Hybrid Formation and Fusion of Cancer Cells In Vitro and In Vivo
Abstract
:Simple Summary
Abstract
1. Introduction
2. Cell Fusion in General
3. DNA Reorganization during Cancer Cell Fusion
3.1. DNA Transfer by Horizontal or Lateral Gene Transfer
3.2. Role of Extracellular Vesicles (EVs) in Cancer Cell Fusion
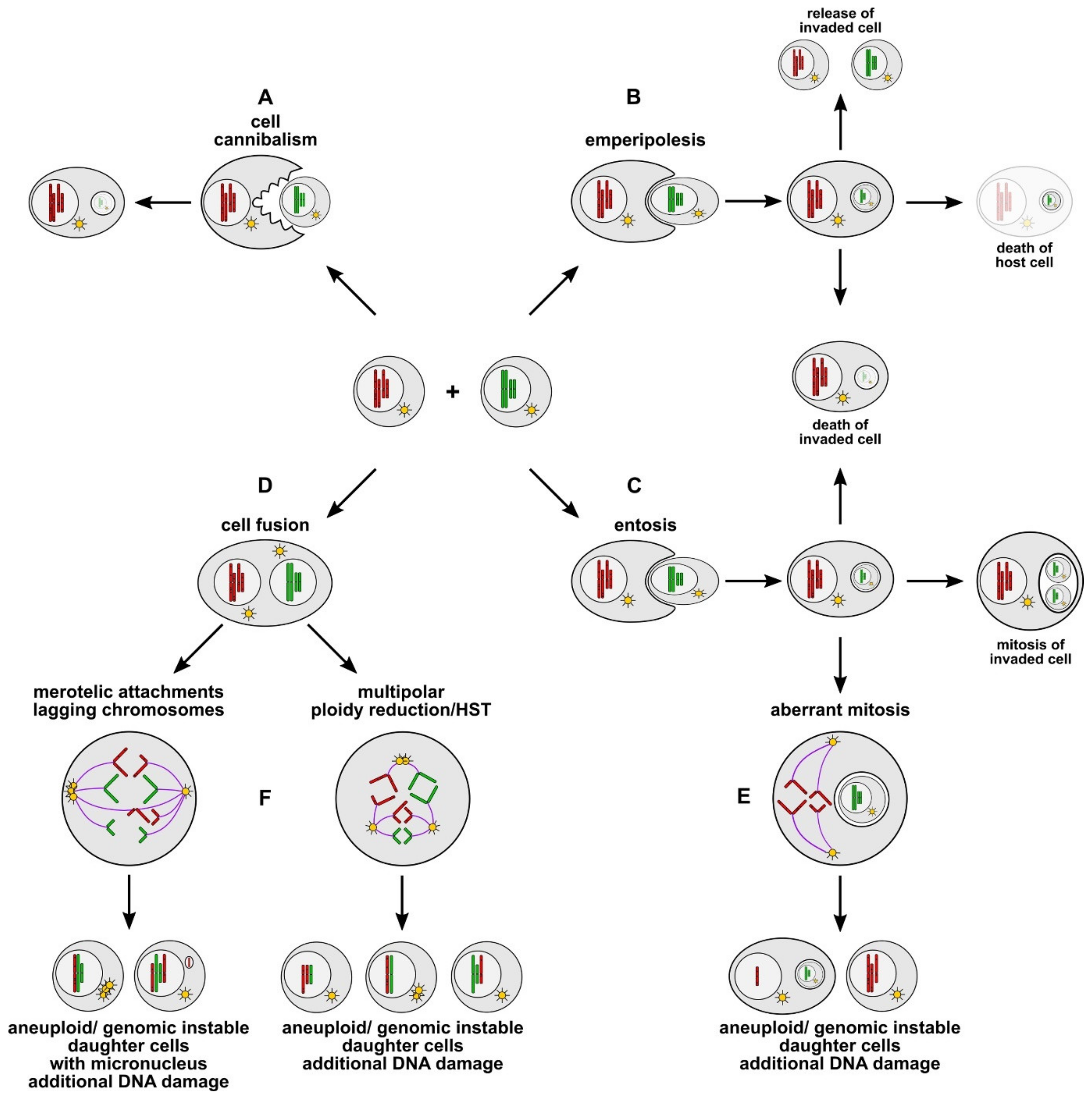
4. Cell Cannibalism, Entosis, and Emperipolesis
4.1. Cell Cannibalism
4.2. Entosis and Emperipolesis
5. Cell Fusion in Tumors
5.1. Evidence for Cancer Cell Fusion by In Vitro Studies
5.2. Cancer Cell Fusion as a Rapid Process and In Vivo Studies
5.3. Indirect Evidence for Cell Fusion by Clinical Observations of Hybrid Cells in Human Tumors
5.4. Direct Evidence for Cell Fusion by Clinical Observations of Hybrid Cells in Human Tumors
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| BMT | bone marrow transplantation |
| CAFs | cancer-associated fibroblasts |
| HGT/LGT | horizontal gene transfer/lateral gene transfer |
| HST | heterokaryon-to-synkaryon transition |
| MSC | mesenchymal stroma-/stem-like cells |
| NSCLC | non-small cell lung cancer |
| PHPP | pre-hybrid preparation process |
| PHSP | post-hybrid selection process |
| PR | ploidy reductions |
| PS | phosphatidylserine |
| TNF-α | tumor necrosis factor-α |
References
- Aguilar, P.S.; Baylies, M.K.; Fleissner, A.; Helming, L.; Inoue, N.; Podbilewicz, B.; Wang, H.; Wong, M. Genetic basis of cell-cell fusion mechanisms. Trends Genet. 2013, 29, 427–437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dittmar, T.; Zänker, K.S. Cell Fusion in Health and Disease: Volume I; Springer: Dordrecht, The Netherlands, 2011; Volume 1. [Google Scholar]
- Dittmar, T.; Zanker, K.S. Cell Fusion in Health and Disease: Volume II; Springer: Dordrecht, The Netherlands, 2011; Volume 2. [Google Scholar]
- Hernandez, J.M.; Podbilewicz, B. The hallmarks of cell-cell fusion. Development 2017, 144, 4481–4495. [Google Scholar] [CrossRef] [Green Version]
- Larsson, L.I.; Bjerregaard, B.; Talts, J.F. Cell fusions in mammals. Histochem. Cell Biol. 2008, 129, 551–561. [Google Scholar] [CrossRef] [Green Version]
- Perez-Vargas, J.; Krey, T.; Valansi, C.; Avinoam, O.; Haouz, A.; Jamin, M.; Raveh-Barak, H.; Podbilewicz, B.; Rey, F.A. Structural basis of eukaryotic cell-cell fusion. Cell 2014, 157, 407–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willkomm, L.; Bloch, W. State of the art in cell-cell fusion. Methods Mol. Biol. 2015, 1313, 1–19. [Google Scholar] [CrossRef]
- Brukman, N.G.; Uygur, B.; Podbilewicz, B.; Chernomordik, L.V. How cells fuse. J. Cell Biol. 2019, 218, 1436–1451. [Google Scholar] [CrossRef] [Green Version]
- Helming, L.; Gordon, S. Molecular mediators of macrophage fusion. Trends Cell Biol. 2009, 19, 514–522. [Google Scholar] [CrossRef] [PubMed]
- Frendo, J.L.; Cronier, L.; Bertin, G.; Guibourdenche, J.; Vidaud, M.; Evain-Brion, D.; Malassine, A. Involvement of connexin 43 in human trophoblast cell fusion and differentiation. J. Cell Sci. 2003, 116, 3413–3421. [Google Scholar] [CrossRef] [Green Version]
- Getsios, S.; MacCalman, C.D. Cadherin-11 modulates the terminal differentiation and fusion of human trophoblastic cells in vitro. Dev. Biol. 2003, 257, 41–54. [Google Scholar] [CrossRef] [Green Version]
- Huppertz, B.; Gauster, M. Trophoblast fusion. Adv. Exp. Med. Biol. 2011, 713, 81–95. [Google Scholar]
- Vargas, A.; Moreau, J.; Landry, S.; LeBellego, F.; Toufaily, C.; Rassart, E.; Lafond, J.; Barbeau, B. Syncytin-2 plays an important role in the fusion of human trophoblast cells. J. Mol. Biol. 2009, 392, 301–318. [Google Scholar] [CrossRef]
- Abmayr, S.M.; Pavlath, G.K. Myoblast fusion: Lessons from flies and mice. Development 2012, 139, 641–656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simionescu, A.; Pavlath, G.K. Molecular mechanisms of myoblast fusion across species. Adv. Exp. Med. Biol. 2011, 713, 113–135. [Google Scholar]
- Hindi, S.M.; Shin, J.; Gallot, Y.S.; Straughn, A.R.; Simionescu-Bankston, A.; Hindi, L.; Xiong, G.; Friedland, R.P.; Kumar, A. MyD88 promotes myoblast fusion in a cell-autonomous manner. Nat. Commun. 2017, 8, 1624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pizza, F.X.; Martin, R.A.; Springer, E.M.; Leffler, M.S.; Woelmer, B.R.; Recker, I.J.; Leaman, D.W. Intercellular adhesion molecule-1 augments myoblast adhesion and fusion through homophilic trans-interactions. Sci. Rep. 2017, 7, 5094. [Google Scholar] [CrossRef] [Green Version]
- Vignery, A. Osteoclasts and giant cells: Macrophage-macrophage fusion mechanism. Int. J. Exp. Pathol. 2000, 81, 291–304. [Google Scholar] [CrossRef] [PubMed]
- Vignery, A. Macrophage fusion: The making of osteoclasts and giant cells. J. Exp. Med. 2005, 202, 337–340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melzer, C.; Ohe, J.V.; Hass, R. Altered Tumor Plasticity after Different Cancer Cell Fusions with MSC. Int. J. Mol. Sci. 2020, 21, 8347. [Google Scholar] [CrossRef]
- Hass, R.; von der Ohe, J.; Ungefroren, H. Impact of the Tumor Microenvironment on Tumor Heterogeneity and Consequences for Cancer Cell Plasticity and Stemness. Cancers 2020, 12, 3716. [Google Scholar] [CrossRef]
- Moller, A.M.; Delaisse, J.M.; Soe, K. Osteoclast Fusion: Time-Lapse Reveals Involvement of CD47 and Syncytin-1 at Different Stages of Nuclearity. J. Cell. Physiol. 2017, 232, 1396–1403. [Google Scholar] [CrossRef]
- Soe, K. Osteoclast Fusion: Physiological Regulation of Multinucleation through Heterogeneity-Potential Implications for Drug Sensitivity. Int. J. Mol. Sci. 2020, 21, 7717. [Google Scholar] [CrossRef] [PubMed]
- Teitelbaum, S.L. Osteoclasts: What do they do and how do they do it? Am. J. Pathol. 2007, 170, 427–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bjerkvig, R.; Tysnes, B.B.; Aboody, K.S.; Najbauer, J.; Terzis, A.J. Opinion: The origin of the cancer stem cell: Current controversies and new insights. Nat. Rev. 2005, 5, 899–904. [Google Scholar] [CrossRef] [PubMed]
- Duncan, A.W.; Hanlon Newell, A.E.; Bi, W.; Finegold, M.J.; Olson, S.B.; Beaudet, A.L.; Grompe, M. Aneuploidy as a mechanism for stress-induced liver adaptation. J. Clin. Investig. 2012, 122, 3307–3315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duncan, A.W.; Hickey, R.D.; Paulk, N.K.; Culberson, A.J.; Olson, S.B.; Finegold, M.J.; Grompe, M. Ploidy reductions in murine fusion-derived hepatocytes. PLoS Genet. 2009, 5, e1000385. [Google Scholar] [CrossRef] [Green Version]
- Matsumoto, T.; Wakefield, L.; Tarlow, B.D.; Grompe, M. In Vivo Lineage Tracing of Polyploid Hepatocytes Reveals Extensive Proliferation during Liver Regeneration. Cell Stem Cell 2020, 26, 34–47.e3. [Google Scholar] [CrossRef]
- Skinner, A.M.; Grompe, M.; Kurre, P. Intra-hematopoietic cell fusion as a source of somatic variation in the hematopoietic system. J. Cell Sci. 2012, 125, 2837–2843. [Google Scholar]
- Zhou, X.; Merchak, K.; Lee, W.; Grande, J.P.; Cascalho, M.; Platt, J.L. Cell Fusion Connects Oncogenesis with Tumor Evolution. Am. J. Pathol. 2015, 185, 2049–2060. [Google Scholar] [CrossRef]
- Delespaul, L.; Gelabert, C.; Lesluyes, T.; Le Guellec, S.; Perot, G.; Leroy, L.; Baud, J.; Merle, C.; Lartigue, L.; Chibon, F. Cell-cell fusion of mesenchymal cells with distinct differentiations triggers genomic and transcriptomic remodelling toward tumour aggressiveness. Sci. Rep. 2020, 10, 21634. [Google Scholar] [CrossRef]
- Delespaul, L.; Merle, C.; Lesluyes, T.; Lagarde, P.; Le Guellec, S.; Perot, G.; Baud, J.; Carlotti, M.; Danet, C.; Fevre, M.; et al. Fusion-mediated chromosomal instability promotes aneuploidy patterns that resemble human tumors. Oncogene 2019, 38, 6083–6094. [Google Scholar] [CrossRef]
- Duncan, A.W.; Taylor, M.H.; Hickey, R.D.; Hanlon Newell, A.E.; Lenzi, M.L.; Olson, S.B.; Finegold, M.J.; Grompe, M. The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature 2010, 467, 707–710. [Google Scholar] [CrossRef] [Green Version]
- Ganem, N.J.; Godinho, S.A.; Pellman, D. A mechanism linking extra centrosomes to chromosomal instability. Nature 2009, 460, 278–282. [Google Scholar] [CrossRef] [Green Version]
- Godinho, S.A.; Kwon, M.; Pellman, D. Centrosomes and cancer: How cancer cells divide with too many centrosomes. Cancer Metastasis Rev. 2009, 28, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Holland, A.J.; Cleveland, D.W. Boveri revisited: Chromosomal instability, aneuploidy and tumorigenesis. Nat. Rev. Mol. Cell Biol. 2009, 10, 478–487. [Google Scholar] [CrossRef] [Green Version]
- Chunduri, N.K.; Storchova, Z. The diverse consequences of aneuploidy. Nat. Cell Biol. 2019, 21, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Passerini, V.; Ozeri-Galai, E.; de Pagter, M.S.; Donnelly, N.; Schmalbrock, S.; Kloosterman, W.P.; Kerem, B.; Storchova, Z. The presence of extra chromosomes leads to genomic instability. Nat. Commun. 2016, 7, 10754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, X.; Platt, J.L. Molecular and cellular mechanisms of Mammalian cell fusion. Adv. Exp. Med. Biol. 2011, 713, 33–64. [Google Scholar]
- Podbilewicz, B. Virus and cell fusion mechanisms. Annu. Rev. Cell Dev. Biol. 2014, 30, 111–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huppertz, B.; Bartz, C.; Kokozidou, M. Trophoblast fusion: Fusogenic proteins, syncytins and ADAMs, and other prerequisites for syncytial fusion. Micron 2006, 37, 509–517. [Google Scholar] [CrossRef]
- Malassine, A.; Handschuh, K.; Tsatsaris, V.; Gerbaud, P.; Cheynet, V.; Oriol, G.; Mallet, F.; Evain-Brion, D. Expression of HERV-W Env glycoprotein (syncytin) in the extravillous trophoblast of first trimester human placenta. Placenta 2005, 26, 556–562. [Google Scholar] [CrossRef]
- Mi, S.; Lee, X.; Li, X.; Veldman, G.M.; Finnerty, H.; Racie, L.; LaVallie, E.; Tang, X.Y.; Edouard, P.; Howes, S.; et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 2000, 403, 785–789. [Google Scholar] [CrossRef]
- Muir, A.; Lever, A.M.; Moffett, A. Human endogenous retrovirus-W envelope (syncytin) is expressed in both villous and extravillous trophoblast populations. J. Gen. Virol. 2006, 87, 2067–2071. [Google Scholar] [CrossRef] [Green Version]
- Millay, D.P.; O’Rourke, J.R.; Sutherland, L.B.; Bezprozvannaya, S.; Shelton, J.M.; Bassel-Duby, R.; Olson, E.N. Myomaker is a membrane activator of myoblast fusion and muscle formation. Nature 2013, 499, 301–305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leikina, E.; Gamage, D.G.; Prasad, V.; Goykhberg, J.; Crowe, M.; Diao, J.; Kozlov, M.M.; Chernomordik, L.V.; Millay, D.P. Myomaker and Myomerger Work Independently to Control Distinct Steps of Membrane Remodeling during Myoblast Fusion. Dev. Cell 2018, 46, 767–780.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bi, P.; Ramirez-Martinez, A.; Li, H.; Cannavino, J.; McAnally, J.R.; Shelton, J.M.; Sanchez-Ortiz, E.; Bassel-Duby, R.; Olson, E.N. Control of muscle formation by the fusogenic micropeptide myomixer. Science 2017, 356, 323–327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quinn, M.E.; Goh, Q.; Kurosaka, M.; Gamage, D.G.; Petrany, M.J.; Prasad, V.; Millay, D.P. Myomerger induces fusion of non-fusogenic cells and is required for skeletal muscle development. Nat. Commun. 2017, 8, 15665. [Google Scholar] [CrossRef] [Green Version]
- Inoue, N.; Ikawa, M.; Isotani, A.; Okabe, M. The immunoglobulin superfamily protein Izumo is required for sperm to fuse with eggs. Nature 2005, 434, 234–238. [Google Scholar] [CrossRef]
- Chalbi, M.; Barraud-Lange, V.; Ravaux, B.; Howan, K.; Rodriguez, N.; Soule, P.; Ndzoudi, A.; Boucheix, C.; Rubinstein, E.; Wolf, J.P.; et al. Binding of sperm protein Izumo1 and its egg receptor Juno drives Cd9 accumulation in the intercellular contact area prior to fusion during mammalian fertilization. Development 2014, 141, 3732–3739. [Google Scholar] [CrossRef] [Green Version]
- Bianchi, E.; Wright, G.J. Cross-species fertilization: The hamster egg receptor, Juno, binds the human sperm ligand, Izumo1. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20140101. [Google Scholar] [CrossRef] [Green Version]
- Kato, K.; Satouh, Y.; Nishimasu, H.; Kurabayashi, A.; Morita, J.; Fujihara, Y.; Oji, A.; Ishitani, R.; Ikawa, M.; Nureki, O. Structural and functional insights into IZUMO1 recognition by JUNO in mammalian fertilization. Nat. Commun. 2016, 7, 12198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soe, K.; Andersen, T.L.; Hobolt-Pedersen, A.S.; Bjerregaard, B.; Larsson, L.I.; Delaisse, J.M. Involvement of human endogenous retroviral syncytin-1 in human osteoclast fusion. Bone 2011, 48, 837–846. [Google Scholar] [CrossRef]
- Chernomordik, L.V.; Zimmerberg, J.; Kozlov, M.M. Membranes of the world unite! J. Cell Biol. 2006, 175, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Jahn, R.; Lang, T.; Sudhof, T.C. Membrane fusion. Cell 2003, 112, 519–533. [Google Scholar] [CrossRef] [Green Version]
- Ogle, B.M.; Cascalho, M.; Platt, J.L. Biological implications of cell fusion. Nat. Rev. Mol. Cell Biol. 2005, 6, 567–575. [Google Scholar] [CrossRef] [PubMed]
- Whitlock, J.M.; Chernomordik, L.V. Flagging fusion: Phosphatidylserine signaling in cell-cell fusion. J. Biol. Chem. 2021, 296, 100411. [Google Scholar] [CrossRef]
- Melzer, C.; von der Ohe, J.; Hass, R. Involvement of Actin Cytoskeletal Components in Breast Cancer Cell Fusion with Human Mesenchymal Stroma/Stem-Like Cells. Int. J. Mol. Sci. 2019, 20, 876. [Google Scholar] [CrossRef] [Green Version]
- Bevers, E.M.; Williamson, P.L. Getting to the Outer Leaflet: Physiology of Phosphatidylserine Exposure at the Plasma Membrane. Physiol. Rev. 2016, 96, 605–645. [Google Scholar] [CrossRef]
- Whitlock, J.M.; Yu, K.; Cui, Y.Y.; Hartzell, H.C. Anoctamin 5/TMEM16E facilitates muscle precursor cell fusion. J. Gen. Physiol. 2018, 150, 1498–1509. [Google Scholar] [CrossRef] [Green Version]
- Griffin, D.A.; Johnson, R.W.; Whitlock, J.M.; Pozsgai, E.R.; Heller, K.N.; Grose, W.E.; Arnold, W.D.; Sahenk, Z.; Hartzell, H.C.; Rodino-Klapac, L.R. Defective membrane fusion and repair in Anoctamin5-deficient muscular dystrophy. Hum. Mol. Genet. 2016, 25, 1900–1911. [Google Scholar] [CrossRef] [Green Version]
- Verma, S.K.; Leikina, E.; Melikov, K.; Gebert, C.; Kram, V.; Young, M.F.; Uygur, B.; Chernomordik, L.V. Cell-surface phosphatidylserine regulates osteoclast precursor fusion. J. Biol. Chem. 2018, 293, 254–270. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Le, T.; Grabau, R.; Mohseni, Z.; Kim, H.; Natale, D.R.; Feng, L.; Pan, H.; Yang, H. TMEM16F phospholipid scramblase mediates trophoblast fusion and placental development. Sci. Adv. 2020, 6, eaba0310. [Google Scholar] [CrossRef]
- Hochreiter-Hufford, A.E.; Lee, C.S.; Kinchen, J.M.; Sokolowski, J.D.; Arandjelovic, S.; Call, J.A.; Klibanov, A.L.; Yan, Z.; Mandell, J.W.; Ravichandran, K.S. Phosphatidylserine receptor BAI1 and apoptotic cells as new promoters of myoblast fusion. Nature 2013, 497, 263–267. [Google Scholar] [CrossRef] [Green Version]
- Park, D.; Tosello-Trampont, A.C.; Elliott, M.R.; Lu, M.; Haney, L.B.; Ma, Z.; Klibanov, A.L.; Mandell, J.W.; Ravichandran, K.S. BAI1 is an engulfment receptor for apoptotic cells upstream of the ELMO/Dock180/Rac module. Nature 2007, 450, 430–434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.Y.; Yun, Y.; Kim, I.S. CD36 is required for myoblast fusion during myogenic differentiation. Biochem. Biophys. Res. Commun. 2012, 427, 705–710. [Google Scholar] [CrossRef] [PubMed]
- Helming, L.; Winter, J.; Gordon, S. The scavenger receptor CD36 plays a role in cytokine-induced macrophage fusion. J. Cell Sci. 2009, 122, 453–459. [Google Scholar] [CrossRef] [Green Version]
- Aguirre, L.A.; Montalban-Hernandez, K.; Avendano-Ortiz, J.; Marin, E.; Lozano, R.; Toledano, V.; Sanchez-Maroto, L.; Terron, V.; Valentin, J.; Pulido, E.; et al. Tumor stem cells fuse with monocytes to form highly invasive tumor-hybrid cells. Oncoimmunology 2020, 9, 1773204. [Google Scholar] [CrossRef] [PubMed]
- Davies, P.S.; Powell, A.E.; Swain, J.R.; Wong, M.H. Inflammation and proliferation act together to mediate intestinal cell fusion. PLoS ONE 2009, 4, e6530. [Google Scholar] [CrossRef] [PubMed]
- Johansson, C.B.; Youssef, S.; Koleckar, K.; Holbrook, C.; Doyonnas, R.; Corbel, S.Y.; Steinman, L.; Rossi, F.M.; Blau, H.M. Extensive fusion of haematopoietic cells with Purkinje neurons in response to chronic inflammation. Nat. Cell Biol. 2008, 10, 575–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nygren, J.M.; Liuba, K.; Breitbach, M.; Stott, S.; Thoren, L.; Roell, W.; Geisen, C.; Sasse, P.; Kirik, D.; Bjorklund, A.; et al. Myeloid and lymphoid contribution to non-haematopoietic lineages through irradiation-induced heterotypic cell fusion. Nat. Cell Biol. 2008, 10, 584–592. [Google Scholar] [CrossRef] [PubMed]
- Hotokezaka, H.; Sakai, E.; Ohara, N.; Hotokezaka, Y.; Gonzales, C.; Matsuo, K.; Fujimura, Y.; Yoshida, N.; Nakayama, K. Molecular analysis of RANKL-independent cell fusion of osteoclast-like cells induced by TNF-alpha, lipopolysaccharide, or peptidoglycan. J. Cell. Biochem. 2007, 101, 122–134. [Google Scholar] [CrossRef] [PubMed]
- Melzer, C.; von der Ohe, J.; Hass, R. In vitro fusion of normal and neoplastic breast epithelial cells with human mesenchymal stroma/stem cells (MSC) partially involves TNF receptor signaling. Stem Cells 2018, 36, 977–989. [Google Scholar] [CrossRef] [Green Version]
- Skokos, E.A.; Charokopos, A.; Khan, K.; Wanjala, J.; Kyriakides, T.R. Lack of TNF-alpha-induced MMP-9 production and abnormal E-cadherin redistribution associated with compromised fusion in MCP-1-null macrophages. Am. J. Pathol. 2011, 178, 2311–2321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weiler, J.; Dittmar, T. Minocycline impairs TNF-alpha-induced cell fusion of M13SV1-Cre cells with MDA-MB-435-pFDR1 cells by suppressing NF-kappaB transcriptional activity and its induction of target-gene expression of fusion-relevant factors. Cell Commun. Signal. 2019, 17, 71. [Google Scholar] [CrossRef] [Green Version]
- Weiler, J.; Mohr, M.; Zanker, K.S.; Dittmar, T. Matrix metalloproteinase-9 (MMP9) is involved in the TNF-alpha-induced fusion of human M13SV1-Cre breast epithelial cells and human MDA-MB-435-pFDR1 cancer cells. Cell Commun. Signal. 2018, 16, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, T.L.; Wang, M.; Xu, Z.; Huang, C.M.; Zhou, X.C.; Jiang, E.H.; Zhao, X.P.; Song, Y.; Song, K.; Shao, Z.; et al. Up-regulation of syncytin-1 contributes to TNF-alpha-enhanced fusion between OSCC and HUVECs partly via Wnt/beta-catenin-dependent pathway. Sci. Rep. 2017, 7, 40983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, K.; Zhu, F.; Zhang, H.Z.; Shang, Z.J. Tumor necrosis factor-alpha enhanced fusions between oral squamous cell carcinoma cells and endothelial cells via VCAM-1/VLA-4 pathway. Exp. Cell Res. 2012, 318, 1707–1715. [Google Scholar] [CrossRef]
- Sinkovics, J.G. Horizontal Gene Transfers with or without Cell Fusions in All Categories of the Living Matter. Cell Fusion Health Dis. 2011, 714, 5–89. [Google Scholar]
- Emamalipour, M.; Seidi, K.; Zununi Vahed, S.; Jahanban-Esfahlan, A.; Jaymand, M.; Majdi, H.; Amoozgar, Z.; Chitkushev, L.T.; Javaheri, T.; Jahanban-Esfahlan, R.; et al. Horizontal Gene Transfer: From Evolutionary Flexibility to Disease Progression. Front. Cell Dev. Biol. 2020, 8, 229. [Google Scholar] [CrossRef]
- Husnik, F.; McCutcheon, J.P. Functional horizontal gene transfer from bacteria to eukaryotes. Nat. Rev. Microbiol. 2018, 16, 67–79. [Google Scholar] [CrossRef]
- Cairns, J.; Ruokolainen, L.; Hultman, J.; Tamminen, M.; Virta, M.; Hiltunen, T. Ecology determines how low antibiotic concentration impacts community composition and horizontal transfer of resistance genes. Commun. Biol. 2018, 1, 35. [Google Scholar] [CrossRef]
- Bronkhorst, A.J.; Ungerer, V.; Holdenrieder, S. The emerging role of cell-free DNA as a molecular marker for cancer management. Biomol. Detect. Quantif. 2019, 17, 100087. [Google Scholar] [CrossRef] [PubMed]
- De la Taille, A.; Chen, M.W.; Burchardt, M.; Chopin, D.K.; Buttyan, R. Apoptotic conversion: Evidence for exchange of genetic information between prostate cancer cells mediated by apoptosis. Cancer Res. 1999, 59, 5461–5463. [Google Scholar] [PubMed]
- Serrano-Heras, G.; Dominguez-Berzosa, C.; Collantes, E.; Guadalajara, H.; Garcia-Olmo, D.; Garcia-Olmo, D.C. NIH-3T3 fibroblasts cultured with plasma from colorectal cancer patients generate poorly differentiated carcinomas in mice. Cancer Lett. 2012, 316, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Olmo, D.C.; Dominguez, C.; Garcia-Arranz, M.; Anker, P.; Stroun, M.; Garcia-Verdugo, J.M.; Garcia-Olmo, D. Cell-free nucleic acids circulating in the plasma of colorectal cancer patients induce the oncogenic transformation of susceptible cultured cells. Cancer Res. 2010, 70, 560–567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bergsmedh, A.; Szeles, A.; Henriksson, M.; Bratt, A.; Folkman, M.J.; Spetz, A.L.; Holmgren, L. Horizontal transfer of oncogenes by uptake of apoptotic bodies. Proc. Natl. Acad. Sci. USA 2001, 98, 6407–6411. [Google Scholar] [CrossRef] [Green Version]
- Holmgren, L.; Bergsmedh, A.; Spetz, A.L. Horizontal transfer of DNA by the uptake of apoptotic bodies. Vox Sang. 2002, 83 (Suppl. 1), 305–306. [Google Scholar] [CrossRef]
- Ehnfors, J.; Kost-Alimova, M.; Persson, N.L.; Bergsmedh, A.; Castro, J.; Levchenko-Tegnebratt, T.; Yang, L.; Panaretakis, T.; Holmgren, L. Horizontal transfer of tumor DNA to endothelial cells in vivo. Cell Death Differ. 2009, 16, 749–757. [Google Scholar] [CrossRef] [Green Version]
- Van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef] [PubMed]
- Yanez-Mo, M.; Siljander, P.R.; Andreu, Z.; Zavec, A.B.; Borras, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colombo, M.; Raposo, G.; Thery, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Han, Y.; Ren, H.; Chen, C.; He, D.; Zhou, L.; Eisner, G.M.; Asico, L.D.; Jose, P.A.; Zeng, C. Extracellular vesicle-mediated transfer of donor genomic DNA to recipient cells is a novel mechanism for genetic influence between cells. J. Mol. Cell Biol. 2013, 5, 227–238. [Google Scholar] [CrossRef] [Green Version]
- Cai, J.; Wu, G.; Tan, X.; Han, Y.; Chen, C.; Li, C.; Wang, N.; Zou, X.; Chen, X.; Zhou, F.; et al. Transferred BCR/ABL DNA from K562 extracellular vesicles causes chronic myeloid leukemia in immunodeficient mice. PLoS ONE 2014, 9, e105200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fischer, S.; Cornils, K.; Speiseder, T.; Badbaran, A.; Reimer, R.; Indenbirken, D.; Grundhoff, A.; Brunswig-Spickenheier, B.; Alawi, M.; Lange, C. Indication of Horizontal DNA Gene Transfer by Extracellular Vesicles. PLoS ONE 2016, 11, e0163665. [Google Scholar] [CrossRef] [Green Version]
- Lian, Q.; Xu, J.; Yan, S.; Huang, M.; Ding, H.; Sun, X.; Bi, A.; Ding, J.; Sun, B.; Geng, M. Chemotherapy-induced intestinal inflammatory responses are mediated by exosome secretion of double-strand DNA via AIM2 inflammasome activation. Cell Res. 2017, 27, 784–800. [Google Scholar] [CrossRef] [PubMed]
- Record, M. Intercellular communication by exosomes in placenta: A possible role in cell fusion? Placenta 2014, 35, 297–302. [Google Scholar] [CrossRef]
- Uygur, B.; Melikov, K.; Arakelyan, A.; Margolis, L.B.; Chernomordik, L.V. Syncytin 1 dependent horizontal transfer of marker genes from retrovirally transduced cells. Sci. Rep. 2019, 9, 17637. [Google Scholar] [CrossRef] [PubMed]
- Howcroft, T.K.; Zhang, H.G.; Dhodapkar, M.; Mohla, S. Vesicle transfer and cell fusion: Emerging concepts of cell-cell communication in the tumor microenvironment. Cancer Biol. Ther. 2011, 12, 159–164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fais, S.; Overholtzer, M. Cell-in-cell phenomena in cancer. Nat. Rev. 2018, 18, 758–766. [Google Scholar] [CrossRef] [PubMed]
- Overholtzer, M.; Brugge, J.S. The cell biology of cell-in-cell structures. Nat. Rev. Mol. Cell Biol. 2008, 9, 796–809. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, Y.; Li, J.; Li, L.; Zhu, H.; Chen, H.; Kong, R.; Wang, G.; Wang, Y.; Hu, J.; et al. Cell-in-Cell Phenomenon and Its Relationship With Tumor Microenvironment and Tumor Progression: A Review. Front. Cell Dev. Biol. 2019, 7, 311. [Google Scholar] [CrossRef] [Green Version]
- Janssen, A.; Medema, R.H. Entosis: Aneuploidy by invasion. Nat. Cell Biol. 2011, 13, 199–201. [Google Scholar] [CrossRef]
- Xia, P.; Wang, S.; Guo, Z.; Yao, X. Emperipolesis, entosis and beyond: Dance with fate. Cell Res. 2008, 18, 705–707. [Google Scholar] [CrossRef] [PubMed]
- Krajcovic, M.; Overholtzer, M. Mechanisms of ploidy increase in human cancers: A new role for cell cannibalism. Cancer Res. 2012, 72, 1596–1601. [Google Scholar] [CrossRef] [Green Version]
- He, M.F.; Wang, S.; Wang, Y.; Wang, X.N. Modeling cell-in-cell structure into its biological significance. Cell Death Dis. 2013, 4, e630. [Google Scholar] [CrossRef] [Green Version]
- Sarode, S.C.; Sarode, G.S. Neutrophil-tumor cell cannibalism in oral squamous cell carcinoma. J. Oral Pathol. Med. 2014, 43, 454–458. [Google Scholar] [CrossRef]
- Sarode, G.S.; Sarode, S.C.; Karmarkar, S. Complex cannibalism: An unusual finding in oral squamous cell carcinoma. Oral Oncol. 2012, 48, e4–e6. [Google Scholar] [CrossRef]
- Caruso, R.A.; Muda, A.O.; Bersiga, A.; Rigoli, L.; Inferrera, C. Morphological evidence of neutrophil-tumor cell phagocytosis (cannibalism) in human gastric adenocarcinomas. Ultrastruct. Pathol. 2002, 26, 315–321. [Google Scholar] [CrossRef]
- Cano, C.E.; Sandi, M.J.; Hamidi, T.; Calvo, E.L.; Turrini, O.; Bartholin, L.; Loncle, C.; Secq, V.; Garcia, S.; Lomberk, G.; et al. Homotypic cell cannibalism, a cell-death process regulated by the nuclear protein 1, opposes to metastasis in pancreatic cancer. EMBO Mol. Med. 2012, 4, 964–979. [Google Scholar] [CrossRef] [PubMed]
- Arya, P.; Khalbuss, W.E.; Monaco, S.E.; Pantanowitz, L. Salivary duct carcinoma with striking neutrophil-tumor cell cannibalism. Cytojournal 2011, 8, 15. [Google Scholar] [CrossRef]
- Kinoshita, M.; Matsuda, Y.; Arai, T.; Soejima, Y.; Sawabe, M.; Honma, N. Cytological diagnostic clues in poorly differentiated squamous cell carcinomas of the breast: Streaming arrangement, necrotic background, nucleolar enlargement and cannibalism of cancer cells. Cytopathology 2018, 29, 22–27. [Google Scholar] [CrossRef] [PubMed]
- Lugini, L.; Matarrese, P.; Tinari, A.; Lozupone, F.; Federici, C.; Iessi, E.; Gentile, M.; Luciani, F.; Parmiani, G.; Rivoltini, L.; et al. Cannibalism of live lymphocytes by human metastatic but not primary melanoma cells. Cancer Res. 2006, 66, 3629–3638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartosh, T.J.; Ullah, M.; Zeitouni, S.; Beaver, J.; Prockop, D.J. Cancer cells enter dormancy after cannibalizing mesenchymal stem/stromal cells (MSCs). Proc. Natl. Acad. Sci. USA 2016, 113, E6447–E6456. [Google Scholar] [CrossRef] [Green Version]
- Lozupone, F.; Fais, S. Cancer Cell Cannibalism: A Primeval Option to Survive. Curr. Mol. Med. 2015, 15, 836–841. [Google Scholar] [CrossRef]
- Sharma, N.; Dey, P. Cell cannibalism and cancer. Diagn. Cytopathol. 2011, 39, 229–233. [Google Scholar] [CrossRef] [PubMed]
- Huber, V.; Fais, S.; Iero, M.; Lugini, L.; Canese, P.; Squarcina, P.; Zaccheddu, A.; Colone, M.; Arancia, G.; Gentile, M.; et al. Human colorectal cancer cells induce T-cell death through release of proapoptotic microvesicles: Role in immune escape. Gastroenterology 2005, 128, 1796–1804. [Google Scholar] [CrossRef] [PubMed]
- Caruso, R.A.; Fedele, F.; Finocchiaro, G.; Arena, G.; Venuti, A. Neutrophil-tumor cell phagocytosis (cannibalism) in human tumors: An update and literature review. Exp. Oncol. 2012, 34, 306–311. [Google Scholar]
- Sierro, F.; Tay, S.S.; Warren, A.; Le Couteur, D.G.; McCaughan, G.W.; Bowen, D.G.; Bertolino, P. Suicidal emperipolesis: A process leading to cell-in-cell structures, T cell clearance and immune homeostasis. Curr. Mol. Med. 2015, 15, 819–827. [Google Scholar] [CrossRef]
- Chen, Y.H.; Wang, S.; He, M.F.; Wang, Y.; Zhao, H.; Zhu, H.Y.; Yu, X.M.; Ma, J.; Che, X.J.; Wang, J.F.; et al. Prevalence of heterotypic tumor/immune cell-in-cell structure in vitro and in vivo leading to formation of aneuploidy. PLoS ONE 2013, 8, e59418. [Google Scholar] [CrossRef] [Green Version]
- Janssen, A.; Medema, R.H. Genetic instability: Tipping the balance. Oncogene 2013, 32, 4459–4470. [Google Scholar] [CrossRef] [Green Version]
- Overholtzer, M.; Mailleux, A.A.; Mouneimne, G.; Normand, G.; Schnitt, S.J.; King, R.W.; Cibas, E.S.; Brugge, J.S. A nonapoptotic cell death process, entosis, that occurs by cell-in-cell invasion. Cell 2007, 131, 966–979. [Google Scholar] [CrossRef] [Green Version]
- Sottile, F.; Aulicino, F.; Theka, I.; Cosma, M.P. Mesenchymal stem cells generate distinct functional hybrids in vitro via cell fusion or entosis. Sci. Rep. 2016, 6, 36863. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, M.N.; Pillat, M.M.; Motaln, H.; Ulrich, H.; Lah, T.T. Kinin-B1 Receptor Stimulation Promotes Invasion and is Involved in Cell-Cell Interaction of Co-Cultured Glioblastoma and Mesenchymal Stem Cells. Sci. Rep. 2018, 8, 1299. [Google Scholar] [CrossRef] [Green Version]
- Miroshnychenko, D.; Baratchart, E.; Ferrall-Fairbanks, M.C.; Velde, R.V.; Laurie, M.A.; Bui, M.M.; Tan, A.C.; Altrock, P.M.; Basanta, D.; Marusyk, A. Spontaneous cell fusions as a mechanism of parasexual recombination in tumour cell populations. Nat. Ecol. Evol. 2021, 5, 379–391. [Google Scholar] [CrossRef]
- Aichel, O. Über Zellverschmelzung mit quantitativ abnormer Chromosomenverteilung als Ursache der Geschwulstbildung. In Vorträge und Aufsätze über Entwicklungsmechanik der Organismen; Roux, W., Ed.; Wilhelm Engelmann: Leipzig, Germany, 1911; pp. 1–115. [Google Scholar]
- Berndt, B.; Haverkampf, S.; Reith, G.; Keil, S.; Niggemann, B.; Zanker, K.S.; Dittmar, T. Fusion of CCL21 non-migratory active breast epithelial and breast cancer cells give rise to CCL21 migratory active tumor hybrid cell lines. PLoS ONE 2013, 8, e63711. [Google Scholar] [CrossRef] [Green Version]
- Dittmar, T.; Nagler, C.; Schwitalla, S.; Reith, G.; Niggemann, B.; Zanker, K.S. Recurrence cancer stem cells--made by cell fusion? Med. Hypotheses 2009, 73, 542–547. [Google Scholar] [CrossRef]
- Dittmar, T.; Schwitalla, S.; Seidel, J.; Haverkampf, S.; Reith, G.; Meyer-Staeckling, S.; Brandt, B.H.; Niggemann, B.; Zanker, K.S. Characterization of hybrid cells derived from spontaneous fusion events between breast epithelial cells exhibiting stem-like characteristics and breast cancer cells. Clin. Exp. Metastasis 2011, 28, 75–90. [Google Scholar] [CrossRef]
- Duelli, D.M.; Padilla-Nash, H.M.; Berman, D.; Murphy, K.M.; Ried, T.; Lazebnik, Y. A virus causes cancer by inducing massive chromosomal instability through cell fusion. Curr. Biol. 2007, 17, 431–437. [Google Scholar] [CrossRef] [Green Version]
- Jacobsen, B.M.; Harrell, J.C.; Jedlicka, P.; Borges, V.F.; Varella-Garcia, M.; Horwitz, K.B. Spontaneous fusion with, and transformation of mouse stroma by, malignant human breast cancer epithelium. Cancer Res. 2006, 66, 8274–8279. [Google Scholar] [CrossRef] [Green Version]
- Lindstrom, A.; Midtbo, K.; Arnesson, L.G.; Garvin, S.; Shabo, I. Fusion between M2-macrophages and cancer cells results in a subpopulation of radioresistant cells with enhanced DNA-repair capacity. Oncotarget 2017, 8, 51370–51386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lizier, M.; Anselmo, A.; Mantero, S.; Ficara, F.; Paulis, M.; Vezzoni, P.; Lucchini, F.; Pacchiana, G. Fusion between cancer cells and macrophages occurs in a murine model of spontaneous neu+ breast cancer without increasing its metastatic potential. Oncotarget 2016, 7, 60793–60806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, F.; Liu, T.; Wang, J.; Li, J.; Ma, P.; Ding, H.; Feng, G.; Lin, D.; Xu, Y.; Yang, K. Bone marrow mesenchymal stem cells participate in prostate carcinogenesis and promote growth of prostate cancer by cell fusion in vivo. Oncotarget 2016, 7, 30924–30934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melzer, C.; von der Ohe, J.; Hass, R. In Vivo Cell Fusion between Mesenchymal Stroma/Stem-Like Cells and Breast Cancer Cells. Cancers 2019, 11, 185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noubissi, F.K.; Harkness, T.; Alexander, C.M.; Ogle, B.M. Apoptosis-induced cancer cell fusion: A mechanism of breast cancer metastasis. FASEB J. 2015, 29, 4036–4045. [Google Scholar] [CrossRef]
- Powell, A.E.; Anderson, E.C.; Davies, P.S.; Silk, A.D.; Pelz, C.; Impey, S.; Wong, M.H. Fusion between Intestinal epithelial cells and macrophages in a cancer context results in nuclear reprogramming. Cancer Res. 2011, 71, 1497–1505. [Google Scholar] [CrossRef] [Green Version]
- Ramakrishnan, M.; Mathur, S.R.; Mukhopadhyay, A. Fusion derived epithelial cancer cells express hematopoietic markers and contribute to stem cell and migratory phenotype in ovarian carcinoma. Cancer Res. 2013, 73, 5360–5370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shabo, I.; Svanvik, J.; Lindstrom, A.; Lechertier, T.; Trabulo, S.; Hulit, J.; Sparey, T.; Pawelek, J. Roles of cell fusion, hybridization and polyploid cell formation in cancer metastasis. World J. Clin. Oncol. 2020, 11, 121–135. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Subedee, A.; Bloushtain-Qimron, N.; Savova, V.; Krzystanek, M.; Li, L.; Marusyk, A.; Tabassum, D.P.; Zak, A.; Flacker, M.J.; et al. Somatic Cell Fusions Reveal Extensive Heterogeneity in Basal-like Breast Cancer. Cell Rep. 2015, 11, 1549–1563. [Google Scholar] [CrossRef] [Green Version]
- Suetsugu, A.; Matsumoto, T.; Hasegawa, K.; Nakamura, M.; Kunisada, T.; Shimizu, M.; Saji, S.; Moriwaki, H.; Bouvet, M.; Hoffman, R.M. Color-coded Live Imaging of Heterokaryon Formation and Nuclear Fusion of Hybridizing Cancer Cells. Anticancer Res. 2016, 36, 3827–3831. [Google Scholar]
- Wang, R.; Lewis, M.S.; Lyu, J.; Zhau, H.E.; Pandol, S.J.; Chung, L.W.K. Cancer-stromal cell fusion as revealed by fluorescence protein tracking. Prostate 2019, 80, 274–283. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.; Sun, X.; Wang, C.Y.; Hu, P.; Chu, C.Y.; Liu, S.; Zhau, H.E.; Chung, L.W. Spontaneous cancer-stromal cell fusion as a mechanism of prostate cancer androgen-independent progression. PLoS ONE 2012, 7, e42653. [Google Scholar] [CrossRef] [Green Version]
- Yao, J.; Zhang, L.; Hu, L.; Guo, B.; Hu, X.; Borjigin, U.; Wei, Z.; Chen, Y.; Lv, M.; Lau, J.T.; et al. Tumorigenic potential is restored during differentiation in fusion-reprogrammed cancer cells. Cell Death Dis. 2016, 7, e2314. [Google Scholar] [CrossRef]
- Yin, L.; Hu, P.; Shi, X.; Qian, W.; Zhau, H.E.; Pandol, S.J.; Lewis, M.S.; Chung, L.W.K.; Wang, R. Cancer cell’s neuroendocrine feature can be acquired through cell-cell fusion during cancer-neural stem cell interaction. Sci. Rep. 2020, 10, 1216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, C.; Zhang, Y.; Park, S.C.; Eun, J.R.; Nguyen, N.T.; Tschudy-Seney, B.; Jung, Y.J.; Theise, N.D.; Zern, M.A.; Duan, Y. CD34 Liver Cancer Stem Cells Were Formed by Fusion of Hepatobiliary Stem/Progenitor Cells with Hematopoietic Precursor-Derived Myeloid Intermediates. Stem Cells Dev. 2015, 24, 2467–2478. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.N.; Kong, C.F.; Zhao, D.; Cong, X.L.; Wang, S.S.; Ma, L.; Huang, Y.H. Fusion with mesenchymal stem cells differentially affects tumorigenic and metastatic abilities of lung cancer cells. J. Cell. Physiol. 2018, 3570–3582. [Google Scholar] [CrossRef]
- Ding, J.; Jin, W.; Chen, C.; Shao, Z.; Wu, J. Tumor associated macrophage x cancer cell hybrids may acquire cancer stem cell properties in breast cancer. PLoS ONE 2012, 7, e41942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukhopadhyay, K.D.; Bandyopadhyay, A.; Chang, T.T.; Elkahloun, A.G.; Cornell, J.E.; Yang, J.; Goins, B.A.; Yeh, I.T.; Sun, L.Z. Isolation and characterization of a metastatic hybrid cell line generated by ER negative and ER positive breast cancer cells in mouse bone marrow. PLoS ONE 2011, 6, e20473. [Google Scholar] [CrossRef] [Green Version]
- Nagler, C.; Zanker, K.S.; Dittmar, T. Murine breast-cancer-cell/mesenchymal-stem-cell hybrids exhibit enhanced drug resistance to different cytostatic drugs. J. Stem Cells Regen. Med. 2010, 6, 133. [Google Scholar]
- Rappa, G.; Mercapide, J.; Lorico, A. Spontaneous Formation of Tumorigenic Hybrids between Breast Cancer and Multipotent Stromal Cells Is a Source of Tumor Heterogeneity. Am. J. Pathol. 2012, 180, 2504–2515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, M.H.; Gao, X.; Luo, D.; Zhou, X.D.; Xiong, W.; Liu, G.X. EMT and acquisition of stem cell-like properties are involved in spontaneous formation of tumorigenic hybrids between lung cancer and bone marrow-derived mesenchymal stem cells. PLoS ONE 2014, 9, e87893. [Google Scholar] [CrossRef] [PubMed]
- Melzer, C.; von der Ohe, J.; Hass, R. Enhanced metastatic capacity of breast cancer cells after interaction and hybrid formation with mesenchymal stroma/stem cells (MSC). Cell Commun. Signal. 2018, 16, 2. [Google Scholar] [CrossRef] [Green Version]
- Melzer, C.; von der Ohe, J.; Hass, R. MSC stimulate ovarian tumor growth during intercellular communication but reduce tumorigenicity after fusion with ovarian cancer cells. Cell Commun. Signal. 2018, 16, 67. [Google Scholar] [CrossRef] [Green Version]
- Melzer, C.; Yang, Y.; Hass, R. Interaction of MSC with tumor cells. Cell Commun. Signal. 2016, 14, 20. [Google Scholar] [CrossRef] [Green Version]
- Chakraborty, A.K.; Sodi, S.; Rachkovsky, M.; Kolesnikova, N.; Platt, J.T.; Bolognia, J.L.; Pawelek, J.M. A spontaneous murine melanoma lung metastasis comprised of host x tumor hybrids. Cancer Res. 2000, 60, 2512–2519. [Google Scholar] [PubMed]
- Goldenberg, D.M.; Pavia, R.A.; Tsao, M.C. In vivo hybridisation of human tumour and normal hamster cells. Nature 1974, 250, 649–651. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Kang, Y. Efficient acquisition of dual metastasis organotropism to bone and lung through stable spontaneous fusion between MDA-MB-231 variants. Proc. Natl. Acad. Sci. USA 2009, 106, 9385–9390. [Google Scholar] [CrossRef] [Green Version]
- Mekler, L.B.; Drize, O.B.; Osechinskii, I.V.; Shliankevich, M.A. Transformation of a normal differentiated cell of an adult organism, induced by the fusion of this cell with another normal cell of the same organism but with different organ or tissue specificity. Vestn. Akad. Meditsinskikh Nauk. SSSR 1971, 26, 75–80. [Google Scholar]
- Rachkovsky, M.; Sodi, S.; Chakraborty, A.; Avissar, Y.; Bolognia, J.; McNiff, J.M.; Platt, J.; Bermudes, D.; Pawelek, J. Melanoma x macrophage hybrids with enhanced metastatic potential. Clin. Exp. Metastasis 1998, 16, 299–312. [Google Scholar] [CrossRef]
- Van Golen, K.L.; Risin, S.; Staroselsky, A.; Berger, D.; Tainsky, M.A.; Pathak, S.; Price, J.E. Predominance of the metastatic phenotype in hybrids formed by fusion of mouse and human melanoma clones. Clin. Exp. Metastasis 1996, 14, 95–106. [Google Scholar] [CrossRef]
- Yan, B.; Wang, J.; Liu, L. Chemotherapy promotes tumour cell hybridization in vivo. Tumour Biol. 2015, 37, 5025–5030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, G.C.; Ye, Q.H.; Dong, Q.Z.; Ren, N.; Jia, H.L.; Qin, L.X. Mesenchymal stem cells seldomly fuse with hepatocellular carcinoma cells and are mainly distributed in the tumor stroma in mouse models. Oncol. Rep. 2013, 29, 713–719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gast, C.E.; Silk, A.D.; Zarour, L.; Riegler, L.; Burkhart, J.G.; Gustafson, K.T.; Parappilly, M.S.; Roh-Johnson, M.; Goodman, J.R.; Olson, B.; et al. Cell fusion potentiates tumor heterogeneity and reveals circulating hybrid cells that correlate with stage and survival. Sci. Adv. 2018, 4, eaat7828. [Google Scholar] [CrossRef] [Green Version]
- Martin-Padura, I.; Marighetti, P.; Gregato, G.; Agliano, A.; Malazzi, O.; Mancuso, P.; Pruneri, G.; Viale, A.; Bertolini, F. Spontaneous cell fusion of acute leukemia cells and macrophages observed in cells with leukemic potential. Neoplasia 2012, 14, 1057–1066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tal, A.; Tal, R.; Shaikh, S.; Gidicsin, S.; Mamillapalli, R.; Taylor, H.S. Characterization of Cell Fusion in an Experimental Mouse Model of Endometriosis. Biol. Reprod. 2018, 100, 390–397. [Google Scholar] [CrossRef]
- Carloni, V.; Mazzocca, A.; Mello, T.; Galli, A.; Capaccioli, S. Cell fusion promotes chemoresistance in metastatic colon carcinoma. Oncogene 2013, 32, 2649–2660. [Google Scholar] [CrossRef] [Green Version]
- Strick, R.; Ackermann, S.; Langbein, M.; Swiatek, J.; Schubert, S.W.; Hashemolhosseini, S.; Koscheck, T.; Fasching, P.A.; Schild, R.L.; Beckmann, M.W.; et al. Proliferation and cell-cell fusion of endometrial carcinoma are induced by the human endogenous retroviral Syncytin-1 and regulated by TGF-beta. J. Mol. Med. 2007, 85, 23–38. [Google Scholar] [CrossRef]
- Li, M.; Li, X.; Zhao, L.; Zhou, J.; Cheng, Y.; Xu, B.; Wang, J.; Wei, L. Spontaneous formation of tumorigenic hybrids between human omental adipose-derived stromal cells and endometrial cancer cells increased motility and heterogeneity of cancer cells. Cell Cycle 2019, 18, 320–332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kerbel, R.S.; Lagarde, A.E.; Dennis, J.W.; Donaghue, T.P. Spontaneous fusion in vivo between normal host and tumor cells: Possible contribution to tumor progression and metastasis studied with a lectin-resistant mutant tumor. Mol. Cell. Biol. 1983, 3, 523–538. [Google Scholar] [CrossRef] [Green Version]
- Duelli, D.M.; Hearn, S.; Myers, M.P.; Lazebnik, Y. A primate virus generates transformed human cells by fusion. J. Cell Biol. 2005, 171, 493–503. [Google Scholar] [CrossRef] [PubMed]
- Lartigue, L.; Merle, C.; Lagarde, P.; Delespaul, L.; Lesluyes, T.; Le Guellec, S.; Perot, G.; Leroy, L.; Coindre, J.M.; Chibon, F. Genome remodeling upon mesenchymal tumor cell fusion contributes to tumor progression and metastatic spread. Oncogene 2020, 39, 4198–4211. [Google Scholar] [CrossRef]
- Wakeling, W.F.; Greetham, J.; Devlin, L.M.; Bennett, D.C. Suppression of properties associated with malignancy in murine melanoma-melanocyte hybrid cells. Br. J. Cancer 1992, 65, 529–537. [Google Scholar] [CrossRef] [Green Version]
- Spees, J.L.; Olson, S.D.; Ylostalo, J.; Lynch, P.J.; Smith, J.; Perry, A.; Peister, A.; Wang, M.Y.; Prockop, D.J. Differentiation, cell fusion, and nuclear fusion during ex vivo repair of epithelium by human adult stem cells from bone marrow stroma. Proc. Natl. Acad. Sci. USA 2003, 100, 2397–2402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wakeling, W.F.; Greetham, J.; Bennett, D.C. Efficient spontaneous fusion between some co-cultured cells, especially murine melanoma cells. Cell Biol. Int. 1994, 18, 207–210. [Google Scholar] [CrossRef]
- Wang, R.; Chen, S.; Li, C.; Ng, K.T.; Kong, C.W.; Cheng, J.; Cheng, S.H.; Li, R.A.; Lo, C.M.; Man, K.; et al. Fusion with stem cell makes the hepatocellular carcinoma cells similar to liver tumor-initiating cells. BMC Cancer 2016, 16, 56. [Google Scholar] [CrossRef] [Green Version]
- Mohr, M.; Tosun, S.; Arnold, W.H.; Edenhofer, F.; Zanker, K.S.; Dittmar, T. Quantification of cell fusion events human breast cancer cells and breast epithelial cells using a Cre-LoxP-based double fluorescence reporter system. Cell. Mol. Life Sci. 2015, 72, 3769–3782. [Google Scholar] [CrossRef] [PubMed]
- Fahlbusch, S.S.; Keil, S.; Epplen, J.T.; Zanker, K.S.; Dittmar, T. Comparison of hybrid clones derived from human breast epithelial cells and three different cancer cell lines regarding in vitro cancer stem/initiating cell properties. BMC Cancer 2020, 20, 446. [Google Scholar] [CrossRef] [PubMed]
- Uygur, B.; Leikina, E.; Melikov, K.; Villasmil, R.; Verma, S.K.; Vary, C.P.H.; Chernomordik, L.V. Interactions with Muscle Cells Boost Fusion, Stemness, and Drug Resistance of Prostate Cancer Cells. Mol. Cancer Res. 2019, 17, 806–820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larsson, L.I.; Holck, S.; Christensen, I.J. Prognostic role of syncytin expression in breast cancer. Hum. Pathol. 2007, 38, 726–731. [Google Scholar] [CrossRef]
- Shabo, I.; Midtbo, K.; Andersson, H.; Akerlund, E.; Olsson, H.; Wegman, P.; Gunnarsson, C.; Lindstrom, A. Macrophage traits in cancer cells are induced by macrophage-cancer cell fusion and cannot be explained by cellular interaction. BMC Cancer 2015, 15, 922. [Google Scholar] [CrossRef] [Green Version]
- Shabo, I.; Olsson, H.; Stal, O.; Svanvik, J. Breast cancer expression of DAP12 is associated with skeletal and liver metastases and poor survival. Clin. Breast Cancer 2013, 13, 371–377. [Google Scholar] [CrossRef]
- Shabo, I.; Olsson, H.; Sun, X.F.; Svanvik, J. Expression of the macrophage antigen CD163 in rectal cancer cells is associated with early local recurrence and reduced survival time. Int. J. Cancer 2009, 125, 1826–1831. [Google Scholar] [CrossRef]
- Shabo, I.; Stal, O.; Olsson, H.; Dore, S.; Svanvik, J. Breast cancer expression of CD163, a macrophage scavenger receptor, is related to early distant recurrence and reduced patient survival. Int. J. Cancer 2008, 123, 780–786. [Google Scholar] [CrossRef]
- LaBerge, G.S.; Duvall, E.; Grasmick, Z.; Haedicke, K.; Pawelek, J. A Melanoma Lymph Node Metastasis with a Donor-Patient Hybrid Genome following Bone Marrow Transplantation: A Second Case of Leucocyte-Tumor Cell Hybridization in Cancer Metastasis. PLoS ONE 2017, 12, e0168581. [Google Scholar] [CrossRef]
- Lazova, R.; Laberge, G.S.; Duvall, E.; Spoelstra, N.; Klump, V.; Sznol, M.; Cooper, D.; Spritz, R.A.; Chang, J.T.; Pawelek, J.M. A Melanoma Brain Metastasis with a Donor-Patient Hybrid Genome following Bone Marrow Transplantation: First Evidence for Fusion in Human Cancer. PLoS ONE 2013, 8, e66731. [Google Scholar] [CrossRef]
- Andersen, T.L.; Boissy, P.; Sondergaard, T.E.; Kupisiewicz, K.; Plesner, T.; Rasmussen, T.; Haaber, J.; Kolvraa, S.; Delaisse, J.M. Osteoclast nuclei of myeloma patients show chromosome translocations specific for the myeloma cell clone: A new type of cancer-host partnership? J. Pathol. 2007, 211, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Andersen, T.L.; Soe, K.; Sondergaard, T.E.; Plesner, T.; Delaisse, J.M. Myeloma cell-induced disruption of bone remodelling compartments leads to osteolytic lesions and generation of osteoclast-myeloma hybrid cells. Br. J. Haematol. 2010, 148, 551–561. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, A.; Lazova, R.; Davies, S.; Backvall, H.; Ponten, F.; Brash, D.; Pawelek, J. Donor DNA in a renal cell carcinoma metastasis from a bone marrow transplant recipient. Bone Marrow Transplant. 2004, 34, 183–186. [Google Scholar] [CrossRef] [PubMed]
- Kurgyis, Z.; Kemeny, L.V.; Buknicz, T.; Groma, G.; Olah, J.; Jakab, A.; Polyanka, H.; Zanker, K.; Dittmar, T.; Kemeny, L.; et al. Melanoma-Derived BRAF(V600E) Mutation in Peritumoral Stromal Cells: Implications for in Vivo Cell Fusion. Int. J. Mol. Sci. 2016, 17, 980. [Google Scholar] [CrossRef] [Green Version]
- Clawson, G.A.; Kimchi, E.; Patrick, S.D.; Xin, P.; Harouaka, R.; Zheng, S.; Berg, A.; Schell, T.; Staveley-O’Carroll, K.F.; Neves, R.I.; et al. Circulating tumor cells in melanoma patients. PLoS ONE 2012, 7, e41052. [Google Scholar] [CrossRef] [Green Version]
- Clawson, G.A.; Matters, G.L.; Xin, P.; Imamura-Kawasawa, Y.; Du, Z.; Thiboutot, D.M.; Helm, K.F.; Neves, R.I.; Abraham, T. Macrophage-Tumor Cell Fusions from Peripheral Blood of Melanoma Patients. PLoS ONE 2015, 10, e0134320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clawson, G.A.; Matters, G.L.; Xin, P.; McGovern, C.; Wafula, E.; dePamphilis, C.; Meckley, M.; Wong, J.; Stewart, L.; D’Jamoos, C.; et al. Stealth dissemination of macrophage-tumor cell fusions cultured from blood of patients with pancreatic ductal adenocarcinoma. PLoS ONE 2017, 12, e0184451. [Google Scholar] [CrossRef] [Green Version]
- Manjunath, Y.; Mitchem, J.B.; Suvilesh, K.N.; Avella, D.M.; Kimchi, E.T.; Staveley-O’Carroll, K.F.; Deroche, C.B.; Pantel, K.; Li, G.; Kaifi, J.T. Circulating giant tumor-macrophage fusion cells are independent prognosticators in non-small cell lung cancer patients. J. Thorac. Oncol. 2020, 15, 1460–1471. [Google Scholar] [CrossRef] [PubMed]
- Manjunath, Y.; Porciani, D.; Mitchem, J.B.; Suvilesh, K.N.; Avella, D.M.; Kimchi, E.T.; Staveley-O’Carroll, K.F.; Burke, D.H.; Li, G.; Kaifi, J.T. Tumor-Cell-Macrophage Fusion Cells as Liquid Biomarkers and Tumor Enhancers in Cancer. Int. J. Mol. Sci. 2020, 21, 1872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yilmaz, Y.; Lazova, R.; Qumsiyeh, M.; Cooper, D.; Pawelek, J. Donor Y chromosome in renal carcinoma cells of a female BMT recipient: Visualization of putative BMT-tumor hybrids by FISH. Bone Marrow Transplant. 2005, 35, 1021–1024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shabo, I.; Svanvik, J. Expression of macrophage antigens by tumor cells. Adv. Exp. Med. Biol. 2011, 714, 141–150. [Google Scholar]
- Sprangers, A.J.; Freeman, B.T.; Kouris, N.A.; Ogle, B.M. A Cre-Lox P recombination approach for the detection of cell fusion in vivo. J. Vis. Exp. JoVE 2012, 59, e3581. [Google Scholar] [CrossRef] [Green Version]
- Koulakov, A.A.; Lazebnik, Y. The Problem of Colliding Networks and its Relation to Cell Fusion and Cancer. Biophys. J. 2012, 103, 2011–2020. [Google Scholar] [CrossRef] [Green Version]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Bjerregaard, B.; Holck, S.; Christensen, I.J.; Larsson, L.I. Syncytin is involved in breast cancer-endothelial cell fusions. Cell. Mol. Life Sci. 2006, 63, 1906–1911. [Google Scholar] [CrossRef] [Green Version]
- Larsson, L.I.; Bjerregaard, B.; Wulf-Andersen, L.; Talts, J.F. Syncytin and cancer cell fusions. Sci. World J. 2007, 7, 1193–1197. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Liu, T.; Zhao, Z.; Chen, Y.; Zeng, J.; Liu, S.; Zhu, F. Mutations in 3′-long terminal repeat of HERV-W family in chromosome 7 upregulate syncytin-1 expression in urothelial cell carcinoma of the bladder through interacting with c-Myb. Oncogene 2014, 33, 3947–3958. [Google Scholar] [CrossRef] [PubMed]
- Hass, R.; Otte, A. Mesenchymal stem cells as all-round supporters in a normal and neoplastic microenvironment. Cell Commun. Signal. 2012, 10, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melzer, C.; von der Ohe, J.; Hass, R. Concise Review: Crosstalk of Mesenchymal Stroma/Stem-Like Cells with Cancer Cells Provides Therapeutic Potential. Stem Cells 2018, 36, 951–968. [Google Scholar] [CrossRef] [Green Version]
- Hass, R. Role of MSC in the Tumor Microenvironment. Cancers 2020, 12, 2107. [Google Scholar] [CrossRef] [PubMed]
- Muller, A.; Homey, B.; Soto, H.; Ge, N.; Catron, D.; Buchanan, M.E.; McClanahan, T.; Murphy, E.; Yuan, W.; Wagner, S.N.; et al. Involvement of chemokine receptors in breast cancer metastasis. Nature 2001, 410, 50–56. [Google Scholar] [CrossRef]
- Gauck, D.; Keil, S.; Niggemann, B.; Zanker, K.S.; Dittmar, T. Hybrid clone cells derived from human breast epithelial cells and human breast cancer cells exhibit properties of cancer stem/initiating cells. BMC Cancer 2017, 17, 515. [Google Scholar] [CrossRef] [Green Version]
- Melzer, C.; Ohe, J.V.; Luo, T.; Hass, R. Spontaneous Fusion of MSC with Breast Cancer Cells Can Generate Tumor Dormancy. Int. J. Mol. Sci. 2021, 22, 5930. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, H.; Meng, F.; Kondo, N.; Iwamoto, A.; Matsuda, Z. Generation of a dual-functional split-reporter protein for monitoring membrane fusion using self-associating split GFP. Protein Eng. Des. Sel. 2012, 25, 813–820. [Google Scholar] [CrossRef] [Green Version]
- Weiler, J.; Dittmar, T. Cell Fusion in Human Cancer: The Dark Matter Hypothesis. Cells 2019, 8, 132. [Google Scholar] [CrossRef] [Green Version]
- Trejo-Becerril, C.; Perez-Cardenas, E.; Taja-Chayeb, L.; Anker, P.; Herrera-Goepfert, R.; Medina-Velazquez, L.A.; Hidalgo-Miranda, A.; Perez-Montiel, D.; Chavez-Blanco, A.; Cruz-Velazquez, J.; et al. Cancer progression mediated by horizontal gene transfer in an in vivo model. PLoS ONE 2012, 7, e52754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hass, R.; von der Ohe, J.; Ungefroren, H. Potential Role of MSC/Cancer Cell Fusion and EMT for Breast Cancer Stem Cell Formation. Cancers 2019, 11, 1432. [Google Scholar] [CrossRef] [Green Version]
- Bracq, L.; Xie, M.; Lambele, M.; Vu, L.T.; Matz, J.; Schmitt, A.; Delon, J.; Zhou, P.; Randriamampita, C.; Bouchet, J.; et al. T Cell-Macrophage Fusion Triggers Multinucleated Giant Cell Formation for HIV-1 Spreading. J. Virol. 2017, 91, e01237-17. [Google Scholar] [CrossRef] [Green Version]
- Kemeny, L.V.; Kurgyis, Z.; Buknicz, T.; Groma, G.; Jakab, A.; Zanker, K.; Dittmar, T.; Kemeny, L.; Nemeth, I.B. Melanoma Cells Can Adopt the Phenotype of Stromal Fibroblasts and Macrophages by Spontaneous Cell Fusion in Vitro. Int. J. Mol. Sci. 2016, 17, 826. [Google Scholar] [CrossRef] [Green Version]
- Streubel, B.; Chott, A.; Huber, D.; Exner, M.; Jager, U.; Wagner, O.; Schwarzinger, I. Lymphoma-specific genetic aberrations in microvascular endothelial cells in B-cell lymphomas. N. Engl. J. Med. 2004, 351, 250–259. [Google Scholar] [CrossRef]
- Duelli, D.; Lazebnik, Y. Cell fusion: A hidden enemy? Cancer Cell 2003, 3, 445–448. [Google Scholar] [CrossRef] [Green Version]
- LaBerge, G.; Duvall, E.; Grasmick, Z.; Haedicke, K.; Galan, A.; Pawelek, J. A melanoma patient with macrophage-cancer cell hybrids in the primary tumor, a lymph node metastasis and a brain metastasis. Cancer Genet. 2021, 256–257, 162–164. [Google Scholar] [CrossRef] [PubMed]
- Silvestris, F.; Ciavarella, S.; Strippoli, S.; Dammacco, F. Cell fusion and hyperactive osteoclastogenesis in multiple myeloma. Adv. Exp. Med. Biol. 2011, 714, 113–128. [Google Scholar]

| Tumor Type | Normal Cells | Marker | Reference |
|---|---|---|---|
| Breast cancer | Macrophages | CD163, MAC387, DAP12 | [182,184,197] |
| Colon/colorectal cancer | Macrophages | CD163, MAC387, DAP12, CD45, CD14, Cytokeratin | [183,192,197] |
| Epithelial ovarian carcinoma | BMDCs | CD45, CXCR4 | [138] |
| Melanoma | Macrophages | CD14, CD45, Cytokeratin | [192] |
| Stromal cells | BRAF (V600E) mutation | [190] | |
| BMDCs | STR analysis * | [185,186] | |
| Multiple myeloma | Osteoclasts | Myeloma specific translocations | [187,188,219] |
| Non-small cell lung cancer | Macrophages | CD14, CD45, Cytokeratin, EpCAM | [194] |
| Pancreatic ductal adenocarcinoma | Macrophages | CD45, Cytokeratin | [191] |
| BMDCs | CD45, EpCAM, | [164] | |
| Y chromosome * | |||
| Renal cell carcinoma | BMDCs | blood group alleles * | [189] |
| BMDCs | Y chromosome * | [196] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hass, R.; von der Ohe, J.; Dittmar, T. Hybrid Formation and Fusion of Cancer Cells In Vitro and In Vivo. Cancers 2021, 13, 4496. https://doi.org/10.3390/cancers13174496
Hass R, von der Ohe J, Dittmar T. Hybrid Formation and Fusion of Cancer Cells In Vitro and In Vivo. Cancers. 2021; 13(17):4496. https://doi.org/10.3390/cancers13174496
Chicago/Turabian StyleHass, Ralf, Juliane von der Ohe, and Thomas Dittmar. 2021. "Hybrid Formation and Fusion of Cancer Cells In Vitro and In Vivo" Cancers 13, no. 17: 4496. https://doi.org/10.3390/cancers13174496
APA StyleHass, R., von der Ohe, J., & Dittmar, T. (2021). Hybrid Formation and Fusion of Cancer Cells In Vitro and In Vivo. Cancers, 13(17), 4496. https://doi.org/10.3390/cancers13174496







