Mutations in Epigenetic Regulation Genes in Gastric Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Tumor Samples
2.2. Mutation Screening by NGS
2.3. Sanger Sequencing
2.4. Statistical Analysis
2.5. Pathogenicity Prediction for Novel Mutations
3. Results
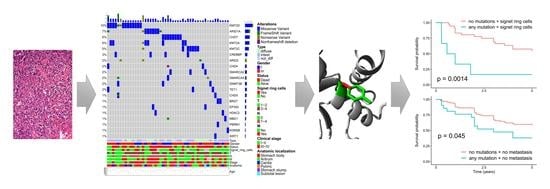
3.1. The Spectrum of Detected Somatic Mutations
3.2. Pathogenicity Analysis of the Detected Mutations by Prediction Programs
3.3. Analysis of Clinical Significance of Mutations in Epigenetic Regulation Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Cancer Genome Atlas Research Network. Comprehensive Molecular Characterization of Gastric Adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, M.; Marcato, P. Epigenetic Modifications as Biomarkers of Tumor Development, Therapy Response, and Recurrence across the Cancer Care Continuum. Cancers 2018, 10, 101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- You, J.S.; Jones, P.A. Cancer Genetics and Epigenetics: Two Sides of the Same Coin? Cancer Cell 2012, 22, 9–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nemtsova, M.V.; Mikhaylenko, D.S.; Kuznetsova, E.B.; Bykov, I.I.; Zamyatnin, A.A. Inactivation of Epigenetic Regulators Due to Mutations in Solid Tumors. Biochemistry 2020, 85, 735–748. [Google Scholar] [CrossRef] [PubMed]
- Smyth, E.C.; Verheij, M.; Allum, W.; Cunningham, D.; Cervantes, A.; Arnold, D. Gastric Cancer: ESMO Clinical Practice Guidelines for Diagnosis, Treatment and Follow-Up. Ann. Oncol. 2016, 27, v38–v49. [Google Scholar] [CrossRef] [PubMed]
- Scarpa, A.; Sikora, K.; Fassan, M.; Rachiglio, A.M.; Cappellesso, R.; Antonello, D.; Amato, E.; Mafficini, A.; Lambiase, M.; Esposito, C.; et al. Molecular Typing of Lung Adenocarcinoma on Cytological Samples Using a Multigene Next Generation Sequencing Panel. PLoS ONE 2013, 8, e80478. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional Annotation of Genetic Variants from High-Throughput Sequencing Data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative Genomics Viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef] [Green Version]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The CBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.-J.; Ahn, H.; Jeong, D.; Pak, M.; Moon, J.H.; Kim, S. Impact of Mutations in DNA Methylation Modification Genes on Genome-Wide Methylation Landscapes and Downstream Gene Activations in Pan-Cancer. BMC Med. Genom. 2020, 13, 27. [Google Scholar] [CrossRef] [PubMed]
- Siraj, A.K.; Pratheeshkumar, P.; Parvathareddy, S.K.; Bu, R.; Masoodi, T.; Iqbal, K.; Al-Rasheed, M.; Al-Dayel, F.; Al-Sobhi, S.S.; Alzahrani, A.S.; et al. Prognostic Significance of DNMT3A Alterations in Middle Eastern Papillary Thyroid Carcinoma. Eur. J. Cancer 2019, 117, 133–144. [Google Scholar] [CrossRef]
- Fagan, R.J.; Dingwall, A.K. COMPASS Ascending: Emerging Clues Regarding the Roles of MLL3/KMT2C and MLL2/KMT2D Proteins in Cancer. Cancer Lett. 2019, 458, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Herz, H.-M.; Hu, D.; Shilatifard, A. Enhancer Malfunction in Cancer. Mol. Cell 2014, 53, 859–866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.; Liu, Y.; Rappaport, A.R.; Kitzing, T.; Schultz, N.; Zhao, Z.; Shroff, A.S.; Dickins, R.A.; Vakoc, C.R.; Bradner, J.E.; et al. MLL3 Is a Haploinsufficient 7q Tumor Suppressor in Acute Myeloid Leukemia. Cancer Cell 2014, 25, 652–665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rampias, T.; Karagiannis, D.; Avgeris, M.; Polyzos, A.; Kokkalis, A.; Kanaki, Z.; Kousidou, E.; Tzetis, M.; Kanavakis, E.; Stravodimos, K.; et al. The Lysine-specific Methyltransferase KMT 2C/MLL 3 Regulates DNA Repair Components in Cancer. EMBO Rep. 2019, 20, 6821. [Google Scholar] [CrossRef] [PubMed]
- Poreba, E.; Lesniewicz, K.; Durzynska, J. Aberrant Activity of Histone–Lysine N-Methyltransferase 2 (KMT2) Complexes in Oncogenesis. Int. J. Mol. Sci. 2020, 21, 9340. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the CBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [Green Version]
- Ding, B.; Yan, L.; Zhang, Y.; Wang, Z.; Zhang, Y.; Xia, D.; Ye, Z.; Xu, H. Analysis of the Role of Mutations in the KMT 2D Histone Lysine Methyltransferase in Bladder Cancer. FEBS Open Bio 2019, 9, 693–706. [Google Scholar] [CrossRef] [Green Version]
- Manna, P.R.; Dyson, M.T.; Stocco, D.M. Role of Basic Leucine Zipper Proteins in Transcriptional Regulation of the Steroidogenic Acute Regulatory Protein Gene. Mol. Cell Endocrinol. 2009, 302, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Inamoto, I.; Shin, J.A. Peptide Therapeutics That Directly Target Transcription Factors. Pept. Sci. 2019, 111, e24048. [Google Scholar] [CrossRef] [Green Version]
- Mészáros, B.; Hajdu-Soltész, B.; Zeke, A.; Dosztányi, Z. Mutations of Intrinsically Disordered Protein Regions Can Drive Cancer but Lack Therapeutic Strategies. Biomolecules 2021, 11, 381. [Google Scholar] [CrossRef]
- Cho, S.-J.; Yoon, C.; Lee, J.H.; Chang, K.K.; Lin, J.-X.; Kim, Y.-H.; Kook, M.-C.; Aksoy, B.A.; Park, D.J.; Ashktorab, H.; et al. KMT2C Mutations in Diffuse-Type Gastric Adenocarcinoma Promote Epithelial-to-Mesenchymal Transition. Clin. Cancer Res. 2018, 24, 6556–6569. [Google Scholar] [CrossRef] [Green Version]
- Deng, Y.; Luo, S.; Deng, C.; Luo, T.; Yin, W.; Zhang, H.; Zhang, Y.; Zhang, X.; Lan, Y.; Ping, Y.; et al. Identifying Mutual Exclusivity across Cancer Genomes: Computational Approaches to Discover Genetic Interaction and Reveal Tumor Vulnerability. Brief. Bioinform. 2019, 20, 254–266. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.; Baik, B.; Park, T.; Nam, D. Powerful P-Value Combination Methods to Detect Incomplete Association. Sci. Rep. 2021, 11, 6980. [Google Scholar] [CrossRef] [PubMed]
- Dietlein, F.; Weghorn, D.; Taylor-Weiner, A.; Richters, A.; Reardon, B.; Liu, D.; Lander, E.S.; Van Allen, E.M.; Sunyaev, S.R. Identification of Cancer Driver Genes Based on Nucleotide Context. Nat. Genet. 2020, 52, 208–218. [Google Scholar] [CrossRef]
- Schallenberg, S.; Bork, J.; Essakly, A.; Alakus, H.; Buettner, R.; Hillmer, A.M.; Bruns, C.; Schroeder, W.; Zander, T.; Loeser, H.; et al. Loss of the SWI/SNF-ATPase Subunit Members SMARCF1 (ARID1A), SMARCA2 (BRM), SMARCA4 (BRG1) and SMARCB1 (INI1) in Oesophageal Adenocarcinoma. BMC Cancer 2020, 20, 12. [Google Scholar] [CrossRef]
- Ye, F.; Huang, J.; Wang, H.; Luo, C.; Zhao, K. Targeting Epigenetic Machinery: Emerging Novel Allosteric Inhibitors. Pharmacol. Ther. 2019, 204, 107406. [Google Scholar] [CrossRef] [PubMed]
- Peixoto, A.; Relvas-Santos, M.; Azevedo, R.; Santos, L.L.; Ferreira, J.A. Protein Glycosylation and Tumor Microenvironment Alterations Driving Cancer Hallmarks. Front. Oncol. 2019, 9, 380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, S.-C.; Ng, K.-F.; Chang, I.Y.-F.; Chang, C.-J.; Chao, Y.-C.; Chang, S.-C.; Chen, M.-C.; Yeh, T.-S.; Chen, T.-C. The Clinicopathological Significance of SWI/SNF Alterations in Gastric Cancer Is Associated with the Molecular Subtypes. PLoS ONE 2021, 16, e0245356. [Google Scholar] [CrossRef]
- Takeshima, H.; Niwa, T.; Takahashi, T.; Wakabayashi, M.; Yamashita, S.; Ando, T.; Inagawa, Y.; Taniguchi, H.; Katai, H.; Sugiyama, T.; et al. Frequent Involvement of Chromatin Remodeler Alterations in Gastric Field Cancerization. Cancer Lett. 2015, 357, 328–338. [Google Scholar] [CrossRef] [PubMed]
- Katona, B.W.; Rustgi, A.K. Gastric Cancer Genomics: Advances and Future Directions. Cell Mol. Gastroenterol. Hepatol. 2017, 3, 211–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Li, M.; Jiang, Z.; Wang, X. ARID1A Mutations Are Associated with Increased Immune Activity in Gastrointestinal Cancer. Cells 2019, 8, 678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamamoto, H.; Watanabe, Y.; Maehata, T.; Morita, R.; Yoshida, Y.; Oikawa, R.; Ishigooka, S.; Ozawa, S.-I.; Matsuo, Y.; Hosoya, K.; et al. An Updated Review of Gastric Cancer in the Next-Generation Sequencing Era: Insights from Bench to Bedside and Vice Versa. World J. Gastroenterol. 2014, 20, 3927–3937. [Google Scholar] [CrossRef]






| № | Gene/Mutation | Position According to hg19 | rsID | MAF (gnomAD Exomes) | ClinVar | # of Cases |
|---|---|---|---|---|---|---|
| 1 | ARID1A:exon1:c.G544A:p.A182T | chr1:27023438 | - | - | - | 1 |
| 2 | ARID1A:exon16:c.G3902A:p.S1301N | chr1:27100106 | rs989613588 | A = 4 × 10−6 | Not Reported | 1 |
| 3 | ARID1A:exon18:c.G4245C:p.Q1415H | chr1:27100963 | - | - | Not Reported | 1 |
| 4 | ARID1A:exon20:c.C6706T:p.R2236C | chr1:27107095 | rs763691986 | T = 2 × 10−5 | Not Reported | 1 |
| 5 | ARID1A:exon20:c.C5483G:p.S1828X | chr1:27105872 | - | - | Not Reported | 1 |
| 6 | ARID1A:exon20:c.C5129T:p.P1710L | chr1:27105518 | - | - | Not Reported | 1 |
| 7 | ARID1A:exon2:c.G1330A:p.G444S | chr1:27056334 | rs541301347 | A = 2.4 × 10−5 | Not Reported | 1 |
| 8 | ARID1A:exon20:c.5881_5887del:p.S1961fs | chr1:27106270 | - | - | Not Reported | 1 |
| 9 | ARID1A:exon18:c.4713_4714del:p.N1571fs | chr1:27101431 | - | - | Not Reported | 1 |
| 10 | ARID2:exon1:c.53_57del:p.A18fs | chr12:46123672 | - | - | Not Reported | 1 |
| 11 | ARID2:exon8:c.C820T:p.R274X | chr12:46230571 | - | - | Likely pathogenic | 1 |
| 12 | ARID2:exon8:c.C985T:p.Q329X | chr12:46230736 | - | - | Not Reported | 1 |
| 13 | SMARCA2:exon7:c.C734T:p.T245M | chr9:2192726 | rs753433101 | A = 3 × 10−5 | Not Reported | 1 |
| 14 | SMARCA2:exon7:c.A1256G:p.K419R | chr9:2056754 | - | - | Not Reported | 1 |
| 15 | SMARCA2:exon7:c.G1202A:p.R401H | chr9:2056700 | rs745500947 | T = 8 × 10−6 | Not Reported | 1 |
| 16 | SMARCA4:exon18:c.C2738T:p.P913L | chr19:11132522 | rs778175819 | T = 4 × 10−6 | Not Reported | 1 |
| 17 | SMARCA4:exon3:c.C430T:p.Q144X | chr19:11096939 | - | - | Not Reported | 1 |
| 18 | SMARCA4:exon3:c.583delC:p.P195fs | chr19:11097092 | - | - | Not Reported | 1 |
| 19 | KDM5B:exon17:c.C2392T:p.R798W | chr1:202711581 | rs1189771603 | A = 2.8 × 10−5 | Not Reported | 1 |
| 20 | CHD4:exon28:c.C4216T:p.R1406C | chr12:6692034 | - | - | Not Reported | 1 |
| 21 | CHD4:exon4:c.417_419del:p.139_140del | chr12:6711145 | rs71584865 | del(TCC)3 = 8 × 10−6 | Not Reported | 2 |
| 22 | CHD4:exon4:c.C421G:p.P141A | chr12:6710929 | - | - | Not Reported | 1 |
| 23 | CHD5:exon2:c.119delT:p.F40fs | chr1:6228298 | - | - | Not Reported | 1 |
| 24 | CHD5:exon15:c.C2257A:p.L753M | chr1:6202367 | - | - | Not Reported | 1 |
| 25 | CHD5:exon7:c.G910A:p.A304T | chr1:6211176 | rs768430028 | T = 1.3 × 10−4 | Not Reported | 2 |
| 26 | CHD5:exon2:c.A156T:p.K52N | chr1:6228261 | rs964095593 | A = 3 × 10−5 | Not Reported | 1 |
| 27 | CHD7:exon31:c.G6112A:p.D2038N | chr8:61765396 | rs747846723 | A = 6 × 10−5 | Uncertain Significance | 1 |
| 28 | CHD7:exon35:c.A7819G:p.S2607G | chr8:61773673 | rs1424434796 | G = 5 × 10−6 | Not Reported | 2 |
| 29 | CHD7:exon22:c.G4859A:p.R1620Q | chr8:61757431 | rs768497646 | A = 2.9 × 10−5 | Not Reported | 1 |
| 30 | CHD7:exon1:c.G749A:p.R250H | chr8:61654740 | rs767475667 | A = 6 × 10−5 | Not Reported | 1 |
| 31 | CHD7:exon22:c.G5017A:p.D1673N | chr8:61757589 | rs769563309 | A = 2.4 × 10−5 | Not Reported | 1 |
| 32 | CHD7:exon29:c.G5828A:p.R1943Q | chr8:61764740 | rs753723769 | A = 2.8 × 10−5 | Uncertain Significance | 1 |
| 33 | EP300:exon3:c.A752G:p.N251S | chr22:41521890 | rs142009367 | G = 2.2 × 10−4 | Benign | 2 |
| 34 | HDAC2:exon13:c.C1430A:p.T477N | chr6:114262878 | rs1341257540 | - | Not Reported | 1 |
| 35 | HDAC2:exon6:c.G511A:p.V171I | chr6:114274569 | - | - | Not Reported | 1 |
| 36 | CREBBP:exon30:c.C6335T:p.P2112L | chr16:3778599 | rs587783512 | A = 1.6 × 10−5 | Uncertain Significance | 1 |
| 37 | CREBBP:exon2:c.C458T:p.P153L | chr16:3900638 | rs146538907 | A = 3.35 × 10−4 | Likely Benign | 1 |
| 38 | CREBBP:exon3:c.C922T:p.P308S | chr16:3860657 | - | - | Not Reported | 1 |
| 39 | CREBBP:exon18:c.A3421C:p.K1141Q | chr16:3807998 | - | - | Not Reported | 1 |
| 40 | BRD7:exon7:c.A871G:p.S291G | chr16:50368638 | rs200218240 | C = 1.6 × 10−4 | Not Reported | 1 |
| 41 | BRD7:exon5:c.C571T:p.Q191X | chr16:50383954 | - | - | Not Reported | 1 |
| 42 | PBRM1:exon17:c.C2032T:p.R678C | chr3:52643864 | rs1422119249 | - | Not Reported | 1 |
| 43 | KMT2A:exon27:c.T9737C:p.I3246T | chr11:118376344 | rs1259638674 | C = 3 × 10−5 | Not Reported | 1 |
| 44 | KMT2A:exon27:c.C9947T:p.A3316V | chr11:118376554 | rs201447376 | T = 1.3 × 10−4 | Not Reported | 1 |
| 45 | KMT2A:exon27:c.G10181A:p.G3394E | chr11:118376788 | rs782460936 | A = 3 × 10−5 | Not Reported | 1 |
| 46 | KMT2A:exon21:c.G5726A:p.W1909X | chr11:118368712 | - | - | Not Reported | 1 |
| 47 | KMT2A:exon27:c.G9247A:p.V3083I | chr11:118375854 | - | - | Not Reported | 1 |
| 48 | KMT2A:exon30:c.A10984G:p.S3662G | chr11:118380746 | rs201724738 | G = 5.6 × 10−5 | Not Reported | 1 |
| 49 | KMT2A:exon36:c.G11903A:p.R3968Q | chr11:118392871 | rs369182428 | A = 3 × 10−5 | Not Reported | 1 |
| 50 | KMT2A:exon30:c.A10975G:p.S3659G | chr11:118380746 | rs201724738 | G = 1 × 10−4 | Not Reported | 1 |
| 51 | KMT2C:exon43:c.G9987A:p.M3329I | chr7:151860675 | rs200804156 | T = 6 × 10−5 | Not Reported | 1 |
| 52 | KMT2D:exon10:c.T2284C:p.S762P | chr12:49445182 | - | - | Not Reported | 1 |
| 53 | KMT2C:exon10:c.C1384T:p.Q462X | chr7:151949716 | - | - | Not Reported | 1 |
| 54 | KMT2C:exon36:c.C6836T:p.P2279L | chr7:151878109 | rs150844259 | A = 6 × 10−5 | Not Reported | 1 |
| 55 | KMT2C:exon36:c.A6919G:p.R2307G | chr7:151878026 | rs772283102 | C = 2.4 × 10−5 | Not Reported | 1 |
| 56 | KMT2C:exon36:c.G5858A:p.C1953Y | chr7:151879087 | - | - | Not Reported | 1 |
| 57 | KMT2C:exon18:c.A2917G:p.R973G | chr7:151927067 | rs60244562 | - | Not Reported | 2 |
| 58 | KMT2C:exon18:c.G2877A:p.M959I | chr7:151927107 | rs4024402 | - | Not Reported | 1 |
| 59 | KMT2D:exon31:c.A7954C:p.M2652L | chr12:49433599 | rs147706410 | G = 4.5 × 10−4 | Likely Benign | 1 |
| 60 | KMT2D:exon41:c.G13780C:p.A4594P | chr12:49424443 | rs545972414 | G = 2.5 × 10−4 | Not Reported | 2 |
| 61 | KMT2D:exon28:c.C5921T:p.T1974M | chr12:49436060 | rs777415982 | A = 6 × 10−5 | Not Reported | 1 |
| 62 | KMT2D:exon48:c.G14893A:p.A4965T | chr12:49420856 | rs200747934 | T = 1.53 × 10−4 | Uncertain Significance | 2 |
| 63 | KMT2D:exon39:c.G12686A:p.R4229Q | chr12:49445262 | rs753607446 | T = 3.2 × 10−5 | Not Reported | 1 |
| 64 | KMT2D:exon31:c.T7829C:p.L2610P | chr12:49433724 | rs200998047 | G = 1.85 × 10−4 | Uncertain Significance | 1 |
| 65 | KMT2D:exon31:c.6673delG:p.E2225fs | chr12:49434880 | - | - | Not Reported | 1 |
| 66 | KMT2D:exon31:c.C7516T:p.L2506F | chr12:49434037 | rs749670394 | A = 4 × 10−6 | Not Reported | 1 |
| 67 | KMT2D:exon39:c.C11179T:p.R3727C | chr12:49427309 | rs566069597 | A = 3.6 × 10−5 | Not Reported | 1 |
| 68 | KMT2D:exon10:c.C1628T:p.S543L | chr12:49445838 | rs776242478 | A = 8 × 10−6 | Not Reported | 1 |
| 69 | KMT2D:exon31:c.G8026T:p.E2676X | chr12:49433527 | - | - | Not Reported | 1 |
| 70 | KMT2D:exon31:c.C7136T:p.A2379V | chr12:49434417 | rs200842315 | A = 6 × 10−5 | Likely benign | 1 |
| 71 | KMT2D:exon39:c.C11495G:p.S3832C | chr12:49426993 | - | - | Not Reported | 1 |
| 72 | MBD1:exon9:c.G796A:p.E266K | chr18:47801382 | rs142015383 | T = 5 × 10−4 | Not Reported | 1 |
| 73 | MBD1:exon8:c.734delC:p.P245fs | chr18:47801527 | rs1173827934 | - | Not Reported | 1 |
| 74 | TET1:exon4:c.G2407A:p.A803T | chr10:70404893 | rs765094207 | A = 2.4 × 10−5 | Not Reported | 1 |
| 75 | TET1:exon2:c.G320A:p.R107Q | chr10:70332415 | rs1419371452 | A = 8 × 10−6 | Not Reported | 1 |
| 76 | TET1:exon4:c.G3476A:p.R1159Q | chr10:70405962 | rs140289196 | A = 2.2 × 10−4 | Not Reported | 1 |
| 77 | DNMT3B:exon19:c.G2138A:p.R713Q | chr20:31390243 | rs747182299 | A = 3 × 10−5 | Likely Pathogenic | 1 |
| 78 | DNMT3B:exon6:c.A680G:p.Y227C | chr20:31379501 | - | - | Not Reported | 1 |
| 79 | DNMT3B:exon17:c.G1855A:p.E619K | chr20:31388054 | rs576798456 | A = 8 × 10−6 | Not Reported | 1 |
| Parameters | Total Cases | Mut− | Mut+ | p-Value * | |
|---|---|---|---|---|---|
| 135 | 86 | 49 | |||
| Age | <50 | 35 | 22 | 13 | 1 |
| >50 | 100 | 64 | 36 | ||
| Sex | m | 83 | 51 | 32 | 0.58 |
| f | 52 | 35 | 17 | ||
| Survival status | Alive | 59 | 42 | 17 | 0.14 |
| Dead | 76 | 44 | 32 | ||
| 5-year survival status | Alive | 60 | 42 | 18 | 0.19 |
| Dead | 69 | 40 | 29 | ||
| <5 years follow-up | 6 | 4 | 2 | ||
| T | T1-2 | 49 | 33 | 16 | 0.46 |
| T3-4 | 84 | 51 | 33 | ||
| is | 2 | 2 | - | ||
| N | N0 | 56 | 37 | 19 | 0.71 |
| N1-3 | 79 | 49 | 30 | ||
| M | M0 | 89 | 59 | 30 | 0.55 |
| M1 | 42 | 25 | 17 | ||
| Unknown | 4 | - | - | ||
| Lauren classification | Diffuse | 59 | 41 | 18 | 0.26 |
| Intestinal | 64 | 38 | 26 | ||
| Not Differentiate | 12 | 7 | 5 | ||
| Stage | I-II | 57 | 39 | 18 | 0.36 |
| III-IV | 76 | 45 | 31 | ||
| Unknown | 2 | - | - | ||
| Anatomical localization | Stomach body | 75 | 52 | 23 | 0.10 |
| Antrum | 36 | 22 | 14 | ||
| Cardia | 20 | 11 | 8 | ||
| Stomach stump | 3 | 0 | 3 | ||
| Subtotal lesion | 1 | 0 | 1 | ||
| Pyloric | 1 | 1 | 0 | ||
| Signet ring cells | yes | 42 | 29 | 13 | 0.69 |
| no | 89 | 57 | 32 | ||
| Unknown | 4 | 0 | 4 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nemtsova, M.V.; Kalinkin, A.I.; Kuznetsova, E.B.; Bure, I.V.; Alekseeva, E.A.; Bykov, I.I.; Khorobrykh, T.V.; Mikhaylenko, D.S.; Tanas, A.S.; Strelnikov, V.V. Mutations in Epigenetic Regulation Genes in Gastric Cancer. Cancers 2021, 13, 4586. https://doi.org/10.3390/cancers13184586
Nemtsova MV, Kalinkin AI, Kuznetsova EB, Bure IV, Alekseeva EA, Bykov II, Khorobrykh TV, Mikhaylenko DS, Tanas AS, Strelnikov VV. Mutations in Epigenetic Regulation Genes in Gastric Cancer. Cancers. 2021; 13(18):4586. https://doi.org/10.3390/cancers13184586
Chicago/Turabian StyleNemtsova, Marina V., Alexey I. Kalinkin, Ekaterina B. Kuznetsova, Irina V. Bure, Ekaterina A. Alekseeva, Igor I. Bykov, Tatiana V. Khorobrykh, Dmitry S. Mikhaylenko, Alexander S. Tanas, and Vladimir V. Strelnikov. 2021. "Mutations in Epigenetic Regulation Genes in Gastric Cancer" Cancers 13, no. 18: 4586. https://doi.org/10.3390/cancers13184586