The Unfolded Protein Response Is Associated with Cancer Proliferation and Worse Survival in Hepatocellular Carcinoma
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Unfolded Protein Response (UPR) Was Positively Correlated with Clinical Parameters of Carcinogenesis and Cancer Progression as Well as the AJCC Cancer Stage of HCC Patients
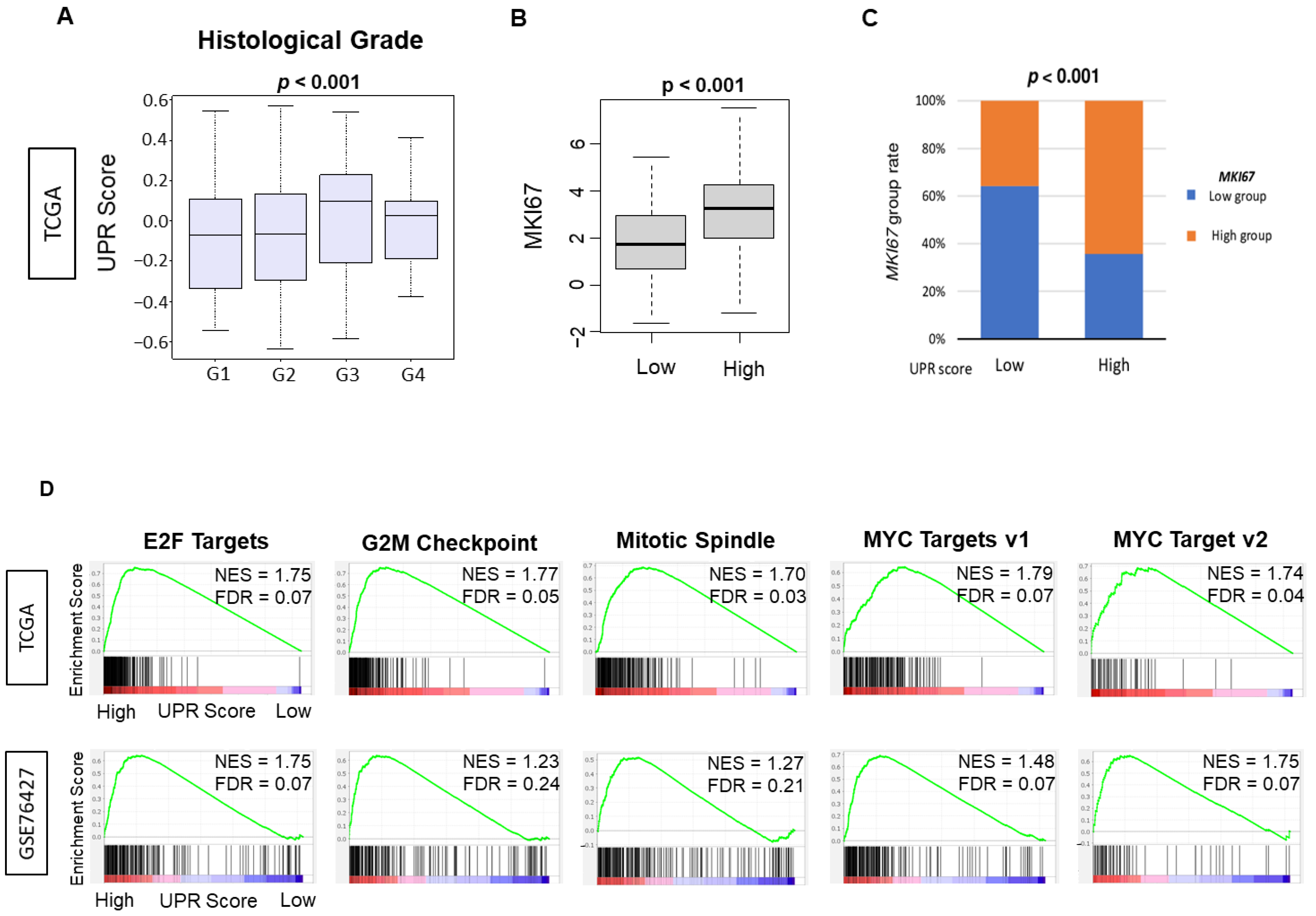
3.2. UPR Was Positively Correlated with Multiple Parameters of Cell Proliferation, including Histological Grade, MKI67 Gene Expression and Enrichment of Cell Proliferation-Related Gene Sets by Gene Set Enrichment Assay (GSEA)
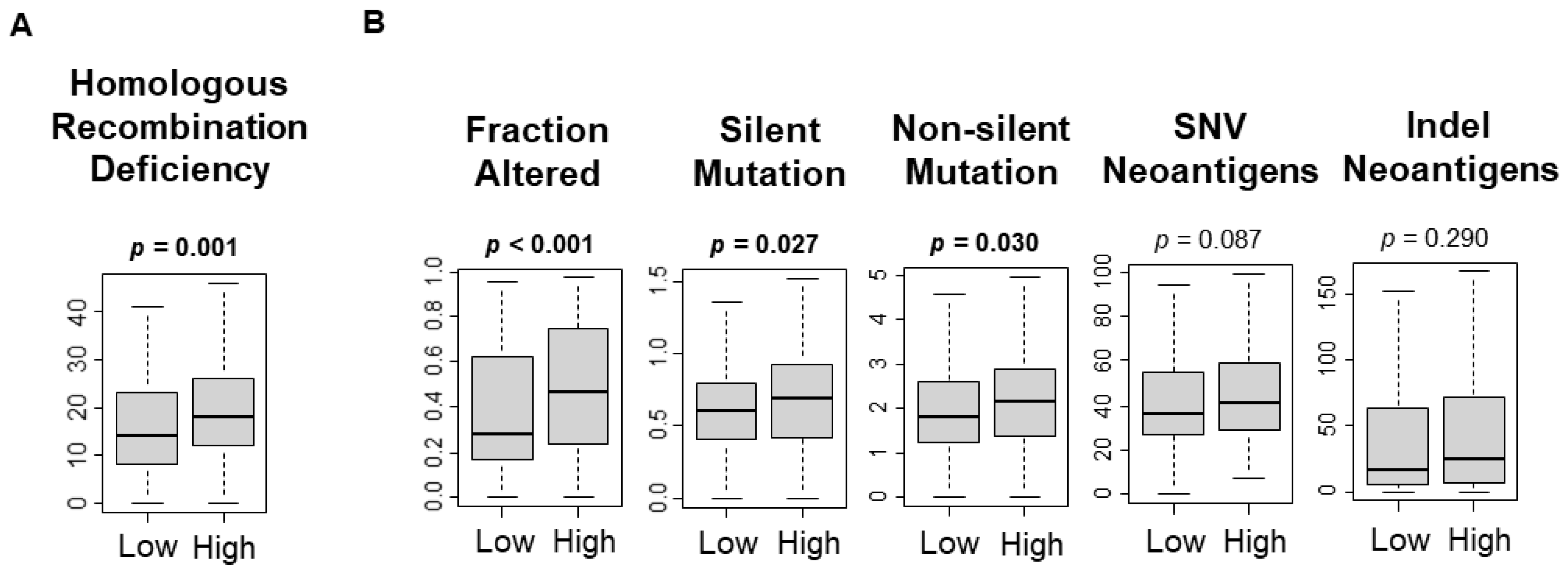
3.3. UPR Was Significantly Associated with Increased Mutational Load
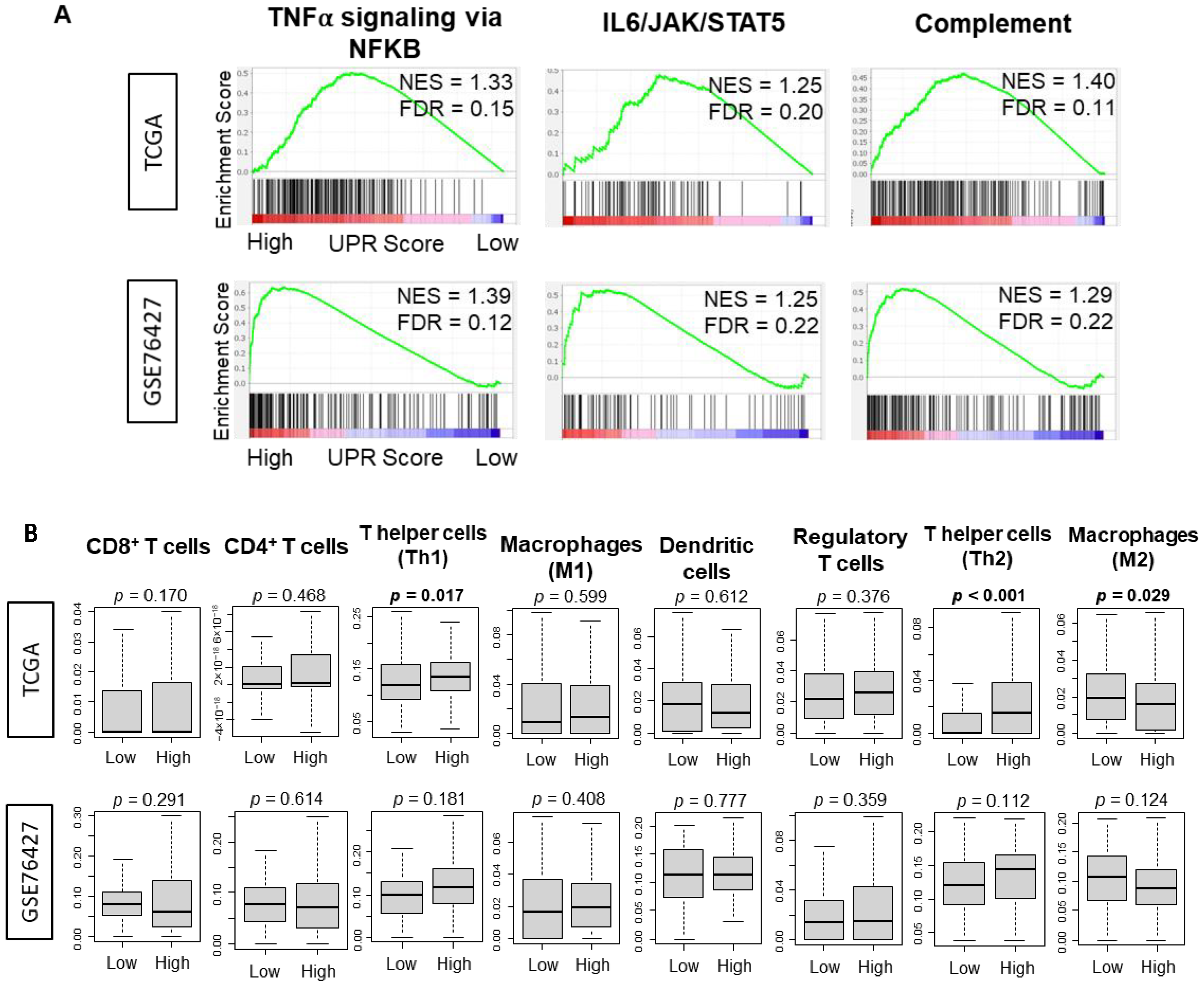
3.4. There Was No Consistently Significant Association between the UPR and Immune Response or Immune Cell Infiltration
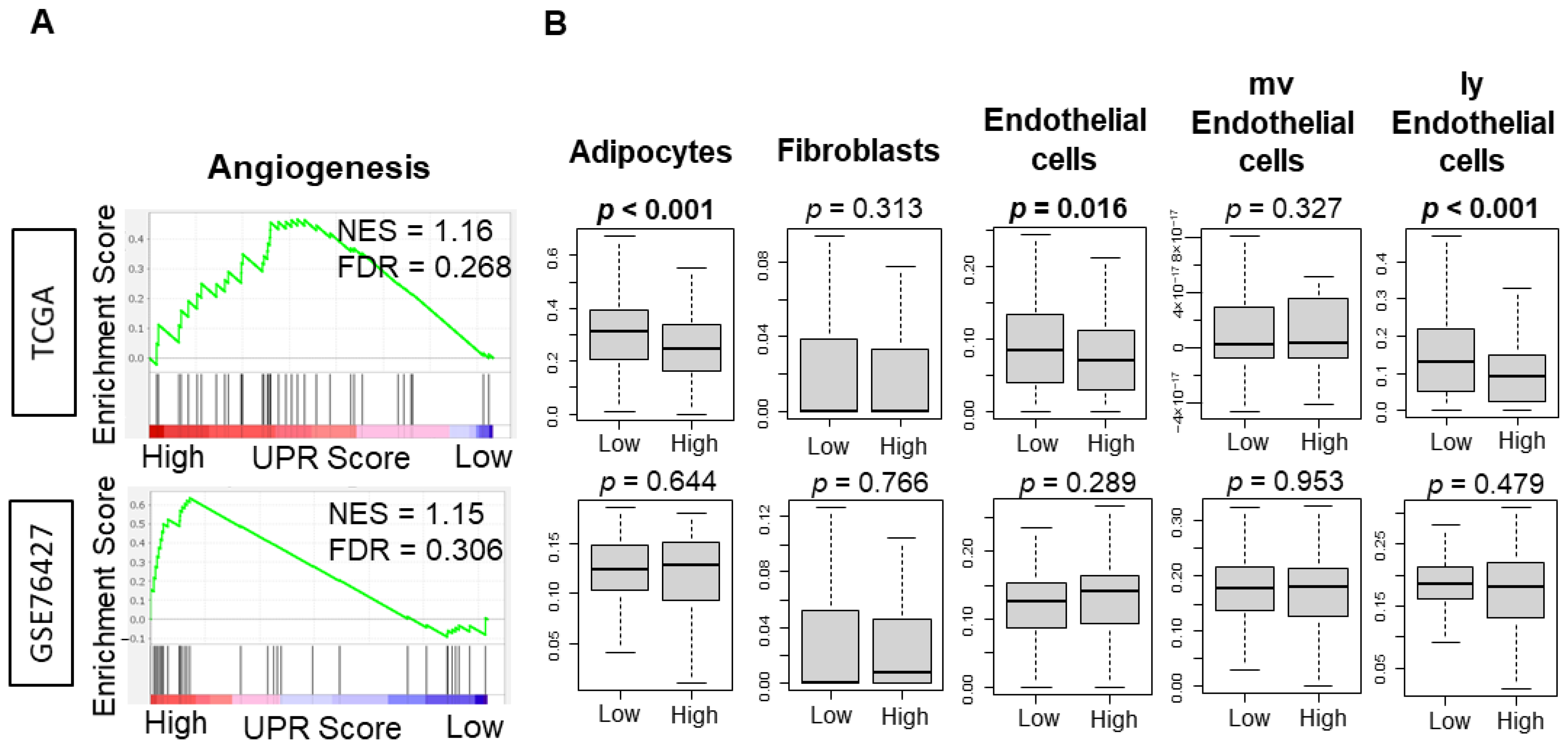
3.5. UPR Was Not Consistently Associated with Angiogeneis across Independent Cohorts of HCC
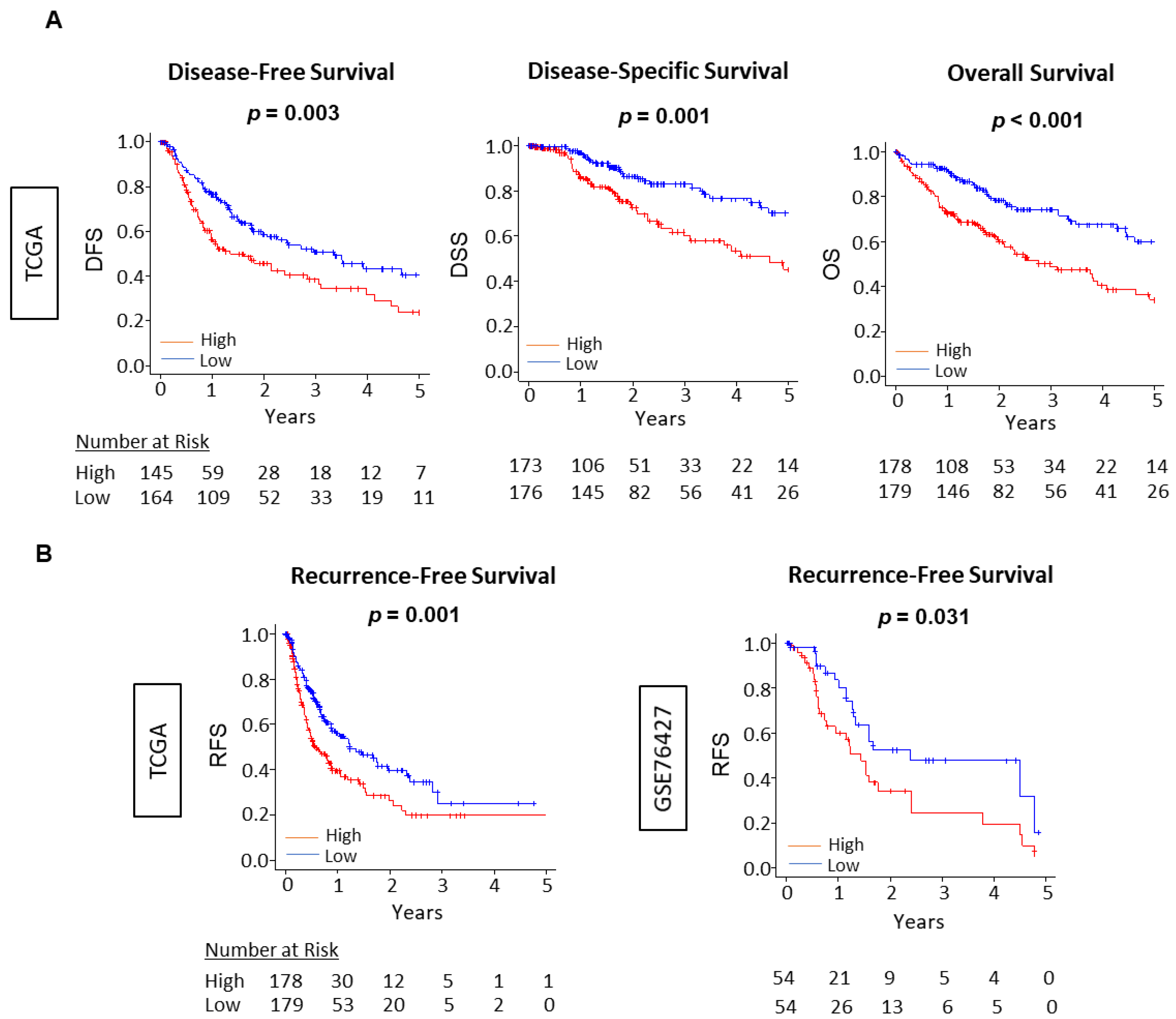
3.6. The Unfolded Protein Response Score Was Consistently Associated with Worse Survival across Independent Cohorts of HCC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Connell, L.C.; Harding, J.J.; Abou-Alfa, G.K. Advanced Hepatocellular Cancer: The Current State of Future Research. Curr. Treat. Options Oncol. 2016, 17, 43. [Google Scholar] [CrossRef]
- Llovet, J.M.; Fuster, J.; Bruix, J. Intention-to-treat analysis of surgical treatment for early hepatocellular carcinoma: Resection versus transplantation. Hepatology 1999, 30, 1434–1440. [Google Scholar] [CrossRef]
- Siwecka, N.; Rozpędek, W.; Pytel, D.; Wawrzynkiewicz, A.; Dziki, A.; Dziki, Ł.; Diehl, J.A.; Majsterek, I. Dual role of endoplasmic reticulum stress-mediated unfolded protein response signaling pathway in carcinogenesis. Int. J. Mol. Sci. 2019, 20, 4354. [Google Scholar] [CrossRef] [Green Version]
- Vandewynckel, Y.-P.; Laukens, D.; Geerts, A.; Bogaerts, E.; Paridaens, A.; Verhelst, X.; Janssens, S.; Heindryckx, F.; Van Vlierberghe, H. The paradox of the unfolded protein response in cancer. Anticancer. Res. 2013, 33, 4683–4694. [Google Scholar]
- Kozutsumi, Y.; Segal, M.R.; Normington, K.; Gething, M.-J.; Sambrook, J. The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature 1988, 332, 462–464. [Google Scholar] [CrossRef]
- Wang, W.-A.; Groenendyk, J.; Michalak, M. Endoplasmic reticulum stress associated responses in cancer. Biochim. Et Biophys. Acta (BBA)-Mol. Cell Res. 2014, 1843, 2143–2149. [Google Scholar] [CrossRef] [Green Version]
- Chakrabarti, A.; Chen, A.W.; Varner, J.D. A review of the mammalian unfolded protein response. Biotechnol. Bioeng. 2011, 108, 2777–2793. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Shan, B.; Dai, J.; Xia, Z.; Cai, J.; Chen, T.; Lv, S.; Feng, Y.; Zheng, L.; Wang, Y.; et al. Dual role for inositol-requiring enzyme 1α in promoting the development of hepatocellular carcinoma during diet-induced obesity in mice. Hepatology 2018, 68, 533–546. [Google Scholar] [CrossRef]
- Bi, M.; Naczki, C.; Koritzinsky, M.; Fels, D.; Blais, J.; Hu, N.; Harding, H.; Novoa, I.; Varia, M.; Raleigh, J.; et al. ER stress-regulated translation increases tolerance to extreme hypoxia and promotes tumor growth. EMBO J. 2005, 24, 3470–3481. [Google Scholar] [CrossRef] [Green Version]
- Ozturk, M. Genetic aspects of hepatocellular carcinogenesis. Semin. Liver Dis. 1999, 19, 235–242. [Google Scholar] [CrossRef]
- Oshi, M.; Newman, S.; Tokumaru, Y.; Yan, L.; Matsuyama, R.; Endo, I.; Katz, M.H.G.; Takabe, K. High G2M Pathway Score Pancreatic Cancer is Associated with Worse Survival, Particularly after Margin-Positive (R1 or R2) Resection. Cancers 2020, 12, 2871. [Google Scholar] [CrossRef]
- Oshi, M.; Takahashi, H.; Tokumaru, Y.; Yan, L.; Rashid, O.M.; Matsuyama, R.; Endo, I.; Takabe, K. G2M Cell Cycle Pathway Score as a Prognostic Biomarker of Metastasis in Estrogen Receptor (ER)-Positive Breast Cancer. Int. J. Mol. Sci. 2020, 21, 2921. [Google Scholar] [CrossRef] [Green Version]
- Oshi, M.; Kim, T.; Tokumaru, Y.; Yan, L.; Matsuyama, R.; Endo, I.; Cherkassky, L.; Takabe, K. Enhanced DNA Repair Pathway is Associated with Cell Proliferation and Worse Survival in Hepatocellular Carcinoma (HCC). Cancers 2021, 13, 323. [Google Scholar] [CrossRef]
- Wurmbach, E.; Chen, Y.-B.; Khitrov, G.; Zhang, W.; Roayaie, S.; Schwartz, M.; Fiel, I.; Thung, S.; Mazzaferro, V.M.; Bruix, J.; et al. Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology 2007, 45, 938–947. [Google Scholar] [CrossRef] [PubMed]
- Eun, J.; Nam, S. Identifying Novel Drivers of Human Hepatocellular Carcinoma and Revealing Clinical Relevance as Early Diagnostic and Prognostic Biomarker; The Catholic University of Korea: Seoul, Korea, 2017; Available online: https://www.omicsdi.org/dataset/geo/GSE89377 (accessed on 27 August 2021).
- Brandon, H.; Shen, D. mRNA Profiling of Hepatocellular Carcinoma (HCC). Gene Expression Omnibus. 2016; GSE56545 (Medimmune). Available online: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE56545 (accessed on 27 August 2021).
- Grinchuk, O.V.; Yenamandra, S.P.; Iyer, R.; Singh, M.; Lee, H.K.; Lim, K.H.; Chow, P.K.; Kuznetsov, V.A. Tumor-adjacent tissue co-expression profile analysis reveals pro-oncogenic ribosomal gene signature for prognosis of resectable hepatocellular carcinoma. Mol. Oncol. 2017, 12, 89–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wanless, I.R.; Party, I.W. Terminology of nodular hepatocellular lesions. Hepatology 1995, 22, 983–993. [Google Scholar]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database Hallmark Gene Set Collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef] [Green Version]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-Seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gandhi, S.; Elkhanany, A.; Oshi, M.; Dai, T.; Opyrchal, M.; Mohammadpour, H.; Repasky, E.A.; Takabe, K. Contribution of Immune Cells to Glucocorticoid Receptor Expression in Breast Cancer. Int. J. Mol. Sci. 2020, 21, 4635. [Google Scholar] [CrossRef]
- Oshi, M.; Asaoka, M.; Tokumaru, Y.; Yan, L.; Matsuyama, R.; Ishikawa, T.; Endo, I.; Takabe, K. CD8 T Cell Score as a Prognostic Biomarker for Triple Negative Breast Cancer. Int. J. Mol. Sci. 2020, 21, 6968. [Google Scholar] [CrossRef]
- Tokumaru, Y.; Oshi, M.; Katsuta, E.; Yan, L.; Huang, J.L.; Nagahashi, M.; Matsuhashi, N.; Futamura, M.; Yoshida, K.; Takabe, K. Intratumoral Adipocyte-High Breast Cancer Enrich for Metastatic and Inflammation-Related Pathways but Associated with Less Cancer Cell Proliferation. Int. J. Mol. Sci. 2020, 21, 5744. [Google Scholar] [CrossRef]
- Oshi, M.; Newman, S.; Murthy, V.; Tokumaru, Y.; Yan, L.; Matsuyama, R.; Endo, I.; Takabe, K. ITPKC as a Prognostic and Predictive Biomarker of Neoadjuvant Chemotherapy for Triple Negative Breast Cancer. Cancers 2020, 12, 2758. [Google Scholar] [CrossRef]
- Oshi, M.; Tokumaru, Y.; Asaoka, M.; Yan, L.; Satyananda, V.; Matsuyama, R.; Matsuhashi, N.; Futamura, M.; Ishikawa, T.; Yoshida, K.; et al. M1 Macrophage and M1/M2 ratio defined by transcriptomic signatures resemble only part of their conventional clinical characteristics in breast cancer. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef]
- Chouliaras, K.; Tokumaru, Y.; Asaoka, M.; Oshi, M.; Attwood, K.M.; Yoshida, K.; Ishikawa, T.; Takabe, K. Prevalence and clinical relevance of tumor-associated tissue eosinophilia (TATE) in breast cancer. Surgery 2020, 169, 1234–1239. [Google Scholar] [CrossRef]
- Oshi, M.; Newman, S.; Tokumaru, Y.; Yan, L.; Matsuyama, R.; Endo, I.; Takabe, K. Inflammation Is Associated with Worse Outcome in the Whole Cohort but with Better Outcome in Triple-Negative Subtype of Breast Cancer Patients. J. Immunol. Res. 2020, 2020, 1–17. [Google Scholar] [CrossRef]
- Oshi, M.; Tokumaru, Y.; Angarita, F.A.; Yan, L.; Matsuyama, R.; Endo, I.; Takabe, K. Degree of Early Estrogen Response Predict Survival after Endocrine Therapy in Primary and Metastatic ER-Positive Breast Cancer. Cancers 2020, 12, 3557. [Google Scholar] [CrossRef]
- Schulze, A.; Oshi, M.; Endo, I.; Takabe, K. MYC Targets Scores Are Associated with Cancer Aggressiveness and Poor Survival in ER-Positive Primary and Metastatic Breast Cancer. Int. J. Mol. Sci. 2020, 21, 8127. [Google Scholar] [CrossRef]
- Oshi, M.; Newman, S.; Tokumaru, Y.; Yan, L.; Matsuyama, R.; Endo, I.; Nagahashi, M.; Takabe, K. Intra-Tumoral Angiogenesis Is Associated with Inflammation, Immune Reaction and Metastatic Recurrence in Breast Cancer. Int. J. Mol. Sci. 2020, 21, 6708. [Google Scholar] [CrossRef]
- Oshi, M.; Takahashi, H.; Tokumaru, Y.; Yan, L.; Rashid, O.M.; Nagahashi, M.; Matsuyama, R.; Endo, I.; Takabe, K. The E2F pathway score as a predictive biomarker of response to neoadjuvant therapy in ER+/HER2− breast cancer. Cells 2020, 9, 1643. [Google Scholar]
- Tokumaru, Y.; Oshi, M.; Katsuta, E.; Yan, L.; Satyananda, V.; Matsuhashi, N.; Futamura, M.; Akao, Y.; Yoshida, K.; Takabe, K. KRAS signaling enriched triple negative breast cancer is associated with favorable tumor immune microenvironment and better survival. Am. J. Cancer Res. 2020, 10, 897–907. [Google Scholar]
- Tokumaru, Y.; Oshi, M.; Patel, A.; Tian, W.; Yan, L.; Matsuhashi, N.; Futamura, M.; Yoshida, K.; Takabe, K. Organoids Are Limited in Modeling the Colon Adenoma–Carcinoma Sequence. Cells 2021, 10, 488. [Google Scholar] [CrossRef]
- Oshi, M.; Tokumaru, Y.; Mukhopadhyay, S.; Yan, L.; Matsuyama, R.; Endo, I.; Takabe, K. Annexin A1 Expression Is Associated with Epithelial–Mesenchymal Transition (EMT), Cell Proliferation, Prognosis, and Drug Response in Pancreatic Cancer. Cells 2021, 10, 653. [Google Scholar] [CrossRef]
- Takahashi, H.; Asaoka, M.; Yan, L.; Rashid, O.M.; Oshi, M.; Ishikawa, T.; Nagahashi, M.; Takabe, K. Biologically Aggressive Phenotype and Anti-cancer Immunity Counterbalance in Breast Cancer with High Mutation Rate. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Oshi, M.; Satyananda, V.; Angarita, F.A.; Kim, T.H.; Tokumaru, Y.; Yan, L.; Matsuyama, R.; Endo, I.; Nagahashi, M.; Takabe, K. Angiogenesis is associated with an attenuated tumor microenvironment, aggressive biology, and worse survival in gastric cancer patients. Am. J. Cancer Res. 2021, 11, 1659–1671. [Google Scholar] [PubMed]
- Oshi, M.; Huyser, M.; Le, L.; Tokumaru, Y.; Yan, L.; Matsuyama, R.; Endo, I.; Takabe, K. Abundance of Microvascular Endothelial Cells Is Associated with Response to Chemotherapy and Prognosis in Colorectal Cancer. Cancers 2021, 13, 1477. [Google Scholar] [CrossRef] [PubMed]
- Okano, M.; Oshi, M.; Butash, A.L.; Katsuta, E.; Tachibana, K.; Saito, K.; Okayama, H.; Peng, X.; Yan, L.; Kono, K.; et al. Triple-Negative Breast Cancer with High Levels of Annexin A1 Expression Is Associated with Mast Cell Infiltration, Inflammation, and Angiogenesis. Int. J. Mol. Sci. 2019, 20, 4197. [Google Scholar] [CrossRef] [Green Version]
- Bild, A.; Febbo, P.G. Application of a priori established gene sets to discover biologically important differential expression in microarray data. Proc. Natl. Acad. Sci. USA 2005, 102, 15278–15279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aran, D.; Hu, Z.; Butte, A.J. xCell: Digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017, 18, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Thorsson, V.; Gibbs, D.; Brown, S.D.; Wolf, D.; Bortone, D.S.; Yang, T.-H.O.; Porta-Pardo, E.; Gao, G.F.; Plaisier, C.L.; Eddy, J.A.; et al. The Immune Landscape of Cancer. Immunity 2019, 51, 411–412. [Google Scholar] [CrossRef] [Green Version]
- Chalmers, Z.R.; Connelly, C.F.; Fabrizio, D.; Gay, L.; Ali, S.M.; Ennis, R.; Schrock, A.; Campbell, B.; Shlien, A.; Chmielecki, J.; et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017, 9, 1–14. [Google Scholar] [CrossRef]
- Ghosh, R.; Lipson, K.L.; Sargent, K.E.; Mercurio, A.M.; Hunt, J.S.; Ron, D.; Urano, F. Transcriptional Regulation of VEGF-A by the Unfolded Protein Response Pathway. PLoS ONE 2010, 5, e9575. [Google Scholar] [CrossRef]
- Lee, A.S. GRP78 Induction in Cancer: Therapeutic and Prognostic Implications: Figure 1. Cancer Res. 2007, 67, 3496–3499. [Google Scholar] [CrossRef] [Green Version]
- Pluquet, O.; Dejeans, N.; Bouchecareilh, M.; Lhomond, S.; Pineau, R.; Higa, A.; Delugin, M.; Combe, C.; Loriot, S.; Cubel, G.; et al. Posttranscriptional Regulation of PER1 Underlies the Oncogenic Function of IREα. Cancer Res. 2013, 73, 4732–4743. [Google Scholar] [CrossRef] [Green Version]
- Jamora, C.; Dennert, G.; Lee, A.S. Inhibition of tumor progression by suppression of stress protein GRP78/BiP induction in fibrosarcoma B/C10ME. Proc. Natl. Acad. Sci. USA 1996, 93, 7690–7694. [Google Scholar] [CrossRef] [Green Version]
- Fernandez, P.M.; Tabbara, S.O.; Jacobs, L.K.; Manning, F.C.R.; Tsangaris, T.N.; Schwartz, A.M.; Kennedy, K.A.; Patierno, S.R. Overexpression of the glucose-regulated stress gene GRP78 in malignant but not benign human breast lesions. Breast Cancer Res. Treat. 2000, 59, 15–26. [Google Scholar] [CrossRef]
- Mantovani, A.; Marchesi, F.; Malesci, A.; Laghi, L.; Allavena, P. Tumour-associated macrophages as treatment targets in oncology. Nat. Rev. Clin. Oncol. 2017, 14, 399–416. [Google Scholar] [CrossRef] [PubMed]
- Mahadevan, N.R.; Rodvold, J.; Sepulveda, H.; Rossi, S.; Drew, A.F.; Zanetti, M. Transmission of endoplasmic reticulum stress and pro-inflammation from tumor cells to myeloid cells. Proc. Natl. Acad. Sci. USA 2011, 108, 6561–6566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aoki, M.; Aoki, H.; Mukhopadhyay, P.; Tsuge, T.; Yamamoto, H.; Matsumoto, N.M.; Toyohara, E.; Okubo, Y.; Ogawa, R.; Takabe, K. Sphingosine-1-Phosphate Facilitates Skin Wound Healing by Increasing Angiogenesis and Inflammatory Cell Recruitment with Less Scar Formation. Int. J. Mol. Sci. 2019, 20, 3381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katsuta, E.; Rashid, O.M.; Takabe, K. Clinical relevance of tumor microenvironment: Immune cells, vessels, and mouse models. Hum. Cell 2020, 33, 930–937. [Google Scholar] [CrossRef]
- Ramanathan, R.; Olex, A.L.; Dozmorov, M.; Bear, H.D.; Fernandez, L.J.; Takabe, K. Angiopoietin pathway gene expression associated with poor breast cancer survival. Breast Cancer Res. Treat. 2017, 162, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Drogat, B.; Auguste, P.; Nguyen, D.T.; Bouchecareilh, M.; Pineau, R.; Nalbantoglu, J.; Kaufman, R.J.; Chevet, E.; Bikfalvi, A.; Moenner, M. IRE1 Signaling Is Essential for Ischemia-Induced Vascular Endothelial Growth Factor-A Expression and Contributes to Angiogenesis and Tumor Growth In vivo. Cancer Res. 2007, 67, 6700–6707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koumenis, C.; Bi, M.; Ye, J.; Feldman, D.; Koong, A.C. Hypoxia and the Unfolded Protein Response. Methods Enzymol. 2007, 435, 275–293. [Google Scholar] [CrossRef]
- Ma, Y.; Hendershot, L.M. The role of the unfolded protein response in tumour development: Friend or foe? Nat. Rev. Cancer 2004, 4, 966–977. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Patel, A.; Oshi, M.; Yan, L.; Matsuyama, R.; Endo, I.; Takabe, K. The Unfolded Protein Response Is Associated with Cancer Proliferation and Worse Survival in Hepatocellular Carcinoma. Cancers 2021, 13, 4443. https://doi.org/10.3390/cancers13174443
Patel A, Oshi M, Yan L, Matsuyama R, Endo I, Takabe K. The Unfolded Protein Response Is Associated with Cancer Proliferation and Worse Survival in Hepatocellular Carcinoma. Cancers. 2021; 13(17):4443. https://doi.org/10.3390/cancers13174443
Chicago/Turabian StylePatel, Ankit, Masanori Oshi, Li Yan, Ryusei Matsuyama, Itaru Endo, and Kazuaki Takabe. 2021. "The Unfolded Protein Response Is Associated with Cancer Proliferation and Worse Survival in Hepatocellular Carcinoma" Cancers 13, no. 17: 4443. https://doi.org/10.3390/cancers13174443






