, N.A.; Lee, W.H.; Min, Y.J.; Ko, B.K.; Lee, B.J.; Lee, A.; Cha, H.J.; Cho, W.J.; et al. Tumor suppressor p53 plays a key role in induction of both tristetraprolin and let-7 in human cancer cells. Nucleic Acids Res. 2013, 41, 5614–5625. [Google Scholar] [CrossRef] [Green Version]
- Hwang, W.-L.; Jiang, J.-K.; Yang, S.-H.; Huang, T.-S.; Lan, H.-Y.; Teng, H.-W.; Yang, C.-Y.; Tsai, Y.-P.; Lin, C.-H.; Wang, H.-W.; et al. MicroRNA-146a directs the symmetric division of Snail-dominant colorectal cancer stem cells. Nat. Cell Biol. 2014, 16, 268–280. [Google Scholar] [CrossRef]
- Li, H.; Li, F. Exosomes from BM-MSCs increase the population of CSCs via transfer of miR-142-3p. Br. J. Cancer 2018, 119, 744–755. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, C.; Liu, X.; Wang, Y.; Zhao, R.; Yang, Y.; Zheng, X.; Zhang, Y.; Zhang, X. circHIPK3 promotes oxaliplatin-resistance in colorectal cancer through autophagy by sponging miR-637. EBioMedicine 2019, 48, 277–288. [Google Scholar] [CrossRef] [Green Version]
- Ren, L.-L.; Yan, T.-T.; Shen, C.-Q.; Tang, J.-Y.; Kong, X.; Wang, Y.-C.; Chen, J.; Liu, Q.; He, J.; Zhong, M.; et al. The distinct role of strand-specific miR-514b-3p and miR-514b-5p in colorectal cancer metastasis. Cell Death Dis. 2018, 9, 687. [Google Scholar] [CrossRef] [Green Version]
- Liu, T.; Zhang, X.; Du, L.; Wang, Y.; Liu, X.; Tian, H.; Wang, L.; Li, P.; Zhao, Y.; Duan, W.; et al. Exosome-transmitted miR-128-3p increase chemosensitivity of oxaliplatin-resistant colorectal cancer. Mol. Cancer 2019, 18, 43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jian, X.; He, H.; Zhu, J.; Zhang, Q.; Zheng, Z.; Liang, X.; Chen, L.; Yang, M.; Peng, K.; Zhang, Z.; et al. Hsa_circ_001680 affects the proliferation and migration of CRC and mediates its chemoresistance by regulating BMI1 through miR-340. Mol. Cancer 2020, 19, 20. [Google Scholar] [CrossRef]
- Kim, C.; Hong, Y.; Lee, H.; Kang, H.; Lee, E.K. MicroRNA-195 desensitizes HCT116 human colon cancer cells to 5-fluorouracil. Cancer Lett. 2018, 412, 264–271. [Google Scholar] [CrossRef]
- Jin, Y.; Wang, M.; Hu, H.; Huang, Q.; Chen, Y.; Wang, G. Overcoming stemness and chemoresistance in colorectal cancer through miR-195-5p-modulated inhibition of notch signaling. Int. J. Biol. Macromol. 2018, 117, 445–453. [Google Scholar] [CrossRef] [PubMed]
- Qu, J.; Zhao, L.; Zhang, P.; Wang, J.; Xu, N.; Mi, W.; Jiang, X.; Zhang, C.; Qu, J. MicroRNA-195 chemosensitizes colon cancer cells to the chemotherapeutic drug doxorubicin by targeting the first binding site of BCL2L2 mRNA. J. Cell Physiol. 2015, 230, 535–545. [Google Scholar] [CrossRef]
- Chang, H.-Y.; Ye, S.-P.; Pan, S.-L.; Kuo, T.-T.; Liu, B.C.; Chen, Y.-L.; Huang, T.-C. Overexpression of miR-194 Reverses HMGA2-driven Signatures in Colorectal Cancer. Theranostics 2017, 7, 3889–3900. [Google Scholar] [CrossRef]
- Weirauch, U.; Beckmann, N.; Thomas, M.; Grünweller, A.; Huber, K.; Bracher, F.; Hartmann, R.K.; Aigner, A. Functional role and therapeutic potential of the pim-1 kinase in colon carcinoma. Neoplasia 2013, 15, 783–794. [Google Scholar] [CrossRef] [Green Version]
- Ji, D.; Zhan, T.; Li, M.; Yao, Y.; Jia, J.; Yi, H.; Qiao, M.; Xia, J.; Zhang, Z.; Ding, H.; et al. Enhancement of Sensitivity to Chemo/Radiation Therapy by Using miR-15b against DCLK1 in Colorectal Cancer. Stem Cell Rep. 2018, 11, 1506–1522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.-Q.; Yu, P.; Li, B.; Guo, Y.-H.; Liang, Z.-R.; Zheng, L.-L.; Yang, J.-H.; Xu, H.; Liu, S.; Zheng, L.-S.; et al. miR-372 and miR-373 enhance the stemness of colorectal cancer cells by repressing differentiation signaling pathways. Mol. Oncol. 2018, 12, 1949–1964. [Google Scholar] [CrossRef]
- Huang, L.; Liu, Z.; Hu, J.; Luo, Z.; Zhang, C.; Wang, L.; Wang, Z. MiR-377-3p suppresses colorectal cancer through negative regulation on Wnt/β-catenin signaling by targeting XIAP and ZEB2. Pharmacol. Res. 2020, 156, 104774. [Google Scholar] [CrossRef]
- Zhang, Y.; Talmon, G.; Wang, J. MicroRNA-587 antagonizes 5-FU-induced apoptosis and confers drug resistance by regulating PPP2R1B expression in colorectal cancer. Cell Death Dis. 2015, 6, e1845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, R.; Cai, W.; Sun, J.; Yu, C.; Li, P.; Zheng, M. Inhibition of KHSRP sensitizes colorectal cancer to 5-fluoruracil through miR-501-5p-mediated ERRFI1 mRNA degradation. J. Cell Physiol. 2020, 235, 1576–1587. [Google Scholar] [CrossRef] [PubMed]
- Cristóbal, I.; Rubio, J.; Santos, A.; Torrejón, B.; Caramés, C.; Imedio, L.; Mariblanca, S.; Luque, M.; Sanz-Alvarez, M.; Zazo, S.; et al. MicroRNA-199b Downregulation Confers Resistance to 5-Fluorouracil Treatment and Predicts Poor Outcome and Response to Neoadjuvant Chemoradiotherapy in Locally Advanced Rectal Cancer Patients. Cancers 2020, 12, 1655. [Google Scholar] [CrossRef]
- Ye, L.; Jiang, T.; Shao, H.; Zhong, L.; Wang, Z.; Liu, Y.; Tang, H.; Qin, B.; Zhang, X.; Fan, J. miR-1290 Is a Biomarker in DNA-Mismatch-Repair-Deficient Colon Cancer and Promotes Resistance to 5-Fluorouracil by Directly Targeting hMSH2. Mol. Ther. Nucleic Acids 2017, 7, 453–464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, B.; Pan, S.; Xiao, Y.; Liu, Q.; Xu, J.; Jia, L. LINC01296/miR-26a/GALNT3 axis contributes to colorectal cancer progression by regulating O-glycosylated MUC1 via PI3K/AKT pathway. J. Exp. Clin. Cancer Res. 2018, 37, 316. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Tang, J.; Li, C.; Kong, J.; Wang, J.; Wu, Y.; Xu, E.; Lai, M. MiR-22 regulates 5-FU sensitivity by inhibiting autophagy and promoting apoptosis in colorectal cancer cells. Cancer Lett. 2015, 356, 781–790. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Zhang, Q.; Yang, X.; Qian, S.Y.; Guo, B. Vitamin D Enhances the Efficacy of Irinotecan through miR-627-Mediated Inhibition of Intratumoral Drug Metabolism. Mol. Cancer Ther. 2016, 15, 2086–2095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kannathasan, T.; Kuo, W.-W.; Chen, M.-C.; Viswanadha, V.P.; Shen, C.-Y.; Tu, C.-C.; Yeh, Y.-L.; Bharath, M.; Shibu, M.A.; Huang, C.-Y. Chemoresistance-Associated Silencing of miR-4454 Promotes Colorectal Cancer Aggression through the GNL3L and NF-κB Pathway. Cancers 2020, 12, 1231. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zhao, X.; Liu, Q.; Li, C.; Graves-Deal, R.; Cao, Z.; Singh, B.; Franklin, J.L.; Wang, J.; Hu, H.; et al. lncRNA MIR100HG-derived miR-100 and miR-125b mediate cetuximab resistance via Wnt/β-catenin signaling. Nat. Med. 2017, 23, 1331–1341. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Fang, Y.; Wang, X.; Han, Y.; Du, F.; Li, C.; Hu, H.; Liu, H.; Liu, Q.; Wang, J.; et al. miR-302a Inhibits Metastasis and Cetuximab Resistance in Colorectal Cancer by Targeting NFIB and CD44. Theranostics 2019, 9, 8409–8425. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Wang, J.; Wang, J.; Yung, V.Y.-W.; Hsu, E.; Li, A.; Kang, Q.; Ma, J.; Han, Q.; Jin, P.; et al. MicroRNA-135b regulates apoptosis and chemoresistance in colorectal cancer by targeting large tumor suppressor kinase 2. Am. J. Cancer Res. 2015, 5, 1382–1395. [Google Scholar] [PubMed]
- Hsu, H.-H.; Kuo, W.-W.; Shih, H.-N.; Cheng, S.-F.; Yang, C.-K.; Chen, M.-C.; Tu, C.-C.; Viswanadha, V.P.; Liao, P.-H.; Huang, C.-Y. FOXC1 Regulation of miR-31-5p Confers Oxaliplatin Resistance by Targeting LATS2 in Colorectal Cancer. Cancers 2019, 11, 1576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, Y.; Zhu, D.; Hou, L.; Wang, Y.; Huang, X.; Zhou, C.; Zhu, L.; Wang, Y.; Li, L.; Gu, Y.; et al. MiR-107 confers chemoresistance to colorectal cancer by targeting calcium-binding protein 39. Br. J. Cancer 2020, 122, 705–714. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Pickard, K.; Jenei, V.; Bullock, M.D.; Bruce, A.; Mitter, R.; Kelly, G.; Paraskeva, C.; Strefford, J.; Primrose, J.; et al. miR-153 supports colorectal cancer progression via pleiotropic effects that enhance invasion and chemotherapeutic resistance. Cancer Res. 2013, 73, 6435–6447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, T.; Ye, L.; Han, Z.; Liu, Y.; Yang, Y.; Peng, Z.; Fan, J. miR-19b-3p promotes colon cancer proliferation and oxaliplatin-based chemoresistance by targeting SMAD4: Validation by bioinformatics and experimental analyses. J. Exp. Clin. Cancer Res. 2017, 36, 131. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Wan, G.; Spizzo, R.; Ivan, C.; Mathur, R.; Hu, X.; Ye, X.; Lu, J.; Fan, F.; Xia, L.; et al. miR-203 induces oxaliplatin resistance in colorectal cancer cells by negatively regulating ATM kinase. Mol. Oncol. 2014, 8, 83–92. [Google Scholar] [CrossRef]
- Bullock, M.D.; Pickard, K.M.; Nielsen, B.S.; Sayan, A.E.; Jenei, V.; Mellone, M.; Mitter, R.; Primrose, J.N.; Thomas, G.J.; Packham, G.K.; et al. Pleiotropic actions of miR-21 highlight the critical role of deregulated stromal microRNAs during colorectal cancer progression. Cell Death Dis. 2013, 4, e684. [Google Scholar] [CrossRef] [Green Version]
- Sun, W.; Li, J.; Zhou, L.; Han, J.; Liu, R.; Zhang, H.; Ning, T.; Gao, Z.; Liu, B.; Chen, X.; et al. The c-Myc/miR-27b-3p/ATG10 regulatory axis regulates chemoresistance in colorectal cancer. Theranostics 2020, 10, 1981–1996. [Google Scholar] [CrossRef]
- Rasmussen, M.H.; Lyskjær, I.; Jersie-Christensen, R.R.; Tarpgaard, L.S.; Primdal-Bengtson, B.; Nielsen, M.M.; Pedersen, J.S.; Hansen, T.P.; Hansen, F.; Olsen, J.V.; et al. miR-625-3p regulates oxaliplatin resistance by targeting MAP2K6-p38 signalling in human colorectal adenocarcinoma cells. Nat. Commun. 2016, 7, 12436. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Zhang, H.; Yang, H.; Bai, M.; Ning, T.; Deng, T.; Liu, R.; Fan, Q.; Zhu, K.; Li, J.; et al. Exosome-delivered circRNA promotes glycolysis to induce chemoresistance through the miR-122-PKM2 axis in colorectal cancer. Mol. Oncol. 2020, 14, 539–555. [Google Scholar] [CrossRef]
- Masciarelli, S.; Fontemaggi, G.; Di Agostino, S.; Donzelli, S.; Carcarino, E.; Strano, S.; Blandino, G. Gain-of-function mutant p53 downregulates miR-223 contributing to chemoresistance of cultured tumor cells. Oncogene 2014, 33, 1601–1608. [Google Scholar] [CrossRef] [Green Version]
- Guo, S.T.; Jiang, C.C.; Wang, G.P.; Li, Y.P.; Wang, C.Y.; Guo, X.Y.; Yang, R.H.; Feng, Y.; Wang, F.H.; Tseng, H.-Y.; et al. MicroRNA-497 targets insulin-like growth factor 1 receptor and has a tumour suppressive role in human colorectal cancer. Oncogene 2013, 32, 1910–1920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, C.; Cai, J.; Xu, Z.; Zhou, S.; Ye, L.; Yan, Q.; Zhang, Y.; Fang, Y.; Liu, Y.; Tu, C.; et al. MiR-532-3p suppresses colorectal cancer progression by disrupting the ETS1/TGM2 axis-mediated Wnt/β-catenin signaling. Cell Death Dis. 2019, 10, 739. [Google Scholar] [CrossRef]
- Yu, T.; Guo, F.; Yu, Y.; Sun, T.; Ma, D.; Han, J.; Qian, Y.; Kryczek, I.; Sun, D.; Nagarsheth, N.; et al. Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell 2017, 170, 548–563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, J.L.; Wang, W.; Lan, X.L.; Zeng, Z.C.; Liang, Y.S.; Yan, Y.R.; Song, F.Y.; Wang, F.F.; Zhu, X.H.; Liao, W.J.; et al. CAFs secreted exosomes promote metastasis and chemotherapy resistance by enhancing cell stemness and epithelial-mesenchymal transition in colorectal cancer. Mol. Cancer 2019, 18, 91. [Google Scholar] [CrossRef] [Green Version]
- Choe, M.H.; Yoon, Y.; Kim, J.; Hwang, S.-G.; Han, Y.-H.; Kim, J.-S. miR-550a-3-5p acts as a tumor suppressor and reverses BRAF inhibitor resistance through the direct targeting of YAP. Cell Death Dis. 2018, 9, 640. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Liu, Y.; Zhang, Q.; Liu, B.; Cheng, Y.; Zhang, Y.; Sun, Y.; Liu, J.; Gen, H. Exosomal Long Non-coding RNA HOTTIP Increases Resistance of Colorectal Cancer Cells to Mitomycin via Impairing MiR-214-Mediated Degradation of KPNA3. Front. Cell Dev. Biol. 2020, 8, 582723. [Google Scholar] [CrossRef]
- Li, P.; Zhang, X.; Wang, H.; Wang, L.; Liu, T.; Du, L.; Yang, Y.; Wang, C. MALAT1 Is Associated with Poor Response to Oxaliplatin-Based Chemotherapy in Colorectal Cancer Patients and Promotes Chemoresistance through EZH2. Mol. Cancer Ther. 2017, 16, 739–751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rokavec, M.; Bouznad, N.; Hermeking, H. Paracrine Induction of Epithelial-Mesenchymal Transition Between Colorectal Cancer Cells and its Suppression by a p53/miR-192/215/NID1 Axis. Cell Mol. Gastroenterol. Hepatol. 2019, 7, 783–802. [Google Scholar] [CrossRef] [Green Version]
- Xu, K.; Zhan, Y.; Yuan, Z.; Qiu, Y.; Wang, H.; Fan, G.; Wang, J.; Li, W.; Cao, Y.; Shen, X.; et al. Hypoxia Induces Drug Resistance in Colorectal Cancer through the HIF-1α/miR-338-5p/IL-6 Feedback Loop. Mol. Ther. 2019, 27, 1810–1824. [Google Scholar] [CrossRef]
- Chen, S.; Bu, D.; Ma, Y.; Zhu, J.; Chen, G.; Sun, L.; Zuo, S.; Li, T.; Pan, Y.; Wang, X.; et al. H19 Overexpression Induces Resistance to 1,25(OH)2D3 by Targeting VDR Through miR-675-5p in Colon Cancer Cells. Neoplasia 2017, 19, 226–236. [Google Scholar] [CrossRef]
- Bamodu, O.A.; Yang, C.-K.; Cheng, W.-H.; Tzeng, D.T.W.; Kuo, K.-T.; Huang, C.-C.; Deng, L.; Hsiao, M.; Lee, W.-H.; Yeh, C.-T. 4-Acetyl-Antroquinonol B Suppresses SOD2-Enhanced Cancer Stem Cell-Like Phenotypes and Chemoresistance of Colorectal Cancer Cells by Inducing hsa-miR-324 re-Expression. Cancers 2018, 10, 269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Y.; Nangia-Makker, P.; Farhana, L.; Rajendra, S.G.; Levi, E.; Majumdar, A.P.N. miR-21 and miR-145 cooperation in regulation of colon cancer stem cells. Mol. Cancer 2015, 14, 98. [Google Scholar] [CrossRef] [Green Version]
- Hartsock, A.; Nelson, W.J. Adherens and Tight Junctions: Structure, Function and Connections to the Actin Cytoskeleton. Available online: https://pubmed.ncbi.nlm.nih.gov/17854762 (accessed on 7 July 2021).
- Arnold, A.; Tronser, M.; Sers, C.; Ahadova, A.; Endris, V.; Mamlouk, S.; Horst, D.; Möbs, M.; Bischoff, P.; Kloor, M.; et al. The Majority of β-Catenin Mutations in Colorectal Cancer Is Homozygous. BMC Cancer 2020, 20, 1038. [Google Scholar] [CrossRef] [PubMed]
- Okada, N.; Lin, C.-P.; Ribeiro, M.C.; Biton, A.; Lai, G.; He, X.; Bu, P.; Vogel, H.; Jablons, D.M.; Keller, A.C.; et al. A Positive Feedback between p53 and miR-34 miRNAs Mediates Tumor Suppression. Genes Dev. 2014, 28, 438–450. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| PMID | miRNA | Target | Drug Name | Ref. |
|---|---|---|---|---|
| 29844307 | miR-550a | YAP1 | vemurafenib | [66] |
| 28327152 | miR-106b, miR-17 | miR-34a, MYCN, TP53, TRIM8 | sorafenib, nutlin-3, axitinib | [22] |
| 33585440 | miR-214 | KPNA3 | mitomycin | [67] |
| 28069878 | miR-218 | MALAT1 | FOLFOX | [68] |
| 30831320 | miR-192, miR-215 | NID1 | doxicyclin | [69] |
| 31208913 | miR-338 | IL6 | cyclophosphamide | [70] |
| 28189050 | miR-675 | VDR | calcitriol | [71] |
| 30103475 | miR-324 | SOD2 | 4-acetylantroquinonol B | [72] |
| 25928322 | miR-145, miR-21 | NUMB, CD44, KRT20, SOX2 | 5-FU and L-OHP mix | [73] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Crudele, F.; Bianchi, N.; Astolfi, A.; Grassilli, S.; Brugnoli, F.; Terrazzan, A.; Bertagnolo, V.; Negrini, M.; Frassoldati, A.; Volinia, S. The Molecular Networks of microRNAs and Their Targets in the Drug Resistance of Colon Carcinoma. Cancers 2021, 13, 4355. https://doi.org/10.3390/cancers13174355
Crudele F, Bianchi N, Astolfi A, Grassilli S, Brugnoli F, Terrazzan A, Bertagnolo V, Negrini M, Frassoldati A, Volinia S. The Molecular Networks of microRNAs and Their Targets in the Drug Resistance of Colon Carcinoma. Cancers. 2021; 13(17):4355. https://doi.org/10.3390/cancers13174355
Chicago/Turabian StyleCrudele, Francesca, Nicoletta Bianchi, Annalisa Astolfi, Silvia Grassilli, Federica Brugnoli, Anna Terrazzan, Valeria Bertagnolo, Massimo Negrini, Antonio Frassoldati, and Stefano Volinia. 2021. "The Molecular Networks of microRNAs and Their Targets in the Drug Resistance of Colon Carcinoma" Cancers 13, no. 17: 4355. https://doi.org/10.3390/cancers13174355
APA StyleCrudele, F., Bianchi, N., Astolfi, A., Grassilli, S., Brugnoli, F., Terrazzan, A., Bertagnolo, V., Negrini, M., Frassoldati, A., & Volinia, S. (2021). The Molecular Networks of microRNAs and Their Targets in the Drug Resistance of Colon Carcinoma. Cancers, 13(17), 4355. https://doi.org/10.3390/cancers13174355








