TCR Clonality and Genomic Instability Signatures as Prognostic Biomarkers in High Grade Serous Ovarian Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.1.1. TCGA Data
2.1.2. CGFL Data
2.2. Methodology for the CGFL Data
2.2.1. Sample Selection
2.2.2. DNA Isolation
2.2.3. Whole Exome Capture and Sequencing
2.2.4. Exome Analysis Pipeline
2.3. Statistical Analysis
3. Results
3.1. Patient Characteristics
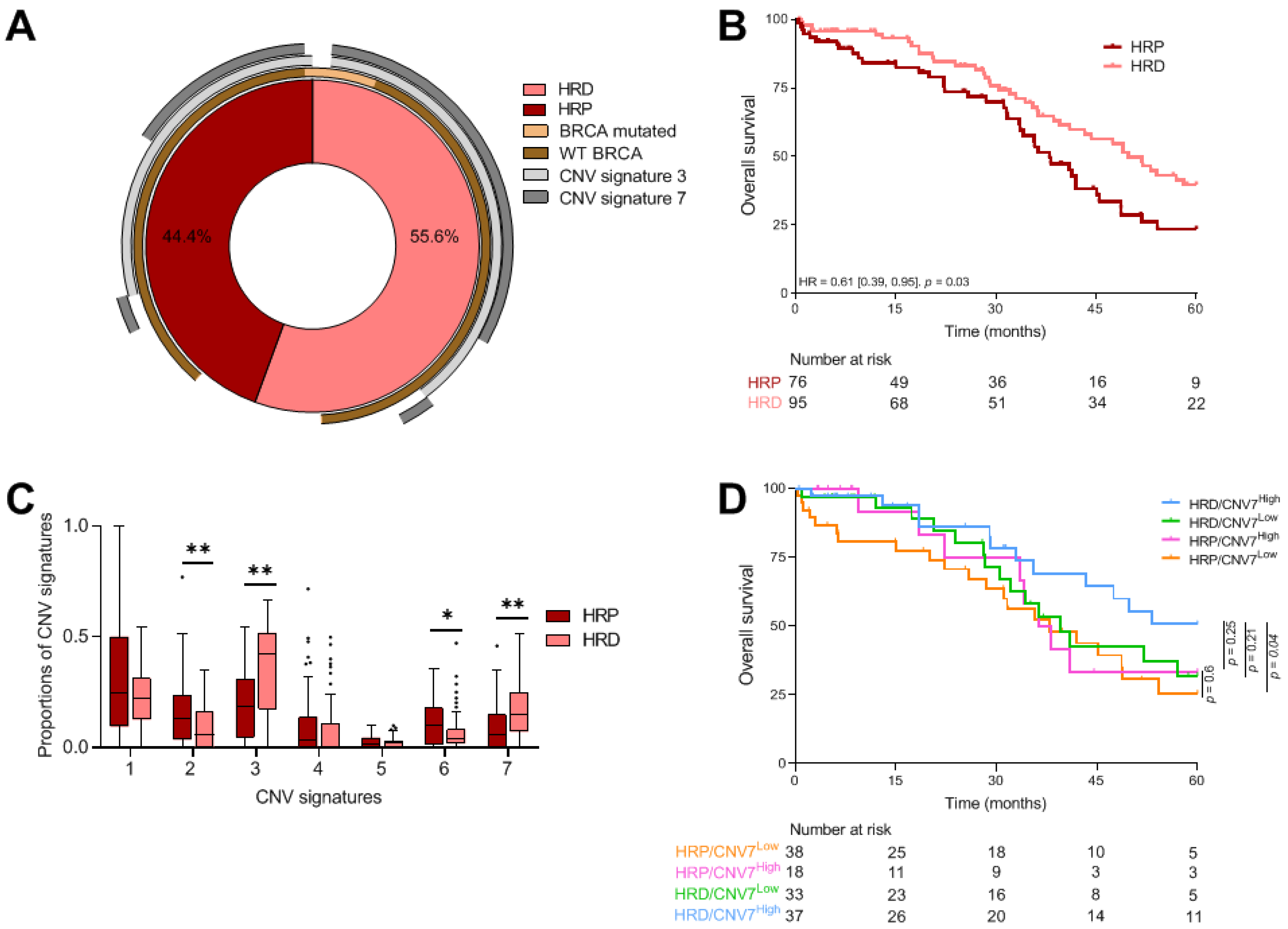
3.2. Prognostic Value of Genomic Instability Signatures
3.2.1. HRD Profile
3.2.2. CNV Signatures
3.2.3. Combined Analysis of HRD Status and CNV Signatures
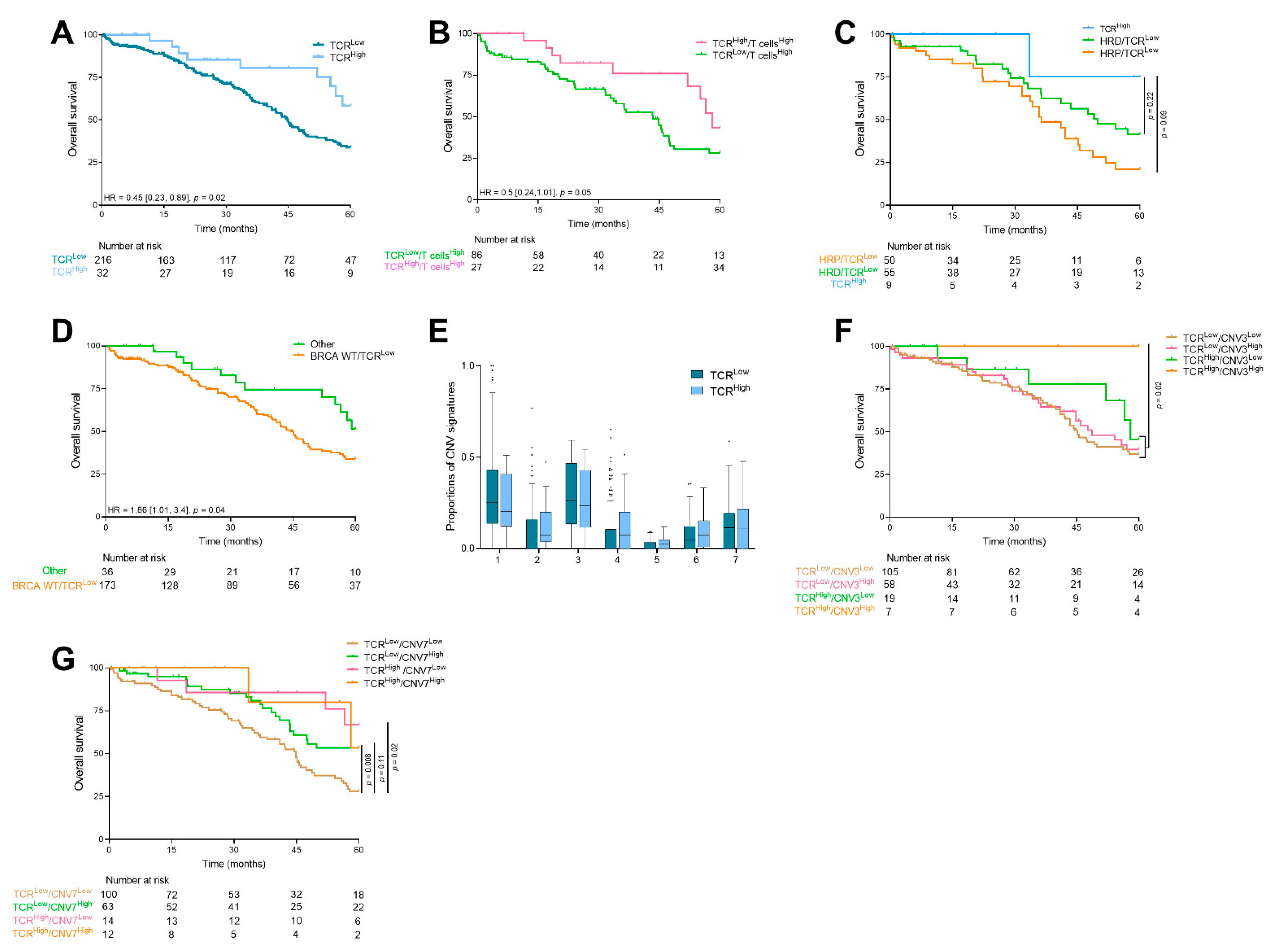
3.3. Prognostic Value and Relationship between Genomic Instability Signatures and Immune Population
3.4. TCR Clonality
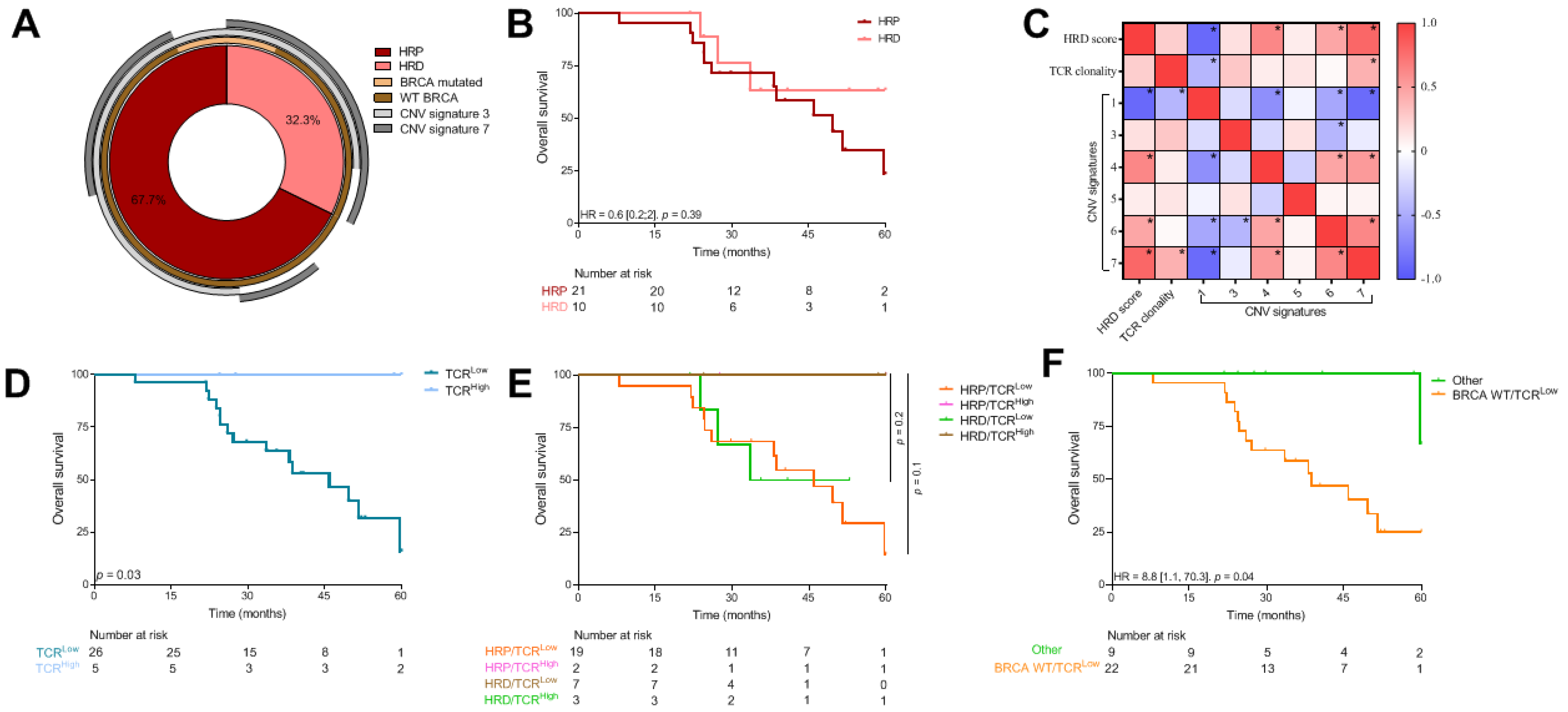
3.5. Same Analysis in an Independent Cohort
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Torre, L.A.; Trabert, B.; DeSantis, C.E.; Miller, K.D.; Samimi, G.; Runowicz, C.D.; Gaudet, M.M.; Jemal, A.; Siegel, R.L. Ovarian cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 284–296. [Google Scholar] [CrossRef]
- Lisio, M.-A.; Fu, L.; Goyeneche, A.; Gao, Z.; Telleria, C. High-Grade Serous Ovarian Cancer: Basic Sciences, Clinical and Therapeutic Standpoints. Int. J. Mol. Sci. 2019, 20, 952. [Google Scholar] [CrossRef] [Green Version]
- Miller, R.E.; Leary, A.; Scott, C.L.; Serra, V.; Lord, C.J.; Bowtell, D.; Chang, D.K.; Garsed, D.W.; Jonkers, J.; Ledermann, J.A.; et al. ESMO recommendations on predictive biomarker testing for homologous recombination deficiency and PARP inhibitor benefit in ovarian cancer. Ann. Oncol. 2020, 31, 1606–1622. [Google Scholar] [CrossRef]
- Ray-Coquard, I.; Pautier, P.; Pignata, S.; Pérol, D.; González-Martín, A.; Berger, R.; Fujiwara, K.; Vergote, I.; Colombo, N.; Mäenpää, J.; et al. Olaparib plus Bevacizumab as First-Line Maintenance in Ovarian Cancer. N. Engl. J. Med. 2019, 381, 2416–2428. [Google Scholar] [CrossRef]
- Weber, J.S.; D’Angelo, S.P.; Minor, D.; Hodi, F.S.; Gutzmer, R.; Neyns, B.; Hoeller, C.; Khushalani, N.I.; Miller, W.H.; Lao, C.D.; et al. Nivolumab versus chemotherapy in patients with advanced melanoma who progressed after anti-CTLA-4 treatment (CheckMate 037): A randomised, controlled, open-label, phase 3 trial. Lancet Oncol. 2015, 16, 375–384. [Google Scholar] [CrossRef]
- Robert, C.; Long, G.V.; Brady, B.; Dutriaux, C.; Maio, M.; Mortier, L.; Hassel, J.C.; Rutkowski, P.; McNeil, C.; Kalinka-Warzocha, E.; et al. Nivolumab in previously untreated melanoma without BRAF mutation. N. Engl. J. Med. 2015, 372, 320–330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weber, J.; Mandala, M.; Del Vecchio, M.; Gogas, H.J.; Arance, A.M.; Cowey, C.L.; Dalle, S.; Schenker, M.; Chiarion-Sileni, V.; Marquez-Rodas, I.; et al. Adjuvant Nivolumab versus Ipilimumab in Resected Stage III or IV Melanoma. N. Engl. J. Med. 2017, 377, 1824–1835. [Google Scholar] [CrossRef] [PubMed]
- Reck, M.; Rodríguez-Abreu, D.; Robinson, A.G.; Hui, R.; Csőszi, T.; Fülöp, A.; Gottfried, M.; Peled, N.; Tafreshi, A.; Cuffe, S.; et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2016, 375, 1823–1833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Motzer, R.J.; Escudier, B.; McDermott, D.F.; George, S.; Hammers, H.J.; Srinivas, S.; Tykodi, S.S.; Sosman, J.A.; Procopio, G.; Plimack, E.R.; et al. Nivolumab versus Everolimus in Advanced Renal-Cell Carcinoma. N. Engl. J. Med. 2015, 373, 1803–1813. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Conejo-Garcia, J.R.; Katsaros, D.; Gimotty, P.A.; Massobrio, M.; Regnani, G.; Makrigiannakis, A.; Gray, H.; Schlienger, K.; Liebman, M.N.; et al. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N. Engl. J. Med. 2003, 348, 203–213. [Google Scholar] [CrossRef] [Green Version]
- Sato, E.; Olson, S.H.; Ahn, J.; Bundy, B.; Nishikawa, H.; Qian, F.; Jungbluth, A.A.; Frosina, D.; Gnjatic, S.; Ambrosone, C.; et al. Intraepithelial CD8+ tumor-infiltrating lymphocytes and a high CD8+/regulatory T cell ratio are associated with favorable prognosis in ovarian cancer. Proc. Natl. Acad. Sci. USA 2005, 102, 18538–18543. [Google Scholar] [CrossRef] [Green Version]
- Pinto, M.P.; Balmaceda, C.; Bravo, M.L.; Kato, S.; Villarroel, A.; Owen, G.I.; Roa, J.C.; Cuello, M.A.; Ibañez, C. Patient inflammatory status and CD4+/CD8+ intraepithelial tumor lymphocyte infiltration are predictors of outcomes in high-grade serous ovarian cancer. Gynecol. Oncol. 2018, 151, 10–17. [Google Scholar] [CrossRef] [Green Version]
- Stanske, M.; Wienert, S.; Castillo-Tong, D.C.; Kreuzinger, C.; Vergote, I.; Lambrechts, S.; Gabra, H.; Gourley, C.; Ganapathi, R.N.; Kolaschinski, I.; et al. Dynamics of the Intratumoral Immune Response during Progression of High-Grade Serous Ovarian Cancer. Neoplasia 2018, 20, 280–288. [Google Scholar] [CrossRef] [PubMed]
- Moore, K.N.; Bookman, M.; Sehouli, J.; Miller, A.; Anderson, C.; Scambia, G.; Myers, T.; Taskiran, C.; Robison, K.; Mäenpää, J.; et al. Atezolizumab, Bevacizumab, and Chemotherapy for Newly Diagnosed Stage III or IV Ovarian Cancer: Placebo-Controlled Randomized Phase III Trial (IMagyn050/GOG 3015/ENGOT-OV39). J. Clin. Oncol. 2021, 39, 1842–1855. [Google Scholar] [CrossRef] [PubMed]
- Hamanishi, J.; Mandai, M.; Ikeda, T.; Minami, M.; Kawaguchi, A.; Murayama, T.; Kanai, M.; Mori, Y.; Matsumoto, S.; Chikuma, S.; et al. Safety and Antitumor Activity of Anti-PD-1 Antibody, Nivolumab, in Patients With Platinum-Resistant Ovarian Cancer. J. Clin. Oncol. 2015, 33, 4015–4022. [Google Scholar] [CrossRef] [PubMed]
- Matulonis, U.A.; Shapira, R.; Santin, A.; Lisyanskaya, A.S.; Pignata, S.; Vergote, I.; Raspagliesi, F.; Sonke, G.S.; Birrer, M.; Sehouli, J.; et al. Final results from the KEYNOTE-100 trial of pembrolizumab in patients with advanced recurrent ovarian cancer. JCO 2020, 38, 6005. [Google Scholar] [CrossRef]
- Ledermann, J.A.; Shapira-Frommer, R.; Santin, A.; Lisyanskaya, A.S.; Pignata, S.; Vergote, I.; Raspagliesi, F.; Sonke, G.S.; Birrer, M.J.; Provencher, D.M.; et al. LBA36—Association of PD-L1 expression and gene expression profiling with clinical response to pembrolizumab in patients with advanced recurrent ovarian cancer: Results from the phase II KEYNOTE-100 study. Ann. Oncol. 2018, 29, viii728. [Google Scholar] [CrossRef]
- Ledermann, J.A.; Shapira-Frommer, R.; Santin, A.D.; Lisyanskaya, A.; Pignata, S.; Vergote, I.B.; Raspagliesi, F.; Sonke, G.S.; Birrer, M.; Provencher, D.; et al. 843P Association of gene expression signatures and TMB with response to pembrolizumab (pembro) in patients (pts) with recurrent ovarian cancer (ROC) enrolled in KEYNOTE-100. Ann. Oncol. 2020, 31, S631–S632. [Google Scholar] [CrossRef]
- McAlpine, J.N.; Porter, H.; Köbel, M.; Nelson, B.H.; Prentice, L.M.; Kalloger, S.E.; Senz, J.; Milne, K.; Ding, J.; Shah, S.P.; et al. BRCA1 and BRCA2 mutations correlate with TP53 abnormalities and presence of immune cell infiltrates in ovarian high-grade serous carcinoma. Mod. Pathol. 2012, 25, 740–750. [Google Scholar] [CrossRef] [Green Version]
- Clarke, B.; Tinker, A.V.; Lee, C.-H.; Subramanian, S.; van de Rijn, M.; Turbin, D.; Kalloger, S.; Han, G.; Ceballos, K.; Cadungog, M.G.; et al. Intraepithelial T cells and prognosis in ovarian carcinoma: Novel associations with stage, tumor type, and BRCA1 loss. Mod. Pathol. 2009, 22, 393–402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Färkkilä, A.; Gulhan, D.C.; Casado, J.; Jacobson, C.A.; Nguyen, H.; Kochupurakkal, B.; Maliga, Z.; Yapp, C.; Chen, Y.-A.; Schapiro, D.; et al. Immunogenomic profiling determines responses to combined PARP and PD-1 inhibition in ovarian cancer. Nat. Commun. 2020, 11, 1459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alexandrov, L.B.; Kim, J.; Haradhvala, N.J.; Huang, M.N.; Tian Ng, A.W.; Wu, Y.; Boot, A.; Covington, K.R.; Gordenin, D.A.; Bergstrom, E.N.; et al. The repertoire of mutational signatures in human cancer. Nature 2020, 578, 94–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macintyre, G.; Goranova, T.E.; De Silva, D.; Ennis, D.; Piskorz, A.M.; Eldridge, M.; Sie, D.; Lewsley, L.-A.; Hanif, A.; Wilson, C.; et al. Copy number signatures and mutational processes in ovarian carcinoma. Nat. Genet. 2018, 50, 1262–1270. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Liu, L.; Zhang, J.; Chen, J.; Ye, J.; Shukla, S.; Qiao, J.; Zhan, X.; Chen, H.; Wu, C.J.; et al. Investigation of Antigen-Specific T-Cell Receptor Clusters in Human Cancers. Clin. Cancer Res. 2020, 26, 1359–1371. [Google Scholar] [CrossRef]
- Knijnenburg, T.A.; Wang, L.; Zimmermann, M.T.; Chambwe, N.; Gao, G.F.; Cherniack, A.D.; Fan, H.; Shen, H.; Way, G.P.; Greene, C.S.; et al. Genomic and Molecular Landscape of DNA Damage Repair Deficiency across The Cancer Genome Atlas. Cell Rep. 2018, 23, 239–254.e6. [Google Scholar] [CrossRef] [Green Version]
- TCGA2STAT. Available online: http://www.liuzlab.org/TCGA2STAT/ (accessed on 20 January 2021).
- Becht, E.; Giraldo, N.A.; Lacroix, L.; Buttard, B.; Elarouci, N.; Petitprez, F.; Selves, J.; Laurent-Puig, P.; Sautès-Fridman, C.; Fridman, W.H.; et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016, 17, 218. [Google Scholar] [CrossRef]
- Korthauer, K.; Kendziorski, K. MADGiC: A model-based approach for identifying driver genes in cancer. Bioinformatics 2015, 31, 1526–1535. [Google Scholar] [CrossRef] [Green Version]
- Koboldt, D.C.; Zhang, Q.; Larson, D.E.; Shen, D.; McLellan, M.D.; Lin, L.; Miller, C.A.; Mardis, E.R.; Ding, L.; Wilson, R.K. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012, 22, 568–576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cibulskis, K.; Lawrence, M.S.; Carter, S.L.; Sivachenko, A.; Jaffe, D.; Sougnez, C.; Gabriel, S.; Meyerson, M.; Lander, E.S.; Getz, G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013, 31, 213–219. [Google Scholar] [CrossRef]
- Kim, S.; Scheffler, K.; Halpern, A.L.; Bekritsky, M.A.; Noh, E.; Källberg, M.; Chen, X.; Kim, Y.; Beyter, D.; Krusche, P.; et al. Strelka2: Fast and accurate calling of germline and somatic variants. Nat. Methods 2018, 15, 591–594. [Google Scholar] [CrossRef]
- Ha, G.; Roth, A.; Khattra, J.; Ho, J.; Yap, D.; Prentice, L.M.; Melnyk, N.; McPherson, A.; Bashashati, A.; Laks, E.; et al. TITAN: Inference of copy number architectures in clonal cell populations from tumor whole-genome sequence data. Genome Res. 2014, 24, 1881–1893. [Google Scholar] [CrossRef]
- Bolotin, D.A.; Poslavsky, S.; Mitrophanov, I.; Shugay, M.; Mamedov, I.Z.; Putintseva, E.V.; Chudakov, D.M. MiXCR: Software for comprehensive adaptive immunity profiling. Nat. Methods 2015, 12, 380–381. [Google Scholar] [CrossRef]
- Migrating the SNP Array-Based Homologous Recombination Deficiency Measures to Next Generation Sequencing Data of Breast Cancer | npj Breast Cancer. Available online: https://www.nature.com/articles/s41523-018-0066-6 (accessed on 9 July 2021).
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Hothorn, T.; Lausen, B. On the exact distribution of maximally selected rank statistics. Comput. Stat. Data Anal. 2003, 43, 121–137. [Google Scholar] [CrossRef]
- Lheureux, S.; Cristea, M.C.; Bruce, J.P.; Garg, S.; Cabanero, M.; Mantia-Smaldone, G.; Olawaiye, A.B.; Ellard, S.L.; Weberpals, J.I.; Wahner Hendrickson, A.E.; et al. Adavosertib plus gemcitabine for platinum-resistant or platinum-refractory recurrent ovarian cancer: A double-blind, randomised, placebo-controlled, phase 2 trial. Lancet 2021, 397, 281–292. [Google Scholar] [CrossRef]
- Rizvi, N.A.; Hellmann, M.D.; Snyder, A.; Kvistborg, P.; Makarov, V.; Havel, J.J.; Lee, W.; Yuan, J.; Wong, P.; Ho, T.S.; et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015, 348, 124–128. [Google Scholar] [CrossRef] [Green Version]
- Bai, X.; Yi, M.; Jiao, Y.; Chu, Q.; Wu, K. Blocking TGF-β Signaling To Enhance The Efficacy Of Immune Checkpoint Inhibitor. Onco Targets Ther. 2019, 12, 9527–9538. [Google Scholar] [CrossRef] [Green Version]
- St-Pierre, F.; Bhatia, S.; Chandra, S. Harnessing Natural Killer Cells in Cancer Immunotherapy: A Review of Mechanisms and Novel Therapies. Cancers 2021, 13, 1988. [Google Scholar] [CrossRef]
- Scheper, W.; Kelderman, S.; Fanchi, L.F.; Linnemann, C.; Bendle, G.; de Rooij, M.A.J.; Hirt, C.; Mezzadra, R.; Slagter, M.; Dijkstra, K.; et al. Low and variable tumor reactivity of the intratumoral TCR repertoire in human cancers. Nat. Med. 2019, 25, 89–94. [Google Scholar] [CrossRef]
- Tsuji, T.; Eng, K.H.; Matsuzaki, J.; Battaglia, S.; Szender, J.B.; Miliotto, A.; Gnjatic, S.; Bshara, W.; Morrison, C.D.; Lele, S.; et al. Clonality and antigen-specific responses shape the prognostic effects of tumor-infiltrating T cells in ovarian cancer. Oncotarget 2020, 11, 2669–2683. [Google Scholar] [CrossRef]

| Variables | CGFL Cohort (n = 31) | TCGA Cohort (n = 578) | p-Value | Adjusted * p-Value | |
|---|---|---|---|---|---|
| Clinical characteristics | Stage, n (%) | 0.005 | 0.01 | ||
| III | 20 (64.5) | 384 (78.5) | |||
| IV | 11 (35.5) | 72 (14.7) | |||
| Other | 0 (0) | 33 (6.7) | |||
| Age at diagnosis, median (IQR) | 64.8 (13.73) | 59 (18) | 0.06 | 0.06 | |
| Age at diagnosis, n (%) | 0.06 | ||||
| ≤60 years | 11 (35.5) | 267 (54) | 0.06 | ||
| >60 years | 20 (64.5) | 225 (46) | |||
| Overall survival (months), median (IQR) | 49.6 (145.8) | 45.5 (49.3) | - | ||
| Genomic characteristics | HRD score, median (IQR) | 34 (35) | 44 (31) | 0.04 | 0.06 |
| HRD score, n (%) | |||||
| <42 | 10 (32.3) | 76 (44.4) | 0.03 | 0.06 | |
| ≥42 | 21 (67.7) | 95 (55.6) | |||
| TCR clones, median (IQR) | 6 (4) | 3 (5) | 0.002 | 0.01 | |
| CNV signature 1, median (IQR) | 0.57 (0.51) | 0.25 (0.28) | <1 × 10−3 | <1 × 10−3 | |
| CNV signature 2, median (IQR) | - | 0.07 (0.17) | - | - | |
| CNV signature 3, median (IQR) | 0.11 (0.17) | 0.26 (0.34) | <1 × 10−3 | <1 × 10−3 | |
| CNV signature 4, median (IQR) | 0.02 (0.15) | 0 (0.12) | 0.56 | 0.56 | |
| CNV signature 5, median (IQR) | 0.05 (0.02) | 0.01 (0.03) | <1 × 10−3 | <1 × 10−3 | |
| CNV signature 6, median (IQR) | 0.03 (0.06) | 0.04 (0.11) | 0.04 | 0.06 | |
| CNV signature 7, median (IQR) | 0.05 (0.25) | 0.11 (0.18) | 0.38 | 0.45 | |
| Variables | HR | 95%CI | p-Value | Adjusted * p-Value |
|---|---|---|---|---|
| CNV signature 1 | 1.3 | [0.71; 2.4] | 0.4 | 0.47 |
| CNV signature 2 | 2.11 | [0.63; 6.99] | 0.22 | 0.31 |
| CNV signature 3 | 0.51 | [0.24; 1.04] | 0.06 | 0.16 |
| CNV signature 4 | 1.94 | [0.77; 4.9] | 0.16 | 0.28 |
| CNV signature 5 | 1.48 | [0.01; 174.54] | 0.87 | 0.87 |
| CNV signature 6 | 4.59 | [0.9; 23.4] | 0.07 | 0.16 |
| CNV signature 7 | 0.21 | [0.06; 0.73] | 0.01 | 0.10 |
| TILS | HR | 95%CI | p-Value | Adjusted * p-Value |
|---|---|---|---|---|
| T Cells | 0.98 | [0.79; 1.21] | 0.85 | 0.95 |
| Cd8 T Cells | 1.02 | [0.89; 1.16] | 0.82 | 0.95 |
| Cytotoxic Lymphocytes | 1.05 | [0.88; 1.25] | 0.61 | 0.95 |
| B Lineage | 0.89 | [0.72; 1.1] | 0.28 | 0.67 |
| Nk Cells | 1.02 | [0.76; 1.37] | 0.89 | 0.95 |
| Monocytic Lineage | 1.08 | [0.92; 1.27] | 0.34 | 0.67 |
| Myeloid Dendritic Cells | 0.99 | [0.85; 1.17] | 0.95 | 0.95 |
| Neutrophils | 1.87 | [1.32; 2.63] | <1 × 10−3 | 0.001 |
| Endothelial Cells | 1.40 | [0.97; 2.02] | 0.07 | 0.24 |
| Fibroblasts | 1.14 | [1.02; 1.28] | 0.02 | 0.09 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lecuelle, J.; Boidot, R.; Mananet, H.; Derangère, V.; Albuisson, J.; Goussot, V.; Arnould, L.; Tharin, Z.; Ray Coquard, I.; Ghiringhelli, F.; et al. TCR Clonality and Genomic Instability Signatures as Prognostic Biomarkers in High Grade Serous Ovarian Cancer. Cancers 2021, 13, 4394. https://doi.org/10.3390/cancers13174394
Lecuelle J, Boidot R, Mananet H, Derangère V, Albuisson J, Goussot V, Arnould L, Tharin Z, Ray Coquard I, Ghiringhelli F, et al. TCR Clonality and Genomic Instability Signatures as Prognostic Biomarkers in High Grade Serous Ovarian Cancer. Cancers. 2021; 13(17):4394. https://doi.org/10.3390/cancers13174394
Chicago/Turabian StyleLecuelle, Julie, Romain Boidot, Hugo Mananet, Valentin Derangère, Juliette Albuisson, Vincent Goussot, Laurent Arnould, Zoé Tharin, Isabelle Ray Coquard, François Ghiringhelli, and et al. 2021. "TCR Clonality and Genomic Instability Signatures as Prognostic Biomarkers in High Grade Serous Ovarian Cancer" Cancers 13, no. 17: 4394. https://doi.org/10.3390/cancers13174394
APA StyleLecuelle, J., Boidot, R., Mananet, H., Derangère, V., Albuisson, J., Goussot, V., Arnould, L., Tharin, Z., Ray Coquard, I., Ghiringhelli, F., Truntzer, C., & Fumet, J.-D. (2021). TCR Clonality and Genomic Instability Signatures as Prognostic Biomarkers in High Grade Serous Ovarian Cancer. Cancers, 13(17), 4394. https://doi.org/10.3390/cancers13174394







