NCOR1 Sustains Colorectal Cancer Cell Growth and Protects against Cellular Senescence
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Cell Culture
2.3. NCOR1 Knockdown with shRNAs
2.4. RNA Isolation and qRT-PCR
2.5. Cell Adhesion and SAβ-Galactosidase Assays
2.6. Soft Agarose and Colony-Forming Cell Assays
2.7. Xenografts into Nude Mice
2.8. Western Blot and ELISA
2.9. RNA-Seq and Bioinformatics
2.10. Statistics
3. Results
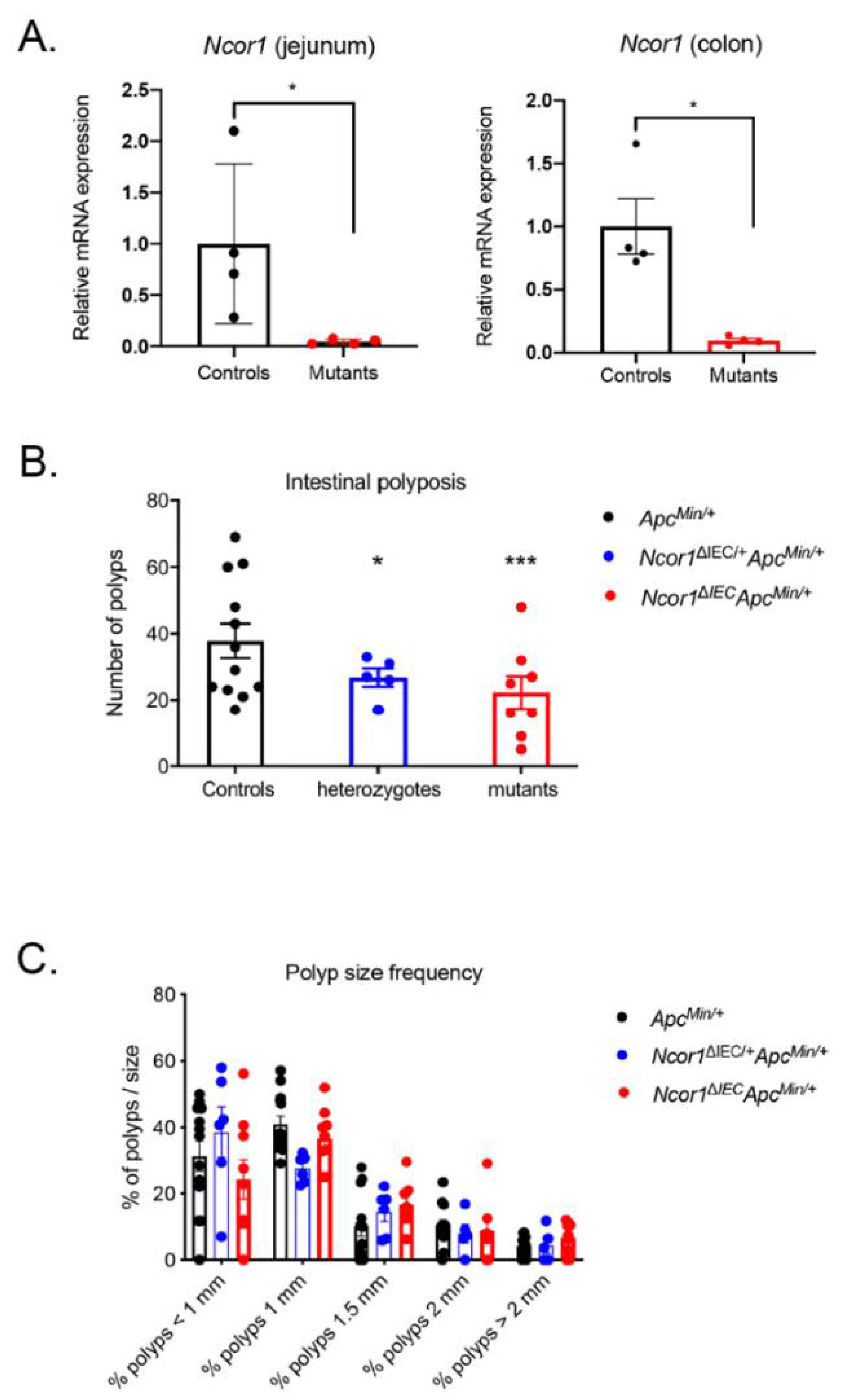
3.1. Loss of Intestinal Epithelial NCOR1 Reduces Polyposis in ApcMin/+ Mice
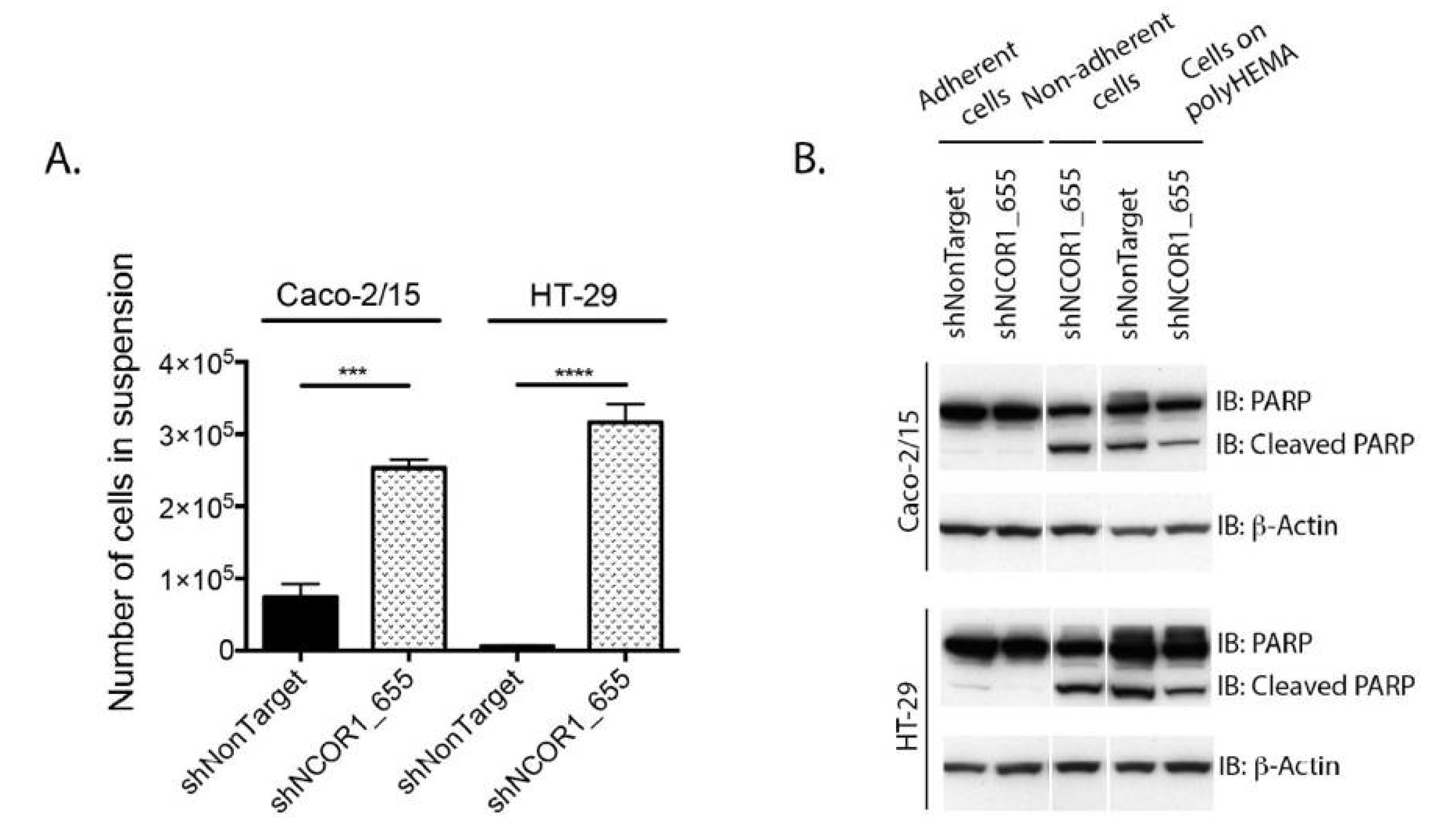
3.2. NCOR1 Is Required for CRC Cell Growth and Adherence
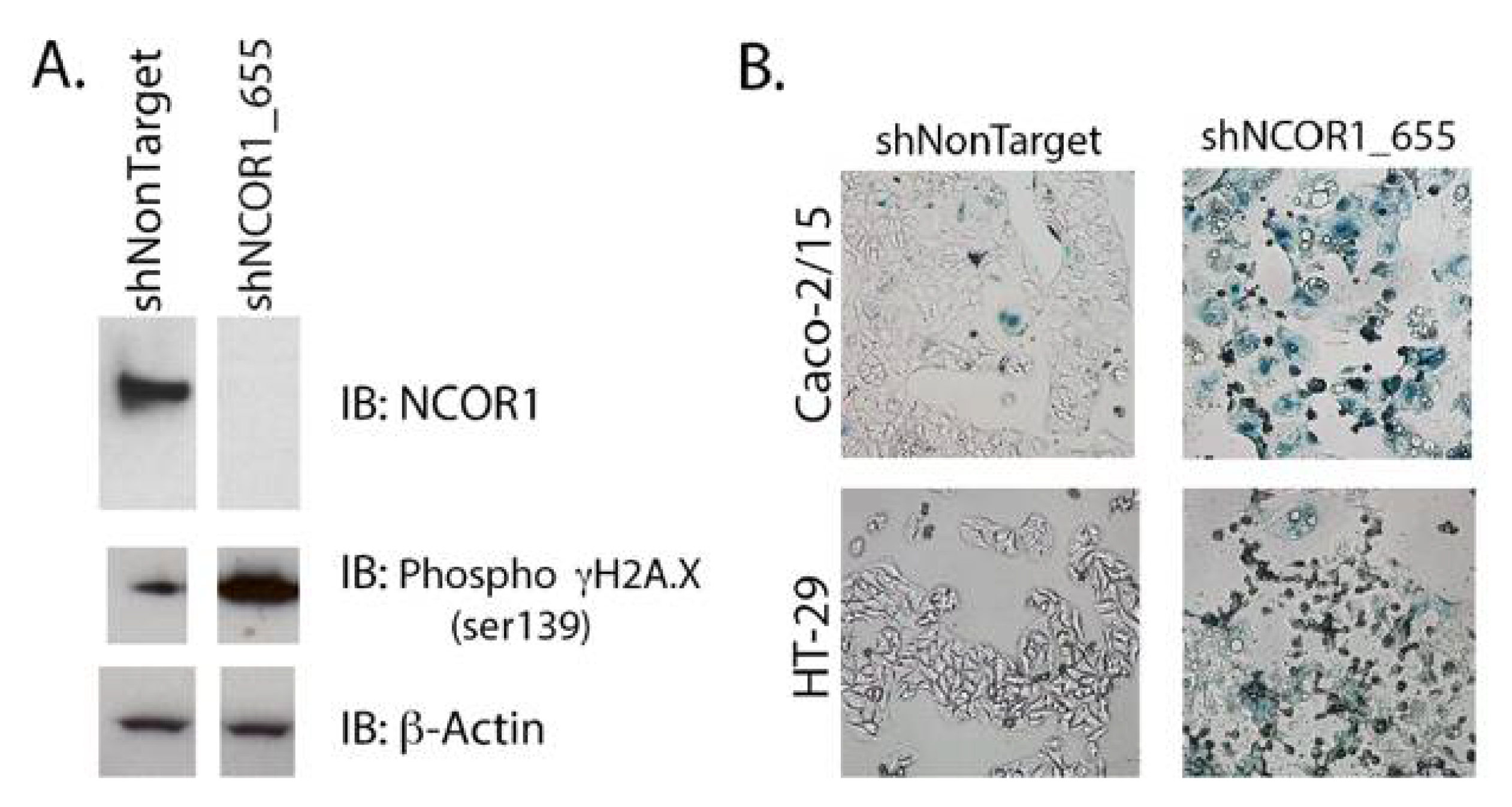
3.3. Loss of NCOR1 Promotes CRC Cell Senescence Associated with a Secretory Phenotype
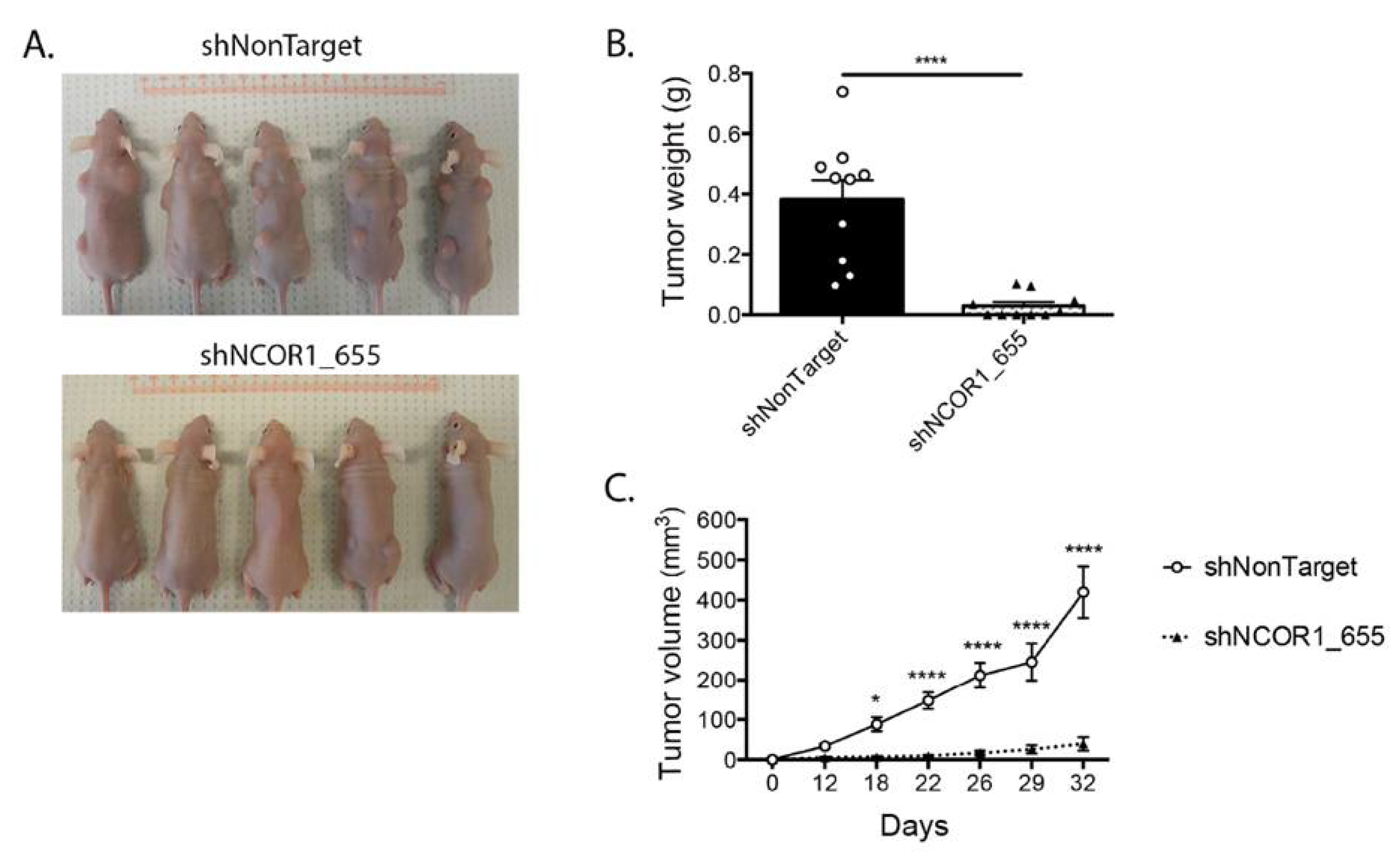
3.4. Loss of NCOR1 Reduces Tumorigenic Growth of HT-29 Cells In Vivo
3.5. Impact of NCOR1 Depletion on CRC Cells Transcriptome
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Horlein, A.J.; Naar, A.M.; Heinzel, T.; Torchia, J.; Gloss, B.; Kurokawa, R.; Ryan, A.; Kamei, Y.; Soderstrom, M.; Glass, C.K.; et al. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature 1995, 377, 397–404. [Google Scholar] [CrossRef]
- Kurokawa, R.; Soderstrom, M.; Horlein, A.; Halachmi, S.; Brown, M.; Rosenfeld, M.G.; Glass, C.K. Polarity-specific activities of retinoic acid receptors determined by a co-repressor. Nature 1995, 377, 451–454. [Google Scholar] [CrossRef]
- Ogawa, S.; Lozach, J.; Jepsen, K.; Sawka-Verhelle, D.; Perissi, V.; Sasik, R.; Rose, D.W.; Johnson, R.S.; Rosenfeld, M.G.; Glass, C.K. A nuclear receptor corepressor transcriptional checkpoint controlling activator protein 1-dependent gene networks required for macrophage activation. Proc. Natl. Acad. Sci. USA 2004, 101, 14461–14466. [Google Scholar] [CrossRef] [Green Version]
- Mottis, A.; Mouchiroud, L.; Auwerx, J. Emerging roles of the corepressors NCoR1 and SMRT in homeostasis. Genes. Dev. 2013, 27, 819–835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- You, S.H.; Lim, H.W.; Sun, Z.; Broache, M.; Won, K.J.; Lazar, M.A. Nuclear receptor co-repressors are required for the histone-deacetylase activity of HDAC3 in vivo. Nat. Struct. Mol. Biol. 2013, 20, 182–187. [Google Scholar] [CrossRef] [Green Version]
- Perissi, V.; Jepsen, K.; Glass, C.K.; Rosenfeld, M.G. Deconstructing repression: Evolving models of co-repressor action. Nat. Rev. Genet. 2010, 11, 109–123. [Google Scholar] [CrossRef]
- Jepsen, K.; Hermanson, O.; Onami, T.M.; Gleiberman, A.S.; Lunyak, V.; McEvilly, R.J.; Kurokawa, R.; Kumar, V.; Liu, F.; Seto, E.; et al. Combinatorial roles of the nuclear receptor corepressor in transcription and development. Cell 2000, 102, 753–763. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, H.; Williams, E.G.; Mouchiroud, L.; Canto, C.; Fan, W.; Downes, M.; Heligon, C.; Barish, G.D.; Desvergne, B.; Evans, R.M.; et al. NCoR1 is a conserved physiological modulator of muscle mass and oxidative function. Cell 2011, 147, 827–839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, P.; Fan, W.; Xu, J.; Lu, M.; Yamamoto, H.; Auwerx, J.; Sears, D.D.; Talukdar, S.; Oh, D.; Chen, A.; et al. Adipocyte NCoR knockout decreases PPARgamma phosphorylation and enhances PPARgamma activity and insulin sensitivity. Cell 2011, 147, 815–826. [Google Scholar] [CrossRef] [Green Version]
- Barish, G.D.; Yu, R.T.; Karunasiri, M.S.; Becerra, D.; Kim, J.; Tseng, T.W.; Tai, L.J.; Leblanc, M.; Diehl, C.; Cerchietti, L.; et al. The Bcl6-SMRT/NCoR cistrome represses inflammation to attenuate atherosclerosis. Cell Metab. 2012, 15, 554–562. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Spann, N.J.; Kaikkonen, M.U.; Lu, M.; Oh, D.Y.; Fox, J.N.; Bandyopadhyay, G.; Talukdar, S.; Xu, J.; Lagakos, W.S.; et al. NCoR repression of LXRs restricts macrophage biosynthesis of insulin-sensitizing omega 3 fatty acids. Cell 2013, 155, 200–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahad, A.; Stevanin, M.; Smita, S.; Mishra, G.P.; Gupta, D.; Waszak, S.; Sarkar, U.A.; Basak, S.; Gupta, B.; Acha-Orbea, H.; et al. NCoR1: Putting the Brakes on the Dendritic Cell Immune Tolerance. iScience 2019, 19, 996–1011. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Lu, W.; Yueh, M.F.; Rettenmeier, E.; Liu, M.; Paszek, M.; Auwerx, J.; Yu, R.T.; Evans, R.M.; Wang, K.; et al. Intestinal NCoR1, a regulator of epithelial cell maturation, controls neonatal hyperbilirubinemia. Proc. Natl. Acad. Sci. USA 2017, 114, E1432–E1440. [Google Scholar] [CrossRef] [Green Version]
- Mennillo, E.; Yang, X.; Paszek, M.; Auwerx, J.; Benner, C.; Chen, S. NCoR1 Protects Mice From Dextran Sodium Sulfate-Induced Colitis by Guarding Colonic Crypt Cells From Luminal Insult. Cell Mol. Gastroenterol. Hepatol. 2020, 10, 133–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doyon, G.; St-Jean, S.; Darsigny, M.; Asselin, C.; Boudreau, F. Nuclear receptor co-repressor is required to maintain proliferation of normal intestinal epithelial cells in culture and down-modulates the expression of pigment epithelium-derived factor. J. Biol. Chem. 2009, 284, 25220–25229. [Google Scholar] [CrossRef] [Green Version]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Fearon, E.R. Molecular genetics of colorectal cancer. Annu. Rev. Pathol. 2011, 6, 479–507. [Google Scholar] [CrossRef] [PubMed]
- Farooqi, A.A.; de la Roche, M.; Djamgoz, M.B.A.; Siddik, Z.H. Overview of the oncogenic signaling pathways in colorectal cancer: Mechanistic insights. Semin. Cancer Biol. 2019, 58, 65–79. [Google Scholar] [CrossRef]
- Penfield, J.D.; Anderson, M.; Lutzke, L.; Wang, K.K. The role of cellular senescence in the gastrointestinal mucosa. Gut. Liver 2013, 7, 270–277. [Google Scholar] [CrossRef]
- Hayflick, L.; Moorhead, P.S. The serial cultivation of human diploid cell strains. Exp. Cell Res. 1961, 25, 585–621. [Google Scholar] [CrossRef]
- Shay, J.W.; Wright, W.E. Telomeres and telomerase: Three decades of progress. Nat. Rev. Genet. 2019, 20, 299–309. [Google Scholar] [CrossRef]
- Rodier, F.; Campisi, J. Four faces of cellular senescence. J. Cell Biol. 2011, 192, 547–556. [Google Scholar] [CrossRef]
- Gorgoulis, V.; Adams, P.D.; Alimonti, A.; Bennett, D.C.; Bischof, O.; Bishop, C.; Campisi, J.; Collado, M.; Evangelou, K.; Ferbeyre, G.; et al. Cellular Senescence: Defining a Path Forward. Cell 2019, 179, 813–827. [Google Scholar] [CrossRef] [PubMed]
- Campisi, J. Aging, cellular senescence, and cancer. Annu. Rev. Physiol. 2013, 75, 685–705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madison, B.B.; Dunbar, L.; Qiao, X.T.; Braunstein, K.; Braunstein, E.; Gumucio, D.L. Cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J. Biol. Chem. 2002, 277, 33275–33283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boudreau, F.; Lussier, C.R.; Mongrain, S.; Darsigny, M.; Drouin, J.L.; Doyon, G.; Suh, E.R.; Beaulieu, J.F.; Rivard, N.; Perreault, N. Loss of cathepsin L activity promotes claudin-1 overexpression and intestinal neoplasia. FASEB J. 2007, 21, 3853–3865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Debacq-Chainiaux, F.; Erusalimsky, J.D.; Campisi, J.; Toussaint, O. Protocols to detect senescence-associated beta-galactosidase (SA-betagal) activity, a biomarker of senescent cells in culture and in vivo. Nat. Protoc. 2009, 4, 1798–1806. [Google Scholar] [CrossRef]
- Jones, C.; St-Jean, S.; Frechette, I.; Bergeron, D.; Rivard, N.; Boudreau, F. Identification of a novel promyelocytic leukemia zinc-finger isoform required for colorectal cancer cell growth and survival. Int. J. Cancer 2013, 133, 58–66. [Google Scholar] [CrossRef] [PubMed]
- Bergeron, S.; Lemieux, E.; Durand, V.; Cagnol, S.; Carrier, J.C.; Lussier, J.G.; Boucher, M.J.; Rivard, N. The serine protease inhibitor serpinE2 is a novel target of ERK signaling involved in human colorectal tumorigenesis. Mol. Cancer 2010, 9, 271. [Google Scholar] [CrossRef] [Green Version]
- Soldani, C.; Scovassi, A.I. Poly(ADP-ribose) polymerase-1 cleavage during apoptosis: An update. Apoptosis 2002, 7, 321–328. [Google Scholar] [CrossRef]
- Hernandez-Segura, A.; Nehme, J.; Demaria, M. Hallmarks of Cellular Senescence. Trends Cell Biol. 2018, 28, 436–453. [Google Scholar] [CrossRef]
- Schmidt, S.; Denk, S.; Wiegering, A. Targeting Protein Synthesis in Colorectal Cancer. Cancers 2020, 12, 1298. [Google Scholar] [CrossRef]
- Steinberg, G.R.; Carling, D. AMP-activated protein kinase: The current landscape for drug development. Nat. Rev. Drug. Discov. 2019, 18, 527–551. [Google Scholar] [CrossRef]
- Haga, R.B.; Ridley, A.J. Rho GTPases: Regulation and roles in cancer cell biology. Small GTPases 2016, 7, 207–221. [Google Scholar] [CrossRef] [Green Version]
- Box, J.K.; Paquet, N.; Adams, M.N.; Boucher, D.; Bolderson, E.; O’Byrne, K.J.; Richard, D.J. Nucleophosmin: From structure and function to disease development. BMC Mol. Biol. 2016, 17, 19. [Google Scholar] [CrossRef] [Green Version]
- Roux, P.P.; Topisirovic, I. Signaling Pathways Involved in the Regulation of mRNA Translation. Mol. Cell. Biol. 2018, 38, e00070-18. [Google Scholar] [CrossRef] [Green Version]
- Sugawara, K.; Shibasaki, T.; Takahashi, H.; Seino, S. Structure and functional roles of Epac2 (Rapgef4). Gene 2016, 575, 577–583. [Google Scholar] [CrossRef]
- Radin, D.P.; Patel, P. BDNF: An Oncogene or Tumor Suppressor? Anticancer Res. 2017, 37, 3983–3990. [Google Scholar] [CrossRef] [Green Version]
- Akakura, S.; Gelman, I.H. Pivotal Role of AKAP12 in the Regulation of Cellular Adhesion Dynamics: Control of Cytoskeletal Architecture, Cell Migration, and Mitogenic Signaling. J. Signal Transduct. 2012, 2012, 529179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merritt, R.C.; Manor, U.; Salles, F.T.; Grati, M.; Dose, A.C.; Unrath, W.C.; Quintero, O.A.; Yengo, C.M.; Kachar, B. Myosin IIIB uses an actin-binding motif in its espin-1 cargo to reach the tips of actin protrusions. Curr. Biol. 2012, 22, 320–325. [Google Scholar] [CrossRef] [Green Version]
- Prakash, S.K.; Paylor, R.; Jenna, S.; Lamarche-Vane, N.; Armstrong, D.L.; Xu, B.; Mancini, M.A.; Zoghbi, H.Y. Functional analysis of ARHGAP6, a novel GTPase-activating protein for RhoA. Hum. Mol. Genet. 2000, 9, 477–488. [Google Scholar] [CrossRef]
- Novak, D.; Huser, L.; Elton, J.J.; Umansky, V.; Altevogt, P.; Utikal, J. SOX2 in development and cancer biology. Semin. Cancer Biol. 2020, 67, 74–82. [Google Scholar] [CrossRef]
- Terao, Y.; Nishida, J.; Horiuchi, S.; Rong, F.; Ueoka, Y.; Matsuda, T.; Kato, H.; Furugen, Y.; Yoshida, K.; Kato, K.; et al. Sodium butyrate induces growth arrest and senescence-like phenotypes in gynecologic cancer cells. Int. J. Cancer 2001, 94, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Rebbaa, A.; Zheng, X.; Chu, F.; Mirkin, B.L. The role of histone acetylation versus DNA damage in drug-induced senescence and apoptosis. Cell Death Differ. 2006, 13, 1960–1967. [Google Scholar] [CrossRef] [Green Version]
- Xu, W.S.; Perez, G.; Ngo, L.; Gui, C.Y.; Marks, P.A. Induction of polyploidy by histone deacetylase inhibitor: A pathway for antitumor effects. Cancer Res. 2005, 65, 7832–7839. [Google Scholar] [CrossRef] [Green Version]
- Eichner, L.J.; Curtis, S.D.; Brun, S.N.; Baumgart, J.T.; Ross, D.S.; Rymoff, T.J.; Shaw, R.J. HDAC3 regulates senescence and lineage specificity in non-small cell lung cancer. bioRxiv 2021. [Google Scholar] [CrossRef]
- Gui, Y.; Guo, G.; Huang, Y.; Hu, X.; Tang, A.; Gao, S.; Wu, R.; Chen, C.; Li, X.; Zhou, L.; et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat. Genet. 2011, 43, 875–878. [Google Scholar] [CrossRef] [PubMed]
- Stephens, P.J.; Tarpey, P.S.; Davies, H.; Van Loo, P.; Greenman, C.; Wedge, D.C.; Nik-Zainal, S.; Martin, S.; Varela, I.; Bignell, G.R.; et al. The landscape of cancer genes and mutational processes in breast cancer. Nature 2012, 486, 400–404. [Google Scholar] [CrossRef]
- Sowalsky, A.G.; Xia, Z.; Wang, L.; Zhao, H.; Chen, S.; Bubley, G.J.; Balk, S.P.; Li, W. Whole transcriptome sequencing reveals extensive unspliced mRNA in metastatic castration-resistant prostate cancer. Mol. Cancer Res. 2015, 13, 98–106. [Google Scholar] [CrossRef] [Green Version]
- Ivanov, I.; Lo, K.C.; Hawthorn, L.; Cowell, J.K.; Ionov, Y. Identifying candidate colon cancer tumor suppressor genes using inhibition of nonsense-mediated mRNA decay in colon cancer cells. Oncogene 2007, 26, 2873–2884. [Google Scholar] [CrossRef] [Green Version]
- Fernandez-Majada, V.; Pujadas, J.; Vilardell, F.; Capella, G.; Mayo, M.W.; Bigas, A.; Espinosa, L. Aberrant cytoplasmic localization of N-CoR in colorectal tumors. Cell Cycle 2007, 6, 1748–1752. [Google Scholar] [CrossRef] [PubMed]
- Nazha, B.; Granner, T.; Maloney, S.; Odashiro, A.N.; Antecka, E.; Burnier, M.N., Jr. Aberrant nuclear repressor coreceptor 1 localization in human retinoblastoma. Ophthalmic. Res. 2013, 49, 171–176. [Google Scholar] [CrossRef]
- Gallardo, F.; Padron, A.; Garcia-Carbonell, R.; Rius, C.; Gonzalez-Perez, A.; Arumi-Uria, M.; Iglesias, M.; Nonell, L.; Bellosillo, B.; Segura, S.; et al. Cytoplasmic accumulation of NCoR in malignant melanoma: Consequences of altered gene repression and prognostic significance. Oncotarget 2015, 6, 9284–9294. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Zhang, F.; Zhang, X.F.; Qi, L.S.; Yang, L.; Guo, H.; Zhang, N. Expression of nucleophosmin/NPM1 correlates with migration and invasiveness of colon cancer cells. J. Biomed. Sci. 2012, 19, 53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brunetto de Farias, C.; Rosemberg, D.B.; Heinen, T.E.; Koehler-Santos, P.; Abujamra, A.L.; Kapczinski, F.; Brunetto, A.L.; Ashton-Prolla, P.; Meurer, L.; Reis Bogo, M.; et al. BDNF/TrkB content and interaction with gastrin-releasing peptide receptor blockade in colorectal cancer. Oncology 2010, 79, 430–439. [Google Scholar] [CrossRef]
- Huang, S.M.; Lin, C.; Lin, H.Y.; Chiu, C.M.; Fang, C.W.; Liao, K.F.; Chen, D.R.; Yeh, W.L. Brain-derived neurotrophic factor regulates cell motility in human colon cancer. Endocr. Relat. Cancer 2015, 22, 455–464. [Google Scholar] [CrossRef] [Green Version]
- Luan, Y.; Li, X.; Luan, Y.; Zhao, R.; Li, Y.; Liu, L.; Hao, Y.; Oleg Vladimir, B.; Jia, L. Circulating lncRNA UCA1 Promotes Malignancy of Colorectal Cancer via the miR-143/MYO6 Axis. Mol. Ther. Nucleic. Acids. 2020, 19, 790–803. [Google Scholar] [CrossRef]
- Blumenthal, R.D.; Hansen, H.J.; Goldenberg, D.M. Inhibition of adhesion, invasion, and metastasis by antibodies targeting CEACAM6 (NCA-90) and CEACAM5 (Carcinoembryonic Antigen). Cancer Res. 2005, 65, 8809–8817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bengs, S.; Becker, E.; Busenhart, P.; Spalinger, M.R.; Raselli, T.; Kasper, S.; Lang, S.; Atrott, K.; Mamie, C.; Vavricka, S.R.; et al. beta6 -integrin serves as a novel serum tumor marker for colorectal carcinoma. Int. J. Cancer 2019, 145, 678–685. [Google Scholar] [CrossRef] [Green Version]
- Guo, F.; Liu, Y.; Huang, J.; Li, Y.; Zhou, G.; Wang, D.; Li, Y.; Wang, J.; Xie, P.; Li, G. Identification of Rho GTPase activating protein 6 isoform 1 variant as a new molecular marker in human colorectal tumors. Pathol. Oncol. Res. 2010, 16, 319–326. [Google Scholar] [CrossRef]
- Ridley, A.J. Rho GTPases and cell migration. J. Cell Sci. 2001, 114, 2713–2722. [Google Scholar] [CrossRef]
- Yildirim, M.; Suren, D.; Yildiz, M.; Sezgin Alikanoglu, A.; Eryilmaz, R.; Goktas, S.; Bulbuller, N.; Sezer, C. AKAP12/Gravin gene expression in colorectal cancer: Clinical importance and review of the literature. J. BUON 2013, 18, 635–640. [Google Scholar] [PubMed]
- Liu, W.; Guan, M.; Hu, T.; Gu, X.; Lu, Y. Re-expression of AKAP12 inhibits progression and metastasis potential of colorectal carcinoma in vivo and in vitro. PLoS ONE 2011, 6, e24015. [Google Scholar] [CrossRef] [Green Version]
- Neumann, J.; Bahr, F.; Horst, D.; Kriegl, L.; Engel, J.; Luque, R.M.; Gerhard, M.; Kirchner, T.; Jung, A. SOX2 expression correlates with lymph-node metastases and distant spread in right-sided colon cancer. BMC Cancer 2011, 11, 518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, J.; Xu, L.; Pan, Y.; Yu, S.; Wang, H.; Kennedy, D.; Zhang, Y. Sox2 modulates motility and enhances progression of colorectal cancer via the Rho-ROCK signaling pathway. Oncotarget 2017, 8, 98635–98645. [Google Scholar] [CrossRef]
- Takeda, K.; Mizushima, T.; Yokoyama, Y.; Hirose, H.; Wu, X.; Qian, Y.; Ikehata, K.; Miyoshi, N.; Takahashi, H.; Haraguchi, N.; et al. Sox2 is associated with cancer stem-like properties in colorectal cancer. Sci. Rep. 2018, 8, 17639. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Du, L.; Wen, Z.; Yang, Y.; Li, J.; Dong, Z.; Zheng, G.; Wang, L.; Zhang, X.; Wang, C. Sex determining region Y-box 2 inhibits the proliferation of colorectal adenocarcinoma cells through the mTOR signaling pathway. Int. J. Mol. Med. 2013, 32, 59–66. [Google Scholar] [CrossRef] [Green Version]
- Cho, Y.Y.; Kim, D.J.; Lee, H.S.; Jeong, C.H.; Cho, E.J.; Kim, M.O.; Byun, S.; Lee, K.Y.; Yao, K.; Carper, A.; et al. Autophagy and cellular senescence mediated by Sox2 suppress malignancy of cancer cells. PLoS ONE 2013, 8, e57172. [Google Scholar] [CrossRef] [Green Version]
- Hu, P.S.; Li, T.; Lin, J.F.; Qiu, M.Z.; Wang, D.S.; Liu, Z.X.; Chen, Z.H.; Yang, L.P.; Zhang, X.L.; Zhao, Q.; et al. VDR-SOX2 signaling promotes colorectal cancer stemness and malignancy in an acidic microenvironment. Signal Transduct. Target. Ther. 2020, 5, 183. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Martinez, R.; Zambrano, A.; Castillo, A.I.; Aranda, A. Vitamin D-dependent recruitment of corepressors to vitamin D/retinoid X receptor heterodimers. Mol. Cell Biol. 2008, 28, 3817–3829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doig, C.L.; Singh, P.K.; Dhiman, V.K.; Thorne, J.L.; Battaglia, S.; Sobolewski, M.; Maguire, O.; O’Neill, L.P.; Turner, B.M.; McCabe, C.J.; et al. Recruitment of NCOR1 to VDR target genes is enhanced in prostate cancer cells and associates with altered DNA methylation patterns. Carcinogenesis 2013, 34, 248–256. [Google Scholar] [CrossRef]
- Kobayashi, A.; Okuda, H.; Xing, F.; Pandey, P.R.; Watabe, M.; Hirota, S.; Pai, S.K.; Liu, W.; Fukuda, K.; Chambers, C.; et al. Bone morphogenetic protein 7 in dormancy and metastasis of prostate cancer stem-like cells in bone. J. Exp. Med. 2011, 208, 2641–2655. [Google Scholar] [CrossRef] [Green Version]
- Triana-Martinez, F.; Loza, M.I.; Dominguez, E. Beyond Tumor Suppression: Senescence in Cancer Stemness and Tumor Dormancy. Cells 2020, 9, 346. [Google Scholar] [CrossRef] [Green Version]



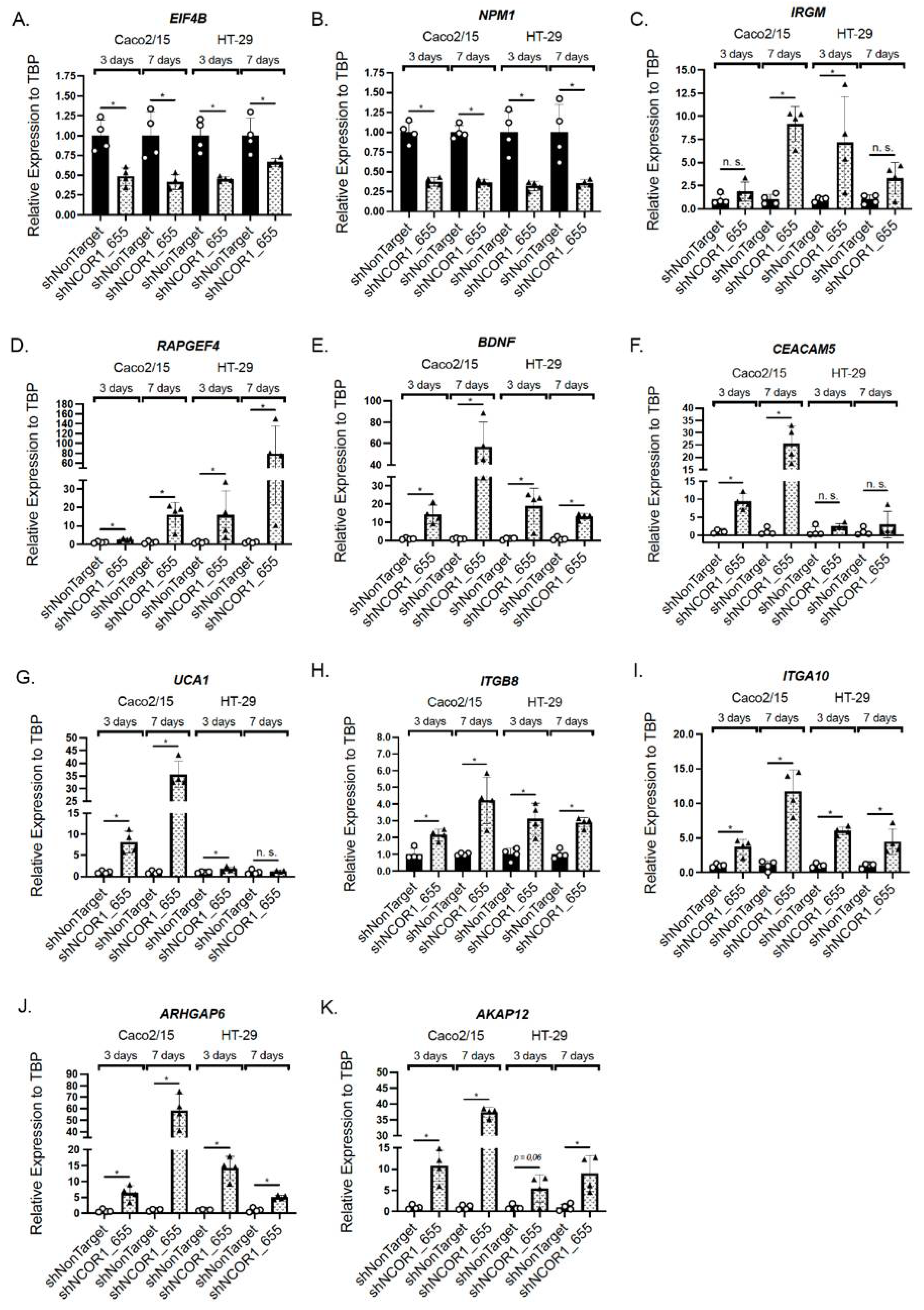
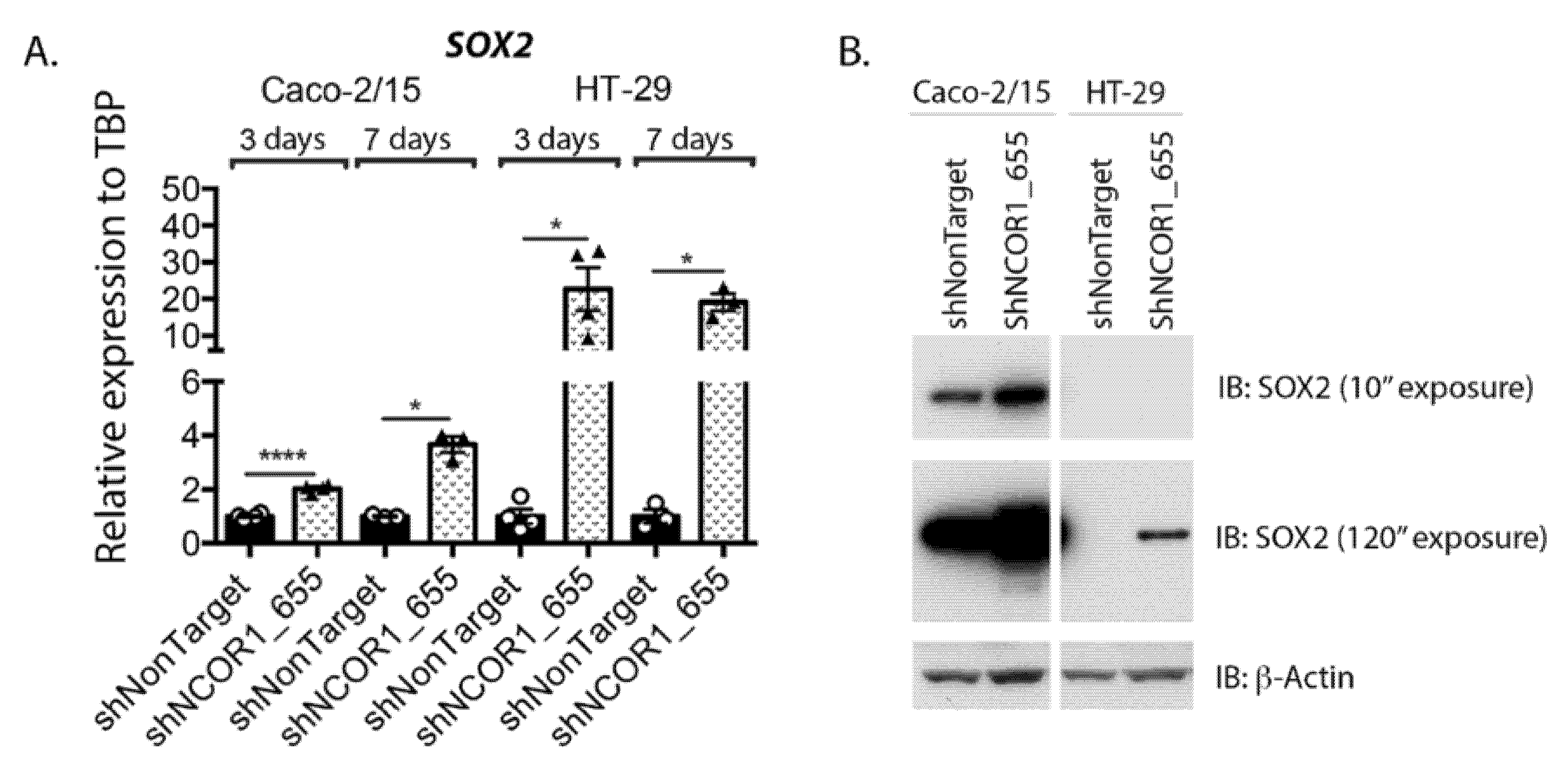
| Symbol | Gene Name | Caco-2/15 | HT-29 | ||
|---|---|---|---|---|---|
| shNCOR1_655 | shNCOR1_655 | ||||
| Ratio | p-Value | Ratio | p-Value | ||
| BHLHE40 | basic helix-loop-helix family, member e40 | 2.955 | 1.10 × 10−46 | 4.211 | 6.70 × 10−10 |
| BMP7 | bone morphogenetic protein 7 | −2.123 | 1.20 × 10−24 | −1.134 | 1.00 × 10+00 |
| BTG3 | BTG family, member 3 | 2.097 | 4.20 × 10−23 | 1.119 | 5.20 × 10−01 |
| CDK6 | cyclin-dependent kinase 6 | 2.563 | 2.90 × 10−40 | 4.179 | 9.10 × 10−23 |
| CDKN1A | cyclin-dependent kinase inhibitor 1A (p21, Cip1) | 3.394 | 2.90 × 10−41 | 1.485 | 5.60 × 10−01 |
| CDKN1C | cyclin-dependent kinase inhibitor 1C (p57, Kip2) | −9.363 | 2.20 × 10−11 | −1.297 | 6.00 × 10−01 |
| CDKN2B | cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) | 2.032 | 6.00 × 10−04 | 3.138 | 1.80 × 10−02 |
| CDKN2D | cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) | 2.521 | 7.80 × 10−04 | 1.397 | 2.90 × 10−02 |
| EREG | epiregulin | 3.961 | 3.00 × 10−51 | 2.328 | 9.70 × 10−02 |
| IFI16 | interferon, gamma-inducible protein 16 | 3.758 | 5.50 × 10−04 | 2.495 | 1.10 × 10−02 |
| JUNB | jun B proto-oncogene | 2.21 | 1.20 × 10−20 | 1.258 | 5.10 × 10−01 |
| NPM1 | nucleophosmin (nucleolar phosphoprotein B23, numatrin) | −2.417 | 2.10 × 10−36 | −3.308 | 1.90 × 10−18 |
| RB1 | retinoblastoma 1 | −3.057 | 1.20 × 10−50 | −4.202 | 1.90 × 10−25 |
| SKP1 | S-phase kinase-associated protein 1 | −3.078 | 6.40 × 10−56 | −3.156 | 6.30 × 10−17 |
| SOX2 | SRY (sex determining region Y)-box 2 | 2.39 | 5.80 × 10−06 | 20.563 | 1.30 × 10−10 |
| TERT | telomerase reverse transcriptase | −3.143 | 2.50 × 10−04 | −2.942 | 2.90 × 10−02 |
| TGFBR1 | transforming growth factor, beta receptor 1 | −2.844 | 1.10 × 10−44 | −4.356 | 2.70 × 10−23 |
| TM4SF1 | transmembrane 4 L six family member 1 | 2.646 | 2.30 × 10−06 | 2.152 | 8.00 × 10−09 |
| TP73 | tumor protein p73 | −4.031 | 2.30 × 10−24 | −4.646 | 5.20 × 10−08 |
| VCAN | versican | 9.747 | 3.70 × 10−15 | 2.944 | 3.60 × 10−02 |
| Symbol | Gene Name | Caco-2/15 | HT-29 | ||
|---|---|---|---|---|---|
| shNCOR1_655 | shNCOR1_655 | ||||
| Ratio | p-Value | Ratio | p-Value | ||
| CSF1 | colony stimulating factor 1 (macrophage) | 3.42 | 6.70 × 10−11 | 7.301 | 2.40 × 10−43 |
| CXCL1 (GRO-a) | chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) | 2.576 | 1.00 × 10−04 | 8.168 | 3.90 × 10−14 |
| CXCL2 (GRO-b) | chemokine (C-X-C motif) ligand 2 | 2.385 | 8.10 × 10−08 | 2.774 | 2.70 × 10−02 |
| CXCL3 | chemokine (C-X-C motif) ligand 3 | 4.124 | 3.10 × 10−05 | 3.88 | 7.20 × 10−05 |
| CXCL5 | chemokine (C-X-C motif) ligand 5 | 26.337 | 7.40 × 10−04 | 15.095 | 6.50 × 10−02 |
| EREG | epiregulin | 3.961 | 3.00 × 10−51 | 2.328 | 9.70 × 10−02 |
| FASLG | Fas ligand (TNF superfamily, member 6) | 36.277 | 2.80 × 10−02 | 1.012 | 1.00 × 10+00 |
| FN1 | fibronectin 1 | 2.418 | 7.80 × 10−13 | 1.421 | 8.20 × 10−01 |
| ICAM1 | intercellular adhesion molecule 1 | 2.299 | 2.10 × 10−16 | 3.304 | 2.60 × 10−10 |
| ICAM3 | intercellular adhesion molecule 3 | −2.218 | 6.20 × 10−14 | −3.247 | 1.20 × 10−08 |
| IGFL2 | IGF-like family member 2 | 4.942 | 4.00 × 10−19 | 4.554 | 2.00 × 10−07 |
| IL15 | interleukin 15 | 5.476 | 7.80 × 10−10 | 2.072 | 1.40 × 10−06 |
| ITPKA | inositol-trisphosphate 3-kinase A | −3.24 | 1.90 × 10−20 | −4.127 | 3.30 × 10−01 |
| MMP13 | matrix metallopeptidase 13 (collagenase 3) | 14.065 | 9.60 × 10−04 | 5.692 | 7.80 × 10−05 |
| MMP14 | matrix metallopeptidase 14 (membrane-inserted) | 2.858 | 1.00 × 10−26 | 1.999 | 2.80 × 10−02 |
| TIMP1 | TIMP metallopeptidase inhibitor 1 | 2.186 | 3.90 × 10−02 | 1.861 | 5.00 × 10−06 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
St-Jean, S.; De Castro, A.C.; Lecours, M.; Jones, C.; Rivard, N.; Rodier, F.; Perreault, N.; Boudreau, F. NCOR1 Sustains Colorectal Cancer Cell Growth and Protects against Cellular Senescence. Cancers 2021, 13, 4414. https://doi.org/10.3390/cancers13174414
St-Jean S, De Castro AC, Lecours M, Jones C, Rivard N, Rodier F, Perreault N, Boudreau F. NCOR1 Sustains Colorectal Cancer Cell Growth and Protects against Cellular Senescence. Cancers. 2021; 13(17):4414. https://doi.org/10.3390/cancers13174414
Chicago/Turabian StyleSt-Jean, Stéphanie, Ariane Cristina De Castro, Mia Lecours, Christine Jones, Nathalie Rivard, Francis Rodier, Nathalie Perreault, and François Boudreau. 2021. "NCOR1 Sustains Colorectal Cancer Cell Growth and Protects against Cellular Senescence" Cancers 13, no. 17: 4414. https://doi.org/10.3390/cancers13174414
APA StyleSt-Jean, S., De Castro, A. C., Lecours, M., Jones, C., Rivard, N., Rodier, F., Perreault, N., & Boudreau, F. (2021). NCOR1 Sustains Colorectal Cancer Cell Growth and Protects against Cellular Senescence. Cancers, 13(17), 4414. https://doi.org/10.3390/cancers13174414






