Simple Summary
Large granular lymphocyte (LGL) leukemia, a lymphoproliferative disease, is characterized by an increased frequency of large-sized lymphocytes with typical expression of T-cell receptor (TCR) αβ, CD3, CD8, CD16, CD45RA, and CD57, and with the expansion of one to three subfamilies of the TCR variable β chain reflecting gene rearrangements. Molecular analysis remains the gold standard for confirmation of TCR clonality; however, flow cytometry is time and labor saving, and can be associated with simultaneous investigation of other surface markers. Moreover, Vβ usage by flow cytometry can be employed for monitoring clonal kinetics during treatment and follow-up of LGL leukemia patients.
Abstract
Large granular lymphocyte (LGL) leukemia is a lymphoproliferative disorder of mature T or NK cells frequently associated with autoimmune disorders and other hematological conditions, such as myelodysplastic syndromes. Immunophenotype of LGL cells is similar to that of effector memory CD8+ T cells with T-cell receptor (TCR) clonality defined by molecular and/or flow cytometric analysis. Vβ usage by flow cytometry can identify clonal TCR rearrangements at the protein level, and is fast, sensitive, and almost always available in every Hematology Center. Moreover, Vβ usage can be associated with immunophenotypic characterization of LGL clone in a multiparametric staining, and clonal kinetics can be easily monitored during treatment and follow-up. Finally, Vβ usage by flow cytometry might identify LGL clones silently underlying other hematological conditions, and routine characterization of Vβ skewing might identify recurrent TCR rearrangements that might trigger aberrant immune responses during hematological or autoimmune conditions.
1. Introduction
Large granular lymphocyte (LGL) leukemia is a chronic lymphoproliferative disorder arising from clonal expansion of mature T or Natural Killer (NK) cells, and accounts for 2–5% of all cases of non-Hodgkin lymphomas (NHL) in Western countries [1]. LGL leukemia is considered a bone marrow failure (BMF) syndrome because of its overlapping pathogenesis with other immune-mediated diseases [2], and because LGLs can be frequently found in patients diagnosed with BMF syndromes, such as pure red cell aplasia (PRCA) and acquired aplastic anemia (AA) [3]. LGL leukemia of mature T cells is the most common entity (85% of cases), usually occurring with an indolent clinical course, while NK-LGL leukemia is rare but aggressive and frequently associated with Epstein–Barr virus (EBV) infection in young Asian adults [1,2,3,4,5,6,7,8]. LGLs are large-sized (15–18 µm) lymphocytes with an abundant cytoplasm containing azurophilic granules, and a round or reniform nucleus with mature chromatin [1]. LGLs can also be found in reactive conditions; however, circulating granular lymphocyte frequency is typically < 300 cells/µL, while LGL count is > 2000 cells/µL during LGL leukemia [1,4,5,9]. In the majority of cases, leukemic cells display a characteristic phenotype with surface expression of TCR αβ, CD3, CD5dim, CD8, CD16, CD45RA, and CD57, while cells are negative for CD4, CD27, CD28, and CD45RO, resembling effector memory and terminal effector memory T cell phenotype [1,7,10]. In other variants, neoplastic cells can be CD3+CD4+CD8dimCD57+ or CD3+CD4+CD8+CD57+ with various expression of CD56, CD57 and CD16, and in fewer cases leukemic cells are TCR γδ+ [11,12,13,14,15]. In NK-LGL leukemia, LGLs are positive for CD2, CD3ε, CD8, CD16, and CD56, while negative for sCD3, CD4, and TCR αβ [16]. T- and NK-LGL cells show restriction (oligoclonality) of T-cell receptor (TCR) or killer immunoglobulin-like receptor (KIR) repertoire that can be identified by flow cytometry, polymerase chain reaction (PCR), or next-generation sequencing (NGS) [17]. Clinical features are not specific to LGL leukemia, as the most common manifestations are neutropenia, anemia, and splenomegaly usually occurring asymptomatically [1,18,19,20]. In case of severe neutropenia (absolute neutrophil count < 0.5 cells/L), patients can experience recurrent oral ulcerations and infections, especially bacterial, less frequently viral and fungal infections [6,19,21]. In 6–22% of cases, anemia can be severe (hemoglobin < 8 g/dL) and require transfusions, and can be associated with autoimmune hemolytic anemia (AIHA) or PRCA [6]. In addition, LGL leukemia patients frequently have a history of autoimmune disorders, such as rheumatoid arthritis (RA), or autoimmune endocrinopathies (e.g., thyroiditis) [19,22,23]. Conversely, CD4+ or CD4+CD8+/− T-LGL leukemia has an indolent course and is frequently characterized by lymphocytosis with normal hemoglobin and platelet count and by the presence of a second neoplasms, especially B-cell NHL [24,25].
In this review, we focus on biological significance of LGL clonality and clinical utility of TCR repertoire investigation by flow cytometry.
2. T-LGL Leukemia Pathogenesis
The exact pathogenesis of LGL leukemia is still unclear. The most accepted hypothesis is a clonal drift of a T cell population after chronic antigen exposure, as suggested by cross-reactivity to human T-cell lymphotropic virus 1 (HTLV-1) epitopes or a frequent concomitant chronic viral infection in LGL leukemia patients, especially EBV, hepatitis C virus (HCV), or cytomegalovirus (CMV), especially in CD4+ T-LGL [1,26,27,28,29]. Despite there being no conclusive evidence that LGLs are activated by known viral antigens, immunohistochemical studies report clusters of LGLs in close contact to bone marrow (BM)-resident dendritic cells (DCs), supporting the hypothesis of a chronic self-antigen stimulation [18,30,31]. Once activated, LGLs expand under interleukin (IL)-15 and platelet-derived growth factor (PDGF) stimulation. IL-15, a proinflammatory cytokine, is overexpressed in LGL leukemia and drives clonal transformation of normal activated cytotoxic T cells, likely through centrosome alterations, aneuploidy, and increased resistance to apoptosis [2,32,33,34]. In addition, IL-15 induces transcription of several anti-apoptotic proteins, such as Bcl-2, while increases proteasome-mediated degradation of pro-apoptotic factors, such as Bid, resulting in increased cell survival [35]. IL-15 also upregulates MYC, AURKA, and AURKB, responsible for centrosome alterations, and DNMT3B, ultimately leading to hypermethylation of tumor suppressor genes [36,37]. However, pathogenesis of LGL leukemia is not driven only by chronic antigen and proinflammatory cytokine stimulation, but also by constitutive activation of Janus kinase (JAK)-signal transducer and activator of transcription (STAT) signaling pathway and resistance to Fas/Fas-ligand (Fas-L)-mediated apoptosis [1,2,38]. IL-6, another proinflammatory cytokine increased in the sera of LGL leukemia patients, is the main activator of STAT3, and is supposed to be mostly released by DCs [39,40]. Upon chronic IL-6 stimulation, LGLs show augmented expression of Mcl-1, an anti-apoptotic protein of the Bcl-2 family [40]. On the other hand, somatic mutations in STAT3 gene can also induce constitutive activation of JAK/STAT pathway, as well as mutations on the STAT5b gene, more frequently described in CD4+ T-LGL leukemia [41,42,43]. Soluble Fas-L (sFas-L), increased in the sera of LGL leukemia patients, especially those with CD8+CD16+CD56− LGLs and STAT3 hyperactivation, functions as a decoy receptor preventing Fas-mediated apoptosis [38,44,45,46]. FasL is also involved in development of neutropenia and its release is STAT3-dependent. Indeed, FasL expression is highly related to STAT3 activation status and CD8 LGL phenotype, while negatively influenced by miR-146b expression, frequently elevated in the plasma of AA and negatively correlated with absolute reticulocyte count [47,48]. In LGL leukemia, the pro-/anti-apoptotic balance is impaired in favor of increased resistance to cell death by neoplastic clones, also mediated by enhanced phosphoinositide 3-kinase (PI3K)/Akt signaling pathway activation through RANTES (also known as chemokine (C-C motif) ligand 5 or CCL5), IL-18, and macrophage inflammatory protein (MIP)-1b stimulation [33,39]. In addition, activation of tumor necrosis factor–related apoptosis-inducing ligand (TRAIL) receptor induces NF-κB signaling pathway, contributing to resistance to apoptosis in leukemic cells [49]. Neutropenia is a frequent clinical finding in LGL leukemia patients; however, pathogenetic mechanisms are still unclear and likely involve peripheral or intramedullary neutrophil destruction, such as FasL-mediated neutrophil apoptosis [5,50]. Indeed, the BM is frequently hypercellular with a left-shifted myeloid maturation, while less often hypocellular with reduced mature neutrophils, suggesting a compensatory myelopoiesis because of increased destruction [50].
3. TCR Clonality
Reactive LGLs can be found at low frequencies in healthy subjects after viral infections; however, in these cases, LGLs are polyclonal while neoplastic clones are oligo- or monoclonal [1]. The terms “polyclonal” or “oligo/monoclonal” refer to the diversity of TCR or B-cell receptor (BCR) repertoire in T- or B-cell pool of an individual, respectively [51,52]. For T lymphocytes, oligoclonality is defined based on the diversity of TCR repertoire based on distribution of variable (V) β and/or α chain rearrangements, or on skewing of a particular region namely the complementarity-determining region (CDR) 3 [53,54,55,56]. In other words, clonality refers to the ability of a random pool of T cells, either CD4+ or CD8+ lymphocytes, to recognize a large number of antigens (polyclonality) or limited ones (oligoclonality) [57]. TCR Vβ repertoire can be infinitely diverse because T cells can theoretically recognize any known or novel self- and non-self-antigens. The majority of T cells carry a TCR composed by an α and a β chain assembled after a complex gene locus rearrangement known as V(D)J rearrangement [57,58,59,60,61,62,63,64]. In this process, a random Vβ locus out of 52 available (grouped in 22 functional families) is sequentially rearranged with the Dβ1 locus, a random Jβ1 out of six, a Cβ1 and a Dβ2 loci, a random Jβ2 region out of seven available, and a Cβ2 locus. After this complex rearrangement, additional random modifications are added by a terminal deoxynucleotidyl transferase (TdT) enzyme between the end of the Vβ1 and the beginning of Jβ1 (the CDR3 region) [62,63,64]. V(D)J recombination and TdT modifications ideally produce up to 1018 diverse TCRs meaning that an individual might recognize up to 1018 diverse antigens [65]. In early phases of infections, polyclonal CD4+CD28+ and CD8+CD28+ T cells are activated and expand to increase the chance of having a functional antigen-specific immune clone that can fight pathogens [66,67]. If this clone can efficiently clear the infectious agent, high antigen-specific CD8+CD28−CD57+ effector memory T cells become immunodominant and constitute an oligoclonal pool of memory T cells [67]. Oligoclonal expansion of CD8+ T lymphocytes is observed in several autoimmune diseases and cancers and is related to poor survival [68,69]. Moreover, oligoclonality strongly suggests an antigen-driven T cell activation, as also described in AA, where CD8+ effector memory cells show oligoclonality with 1–3 immunodominant clones private to the disease [53,70,71,72]. Those clonotypes can be also present at very low frequencies in healthy individuals, supporting the hypothesis of an immune response to common epitopes [53]. Interestingly, LGL phenotype resembles the immunophenotype of effector memory T cells with CD3, CD4 or CD8, CD57 positivity, clonal expansion of a particular immunodominant clone, and gene expression profiling similar to that reported in healthy counterpart [73]. TCR clonality mirrors the oligoclonal drift that characterized T-LGL cell expansion likely driven by chronic antigen stimulation, IL-15 and PDGF overstimulation, and acquisition of somatic mutations in STAT3 or STAT5b genes [1,32,33]. Despite common epitopes are believed to trigger autoimmune responses against hematopoietic stem and progenitor cells (HSPCs) in T-LGL leukemia and other marrow failure syndromes, lack of recurrent Vβ family or CDR3 sequence expansion among patients suggests that clonotypes are private to each disease and that uncommon elusive chronic antigens might drive monoclonal expansion of leukemic cells [74,75]. CDR3 sequences might be shared within patients with marrow failure syndromes and between patients with different diseases and healthy subjects at very low frequencies, as described for paroxysmal nocturnal hemoglobinuria (PNH)-related clonotypes found at very low frequencies in patients with AA and healthy controls [53]. Therefore, TCR clonality in effector memory-like T cells indicates a chronic antigen stimulation in disease initiation; however, whether a common or an elusive infrequent antigen triggers this expansion remains an open question.
4. Flow Cytometric Vβ Usage
The diagnostic definition of clonality is still challenging because current technologies alone cannot conclusively define TCR oligo/monoclonality or cannot be applied in routine diagnostic settings [74,75,76]. For example, Southern blot has been the gold standard until a few decades ago, because it can ideally identify every TCR gene rearrangement if appropriate probes and restriction enzymes are used; however, Southern blot is time and labor consuming and high-quality DNA is required [77,78]. PCR cannot cover all possible TCR αβ gene rearrangements and is usually applied for the detection of a limited number of sequences, or for TCR gamma (TCRG) rearrangements, which are more limited than those of TCRB gene [79,80]. Sanger sequencing and spectratyping of CDR3 are also old-fashioned techniques because of their impracticability to individual sequencing or on a large number of T cell clones [75]. In recent years, deep next-generation sequencing (NGS) has completely changed the analysis of TCR repertoire because a single clone with a unique rearrangement and CDR3 sequence can be detected, even at very low frequency; however, TCR repertoire by NGS is still expensive and requires high-quality DNA and bioinformatics competence for analysis of high-throughput data [53,75,81,82,83]. Therefore, despite its relevance in research settings, TCR repertoire by NGS is not yet applicable to routine diagnostic analysis. The introduction of TCR Vβ monoclonal antibodies has markedly improved diagnostic definition of clonality in LGL leukemia [76,84,85]. Current Vβ antibodies cover more than 65% of all Vβ domains and can be grouped in 22 families (Table 1) [86,87].

Table 1.
Antibodies for each Vβ family.
In available kits, antibodies are conjugated to fluorescein isothiocyanate (FITC), phycoerythrin (PE), or tandem FITC/PE; therefore, sample acquisition requires instruments equipped just with a blue laser (488 nm excitation) and sensors for detection of FITC and PE emission wavelengths (peaks at 525 nm and 574 nm, respectively) [45,68]. Vβ usage by flow cytometry also allows absolute clone count and simultaneous characterization of TCR Vβ distribution in different lymphocyte subsets using appropriate antibody combinations [74]. For example, TCR Vβ usage has been studied on CD3+, CD4+, and CD8+ T cells [74,88,89] in LGL leukemia patients, or in naïve, effector, and memory lymphocytes in healthy subjects [57], or in total CD4+ and CD8+, effector CD4+CD28+ and CD8+CD28+, and effector memory CD4+CD28−CD57+ and CD8+CD28−CD57+ cells in AA [53,70,71]. When a known immunodominant clone has to be monitored (e.g., during treatment or follow-up), immunophenotype of LGLs can be combined with conjugated antibody corresponding to the expanded Vβ family in a multiparametric staining [74]; however, the fluorophore choice for some Vβ family is constrained, as Vβ4 is conjugated only with tandem FITC/PE, Vβ6.1, Vβ6.7, Vβ12, Vβ13.3, and Vβ20 only with FITC, and Vβ13.2 only conjugated with PE (Figure 1). Antibodies conjugated with fluorophores excited by violet (405 nm) and ultraviolet (320 nm) lasers are available for Vβ3, Vβ5.1, and Vβ8, and conjugated in VioBlue (excitation 400 nm/emission 452 nm) for Vβ2, Vβ11, Vβ13.1, and Vβ21.3. Recombinant antibodies are also available for Vβ2, Vβ5.1, Vβ5.3, Vβ7.1, Vβ7.2, Vβ11, Vβ13.1, Vβ13.6, Vβ14, Vβ16, Vβ17, Vβ21.3, and Vβ23. The advantages of recombinant antibodies are reduced lot-to-lot variability and higher purity, because antibodies are produced using a specific DNA sequence for one type of heavy and light chain using mammalian cell lines. Moreover, constant region is the same specifically mutated IgG1 sequence requiring only one type of isotype control [90].
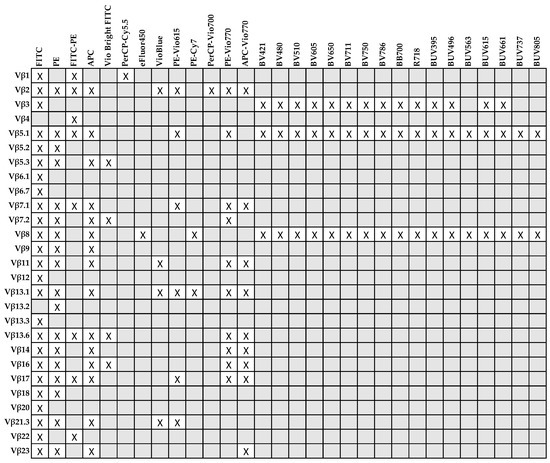
Figure 1.
Conjugated antibodies for each Vβ family available for flow cytometry and relative fluorophores (X).
The presence of 1–3 immunodominant Vβ clones does not prove clonality and the diagnosis of LGL leukemia, because flow cytometry results should be validated by DNA sequencing of both Vα and Vβ regions and supported by clinical manifestations and immunophenotype compatible with LGL leukemia [1,74,91]. Vβ skewing can be also frequently found in elderly (more than 40% of healthy subjects aged 45 years and older) caused by infections occurring during the lifetime without any clinical significance [53,92,93,94]. Therefore, Vβ usage by flow cytometry should be defined based on mean Vβ frequencies detected in a pool of healthy subjects used as a reference range, and skewing should be defined when the frequency of a particular Vβ family is higher than the mean + 3 standard deviations (SDs) in healthy subjects [53,74]. Despite those limitations, flow cytometric Vβ usage analysis could be a rapid and effective screening tool for identification of clonal LGLs, especially for differential diagnosis with other clonal benign disorders or reactive conditions [1,74]. Moreover, once clonality is confirmed by molecular analysis that is time and labor consuming and performed in specialized laboratories, flow cytometry Vβ usage could be employed for monitoring the leukemic clone expansion during treatment and follow-up using a fast and specific technique that is commonly available in the majority of Hematology Centers [74].
5. Literature Search
Relevant literature using the key word “large granular lymphocyte leukemia” was searched in PubMed database from 1977 to July 2021. Limiting factors were “large granular lymphocyte leukemia”, and full text available in English language. Two investigators independently reviewed the reference list for potential eligible manuscripts, and selected articles were then reviewed independently for inclusion in the analysis. Studies were included when: (1) the year of publication was between 2011 and 2021; (2) the studies were conducted on humans and reported clinical data and Vβ usage by flow cytometry; and (3) the studies were on T-LGL leukemia (Figure 2).
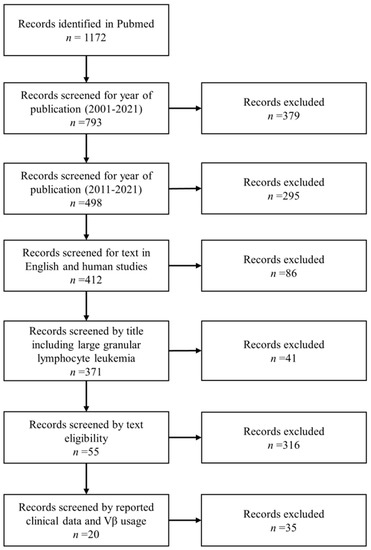
Figure 2.
Literature search strategy using “large granular lymphocyte leukemia” as a keyword in Pubmed. After identification of 1172 articles, records were screened by year of publication, title, and text eligibility. A total of 20 articles were eligible for further analysis.
From selected articles (n = 20), data were collected into a standardized form including publication year, source, number of total patients and divided by sex, number of subjects with autoimmune disorders, hematological malignancies, or cancers, and Vβ usage by flow cytometry (Table 2) [41,42,46,75,76,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107]. A total of 533 T-LGL leukemia patients were evaluable for Vβ usage analysis, and sex was available in 524 subjects: 308 of them were males (58.8%) and 216 females (41.2%) with a male:female ratio of 1.4, as already described in smaller cohorts [1]. Autoimmune disorders were reported in 136 subjects (25.5% of cases) and hematological disorders or cancers in 189 patients (35.5%). RA was diagnosed in 24 subjects (4.5% of total cases and 17.6% of patients with autoimmune disorders), and positivity for anti-nuclear antibodies (ANA) or rheumatoid factor (RF) was reported in 58 (10.9% of total cases and 42.6% of subjects with autoimmune disorders) and 16 subjects (3% of total cases and 11.8% of patients with autoimmune disorders), respectively. Other autoimmune disorders were collagenosis, myositis, autoimmune thyroiditis (n = 6; 1% of total cases), and autoimmune polyglandular syndrome type 1. Among hematological diseases, PRCA was the most frequent disorder associated with T-LGL leukemia (n = 45; 8.4% of total cases), followed by plasma cell disorders (n = 25; 4.7% of total cases), especially monoclonal gammopathy of uncertain significance (MGUS), and myelodysplastic syndromes (MDS; n = 21; 3.9% of total cases). Other reported hematological conditions associated with T-LGL leukemia were: AIHA (n = 10; 1.9% of total cases); T- and B-cell NHL (n = 9; 1.7% of total cases); acute leukemias, both myeloid and lymphoblastic; immune thrombocytopenic purpura (ITP); or AA. Solid neoplasms were also frequently associated with T-LGL leukemia (n = 52; 9.8% of total cases). Reported frequencies are slightly higher especially for AIHA, ITP, and RA likely because Vβ usage by flow cytometry is not routinely performed, and TCR clonality is mostly confirmed by PCR without indicating expanded Vβ family but only reporting LGL phenotype and clinical manifestations [18].

Table 2.
Characteristics of included studies.
6. Vβ Usage in T-LGL Leukemia
TCR clonality is a major criterion for LGL leukemia diagnosis and can be defined using a molecular or flow cytometric analysis, as described above, and clonal kinetics have shown clinical utility in monitoring disease progression and responsiveness to treatment [74]. TCR repertoire is heterogeneous among patients, and there is no recurrent Vβ expansion across studies [53,75]. However, because of the lack of a consensus on TCR clonality detection methodology, comparison of Vβ skewing among cohorts, case series, or case reports is difficult, especially when clonality is reported as the nucleotide length distribution by PCR without reporting specific TCRBV or TCRBJ genes involved. Indeed, among more than 700 studies between 2001 and 2021, only 20 were selected for data comparison of Vβ usage by flow cytometry. When studies were analyzed independently, no recurrent Vβ family was found between LGL leukemia patients. Of total 533 evaluable patients, the presence of at least one immunodominant clone by flow cytometry was described in 265 subjects (49.7%), and 1–3 Vβ subgroups were expanded in 12 of them (4.5% of cases). Among studies, the most frequent expanded Vβ family was the Vβ13.1 (n = 33; 12.5% of cases), followed by Vβ3 (n = 29; 10.9%), Vβ17 (n = 24; 9.1%), and Vβ2 and Vβ14 (both n = 22; 8.3% of total cases) (Figure 3). Vβ8 represented the immunodominant clone in 6.8% of cases (n = 18), and Vβ1 in 5.7% (n = 15). Other Vβ families were expanded in less than 5% of cases, and in less than 1% of subjects (case reports) for Vβ5.2, Vβ5.3, Vβ6.1, Vβ6.7, Vβ11, and Vβ13.3. Of total cases, the immunophenotype of LGLs was detailed only in 520 subjects, and 89 cases (17.1%) were CD4+ T-LGLs, 19 (3.7%) CD4+CD8+/dim, and the remaining cases were CD8+ T-LGLs. Of the total CD4+ T-LGL leukemia cases, in 48 of them, Vβ usage was described in detail, and there was no Vβ skewing in most cases (n = 20; 41.7%) or Vβ13.1 expansion (n = 16; 33.3%). Other reported expanded Vβ families were: Vβ3; Vβ8; Vβ17; and anecdotical Vβ1, Vβ2, Vβ5.2/5.3, Vβ12.2, Vβ14, and Vβ22. Of the total 33 cases with expanded Vβ13.1, 16 of them (48.5%) were CD4+ T-LGLs, three (9.1%) were CD4+CD8+, and the remaining 14 cases (42.4%) were CD8+ T-LGLs. These cumulative data are similar to those reported in single case series showing a preferential Vβ13.1 expansion in CD4+ T-LGL leukemia [46,75]. Vβ13.1 family expansion is recurrent also during infectious diseases, especially CMV and human immunodeficiency virus (HIV) [108,109,110]. In the latter, Vβ13.1 expansion is more frequently found in CD4+CD8dim T cells expressing the killer lectin-like receptor NKG2D, a CD4+ T cell subset present in RA, human T-lymphotropic virus 1-associated myelopathy, and Wegener granulomatosis [108,111]. Interestingly, these CD4+CD8dimNKG2D+ T cells bearing the HLA-DRB1*0701 allele and restricted to Vβ13.1 have a highly homogeneous TCR repertoire [110]. Moreover, CD4+CD8dimNKG2D+ T cells can produce FasL while protected from Fas-mediated growth arrest because of NKG2D protective functions [112]. Therefore, the presence of Vβ13.1 expansion in CD4+ leukemic cells might add evidence to the antigen-driven clonal drift pathogenetic theory in T-LGL leukemia.
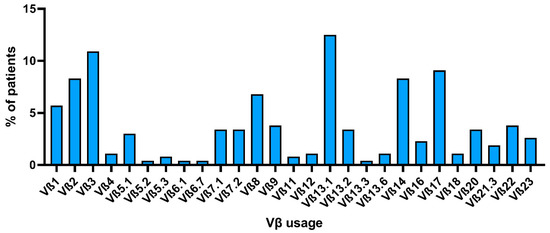
Figure 3.
Vβ usage by flow cytometry among selected studies. The percent of patients across selected articles who showed expansion of an LGLL clone carrying each specific Vβ family is reported.
An exact association between Vβ usage and underlying hematological disease rather than T-LGL leukemia was identified in only 32 subjects out of 189 patients (16.9% of hematological cases). There was no recurrent Vβ expansion, despite Vβ14 represented the immunodominant clone in six cases (18.8% of hematological cases). Vβ14 was not associated with a specific disease, as it was expanded in subjects with MDS, acute leukemias, NHL, or chronic myeloid leukemia; however, the number of evaluable patients was too small to draw any significant conclusion. Conversely, these data could be a starting point for implementing Vβ usage analysis by flow cytometry in subjects with hematological conditions with concomitant increased LGL count. Indeed, case series report concomitant Vβ skewing in MDS patients [107]; however, the number of studied patients is still limited for identification of a recurrent Vβ expansion in a specific MDS group, or if clonal kinetics might be related to disease progression to acute myeloid leukemia (AML). The concept that Vβ skewing is patient- or disease-specific is probably an evolving concept, especially in the NGS era. Sequencing can identify even one TCR rearrangement, and has shown that, despite clonotypes being private to each disease (e.g., T-LGL leukemia or AA), those expanded CDR3 sequences might be found at very low frequencies even in healthy subjects [53,75]. For example, PNH-related clonotypes have been described at low frequencies in both AA and healthy subjects supporting the hypothesis of an autologous immune attack against a self-antigen on hematopoietic stem cells [53,113].
7. Conclusions
LGL leukemia is classified as a lymphoproliferative disorder of mature T or NK cells; however, this clinical entity is considered among other BMF syndromes and is frequently associated with autoimmune disorders and other benign and malignant hematological conditions, such as PRCA or acute leukemias [1,2,3]. The immunophenotype of leukemic cells resembles that typical of effector memory CD8+ T cells with aberrancy and TCR clonality, mostly TCR αβ and less commonly TCR γδ clonal rearrangements [1,73]. Demonstration of TCR clonality by molecular and/or flow cytometric analysis together with clinical manifestations and increased LGL count by cytology is required for the correct diagnostic definition of this disease, as LGLs can also be present in reactive conditions without any clinical significance [74,92,93,94,95,96]. Molecular analysis is more accurate and can identify every TCR rearrangement, even at very low frequencies, by using novel deep sequencing technologies (e.g., NGS or TCR repertoire coupled with single-cell RNA sequencing) [75,114]. However, these methods are time and labor consuming and high-quality DNA is required [74,75]. Vβ usage by flow cytometry can identify clonal TCR rearrangements at protein level by recognition of 22 Vβ protein families without differentiating within multiple rearrangements that can occur in each given family [53,75]. Despite flow cytometry not being explored as deeply in TCR repertoire analysis as NGS, this technique is fast, sensitive, and almost always available in every Hematology Center, and can be associated with immunophenotypic characterization of the neoplastic clone in a multiparametric staining [53,70,71,72,74,88,89]. Therefore, Vβ usage by flow cytometry might help in quickly identifying clonality and confirming clinical, cytological, and molecular findings; however, the absence of clonal expansion by flow cytometry cannot exclude TCR clonality that might be detected with more sensitive techniques, such as NGS. Moreover, a lack of Vβ skewing might be related to a small T-LGL clone and Vβ usage analysis performed on total CD3+CD8+ T cells. Indeed, small T-LGL clonotypes might be diluted on total CD8+ T cells that appear polyclonal, as elegantly described in AA by both flow cytometry and NGS [53]. In addition, once identified, the immunodominant clone can be easily monitored during treatment and follow-up and can identify disease progression or relapse early, even under clonal drift, as NGS or TCR sequencing are still expensive techniques that cannot be routinely and frequently applied in clinical practice [74]. Finally, the presence and frequency of LGLs should be routinely checked in patients with autoimmune and hematological diseases because of their frequent association, and Vβ usage by flow cytometry might identify some subgroups of hematological conditions with a profound immune dysregulation that might benefit from immunomodulatory or immunosuppressive therapy [107]. Therefore, Vβ usage analysis should be implemented to better clarify the role of LGLs, either reactive or clonal, in other benign and malignant conditions, and to possibly identify recurrent TCR rearrangements for prediction of possible self-antigens triggering the autologous T cell expansion during LGL leukemia and other autoimmune and hematological or neoplastic diseases.
Author Contributions
Conceptualization, V.G. and C.S.; literature search, V.G. and M.D.; data curation, V.G. and N.M.; writing—original draft preparation, V.G. and M.D.; writing—review and editing, C.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are available upon request by the authors.
Acknowledgments
This research was supported by the Intramural Program of the Department of Medicine, Surgery and Dentistry “Scuola Medica Salernitana”, University of Salerno, Salerno, Italy.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lamy, T.; Moignet, A.; Loughran, T.P., Jr. LGL leukemia: From pathogenesis to treatment. Blood 2017, 129, 1082–1094. [Google Scholar] [CrossRef]
- Giudice, V.; Cardamone, C.; Triggiani, M.; Selleri, C. Bone Marrow Failure Syndromes, Overlapping Diseases with a Common Cytokine Signature. Int. J. Mol. Sci. 2021, 22, 705. [Google Scholar] [CrossRef]
- Patel, B.A.; Giudice, V.; Young, N.S. Immunologic effects on the haematopoietic stem cell in marrow failure. Best Pract. Res. Clin. Haematol. 2021, 34, 101276. [Google Scholar] [CrossRef] [PubMed]
- Loughran, T.P., Jr. Clonal diseases of large granular lymphocytes. Blood 1993, 82, 1–14. [Google Scholar] [CrossRef]
- Gazitt, T.; Loughran, T.P., Jr. Chronic neutropenia in LGL leukemia and rheumatoid arthritis. Hematol. Am. Soc. Hematol. Educ. Program 2017, 2017, 181–186. [Google Scholar] [CrossRef]
- Bareau, B.; Rey, J.; Hamidou, M.; Donadieu, J.; Morcet, J.; Reman, O.; Schleinitz, N.; Tournilhac, O.; Roussel, M.; Fest, T.; et al. Analysis of a French cohort of patients with large granular lymphocyte leukemia: A report on 229 cases. Haematologica 2010, 95, 1534–1541. [Google Scholar] [CrossRef]
- Swerdlow, S.H.; Campo, E.; Pileri, S.A.; Harris, N.L.; Stein, H.; Siebert, R.; Advani, R.; Ghielmini, M.; Salles, G.A.; Zelenetz, A.D.; et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood 2016, 127, 2375–2390. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, R.; Suzumiya, J.; Nakamura, S.; Aoki, S.; Notoya, A.; Ozaki, S.; Gondo, H.; Hino, N.; Mori, H.; Sugimori, H.; et al. Aggressive natural killer-cell leukemia revisited: Large granular lymphocyte leukemia of cytotoxic NK cells. Leukemia 2004, 18, 763–770. [Google Scholar] [CrossRef] [PubMed]
- Semenzato, G.; Zambello, R.; Starkebaum, G.; Oshimi, K.; Loughran, T.P., Jr. The lymphoproliferative disease of granular lymphocytes: Updated criteria for diagnosis. Blood 1997, 89, 256–260. [Google Scholar] [CrossRef]
- Lundell, R.; Hartung, L.; Hill, S.; Perkins, S.L.; Bahler, D.W. T-cell large granular lymphocyte leukemias have multiple phenotypic abnormalities involving pan-T-cell antigens and receptors for MHC molecules. Am. J. Clin. Pathol. 2005, 124, 937–946. [Google Scholar] [CrossRef]
- Kuwahara, N.; Kodaka, T.; Zushi, Y.; Sasaki, M.; Goka, T.; Maruoka, H.; Aoyama, Y.; Tsunemine, H.; Yamane, T.; Kobayashi, J.; et al. T-cell large granular lymphocytic (LGL) leukemia consists of CD4+/CD8dim and CD4-/CD8+ LGL populations in association with immune thrombocytopenia; autoimmune neutropenia; and monoclonal B-cell lymphocytosis. J. Clin. Exp. Hematop. 2019, 59, 202–206. [Google Scholar] [CrossRef]
- Soliman, D.; Sallam, S.; Akiki, S.; Mudawi, D.; Ibrahim, F. T-Cell Large Granular Lymphocytic Leukemia with Extremely Rare Immunophenotype (CD4/CD8 Double-Positive) Followed by Multiple Myeloma Diagnosis. Case Rep. Hematol. 2020, 2020, 8839144. [Google Scholar] [CrossRef]
- Kallemeijn, M.J.; de Ridder, D.; Schilperoord-Vermeulen, J.; van der Klift, M.Y.; Sandberg, Y.; van Dongen, J.J.; Langerak, A.W. Dysregulated signaling; proliferation and apoptosis impact on the pathogenesis of TCRγδ+ T cell large granular lymphocyte leukemia. PLoS ONE 2017, 12, e0175670. [Google Scholar] [CrossRef]
- De Luca, G.; Trasarti, S.; Bizzoni, L.; Del Giudice, I.; Della Starza, I.; De Propris, M.S.; Gentile, G.; Mancini, F.; Mantovani, S.; Petrucci, L.; et al. Lymphomatoid granulomatosis and large granular lymphocyte leukemia; a rare association of two lymphoproliferative disorders. Leuk. Lymphoma 2018, 59, 2715–2718. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Yu, Y.; Yan, S.; Wang, R.; Liu, X.; Chen, C. Pure Red Cell Aplasia and Autoimmune Hemolytic Anemia Sequentially Occurring in a Patient with Large Granular T-lymphocytic Leukemia. Intern Med. 2016, 55, 1491–1496. [Google Scholar] [CrossRef][Green Version]
- Gentile, T.C.; Uner, A.H.; Hutchison, R.E.; Wright, J.; Ben-Ezra, J.; Russell, E.C.; Loughran, T.P., Jr. CD3+; CD56+ aggressive variant of large granular lymphocyte leukemia. Blood 1994, 84, 2315–2321. [Google Scholar] [CrossRef] [PubMed]
- Zambello, R.; Falco, M.; Della Chiesa, M.; Trentin, L.; Carollo, D.; Castriconi, R.; Cannas, G.; Carlomagno, S.; Cabrelle, A.; Lamy, T.; et al. Expression and function of KIR and natural cytotoxicity receptors in NK-type lymphoproliferative diseases of granular lymphocytes. Blood 2003, 102, 1797–1805. [Google Scholar] [CrossRef]
- Barilà, G.; Calabretto, G.; Teramo, A.; Vicenzetto, C.; Gasparini, V.R.; Semenzato, G.; Zambello, R. T cell large granular lymphocyte leukemia and chronic NK lymphocytosis. Best Pract. Res. Clin. Haematol. 2019, 32, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Moignet, A.; Lamy, T. Latest Advances in the Diagnosis and Treatment of Large Granular Lymphocytic Leukemia. Am. Soc. Clin. Oncol. Educ. Book 2018, 38, 616–625. [Google Scholar] [CrossRef]
- Oshimi, K. Clinical Features; Pathogenesis; and Treatment of Large Granular Lymphocyte Leukemias. Intern. Med. 2017, 56, 1759–1769. [Google Scholar] [CrossRef] [PubMed]
- Sanikommu, S.R.; Clemente, M.J.; Chomczynski, P.; Afable, M.G., 2nd; Jerez, A.; Thota, S.; Patel, B.; Hirsch, C.; Nazha, A.; Desamito, J.; et al. Clinical features and treatment outcomes in large granular lymphocytic leukemia (LGLL). Leuk. Lymphoma 2018, 59, 416–422. [Google Scholar] [CrossRef]
- Lamy, T.; Loughran, T.P., Jr. Clinical features of large granular lymphocyte leukemia. Semin. Hematol. 2003, 40, 185–195. [Google Scholar] [CrossRef]
- Zhang, R.; Shah, M.V.; Loughran, T.P., Jr. The root of many evils: Indolent large granular lymphocyte leukaemia and associated disorders. Hematol. Oncol. 2010, 28, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Lima, M.; Almeida, J.; Dos Anjos Teixeira, M.; Alguero Md Mdel, C.; Santos, A.H.; Balanzategui, A.; Queirós, M.L.; Bárcena, P.; Izarra, A.; Fonseca, S.; et al. TCRalphabeta+/CD4+ large granular lymphocytosis: A new clonal T-cell lymphoproliferative disorder. Am. J. Pathol. 2003, 163, 763–771. [Google Scholar] [CrossRef]
- Rivero, A.; Mozas, P.; Jiménez, L.; López-Guerra, M.; Colomer, D.; Bataller, A.; Correa, J.; Rivas-Delgado, A.; Bastidas, G.; Baumann, T.; et al. Clinicobiological Characteristics and Outcomes of Patients with T-Cell Large Granular Lymphocytic Leukemia and Chronic Lymphoproliferative Disorder of Natural Killer Cells from a Single Institution. Cancers 2021, 13, 3900. [Google Scholar] [CrossRef] [PubMed]
- Poullot, E.; Bouscary, D.; Guyader, D.; Ghandour, C.; Roussel, M.; Fest, T.; Houot, R.; Lamy, T. Large granular lymphocyte leukemia associated with hepatitis C virus infection and B cell lymphoma: Improvement after antiviral therapy. Leuk. Lymphoma 2013, 54, 1797–1799. [Google Scholar] [CrossRef] [PubMed]
- Kimura, H.; Ito, Y.; Kawabe, S.; Gotoh, K.; Takahashi, Y.; Kojima, S.; Naoe, T.; Esaki, S.; Kikuta, A.; Sawada, A.; et al. EBV-associated T/NK-cell lymphoproliferative diseases in nonimmunocompromised hosts: Prospective analysis of 108 cases. Blood 2012, 119, 673–686. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Caballero, A.; García-Montero, A.C.; Bárcena, P.; Almeida, J.; Ruiz-Cabello, F.; Tabernero, M.D.; Garrido, P.; Muñoz-Criado, S.; Sandberg, Y.; Langerak, A.W.; et al. Expanded cells in monoclonal TCR-alphabeta+/CD4+/NKa+/CD8-/+dim T-LGL lymphocytosis recognize hCMV antigens. Blood 2008, 112, 4609–4616. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zambello, R.; Loughran, T.P., Jr.; Trentin, L.; Pontisso, P.; Battistella, L.; Raimondi, R.; Facco, M.; Sancetta, R.; Agostini, C.; Pizzolo, G.; et al. Serologic and molecular evidence for a possible pathogenetic role of viral infection in CD3-negative natural killer-type lymphoproliferative disease of granular lymphocytes. Leukemia 1995, 9, 1207–1211. [Google Scholar]
- Zambello, R.; Semenzato, G. Large granular lymphocytosis. Haematologica 1998, 83, 936–942. [Google Scholar]
- Zambello, R.; Berno, T.; Cannas, G.; Baesso, I.; Binotto, G.; Bonoldi, E.; Bevilacqua, P.; Miorin, M.; Facco, M.; Trentin, L.; et al. Phenotypic and functional analyses of dendritic cells in patients with lymphoproliferative disease of granular lymphocytes (LDGL). Blood 2005, 106, 3926–3931. [Google Scholar] [CrossRef][Green Version]
- Zhang, R.; Shah, M.V.; Yang, J.; Nyland, S.B.; Liu, X.; Yun, J.K.; Albert, R.; Loughran, T.P., Jr. Network model of survival signaling in large granular lymphocyte leukemia. Proc. Natl. Acad. Sci. USA 2008, 105, 16308–16313. [Google Scholar] [CrossRef]
- Kothapalli, R.; Nyland, S.B.; Kusmartseva, I.; Bailey, R.D.; McKeown, T.M.; Loughran, T.P., Jr. Constitutive production of proinflammatory cytokines RANTES.; MIP-1beta and IL-18 characterizes LGL leukemia. Int. J. Oncol. 2005, 26, 529–535. [Google Scholar]
- Hodge, D.L.; Yang, J.; Buschman, M.D.; Schaughency, P.M.; Dang, H.; Bere, W.; Yang, Y.; Savan, R.; Subleski, J.J.; Yin, X.M.; et al. Interleukin-15 enhances proteasomal degradation of bid in normal lymphocytes: Implications for large granular lymphocyte leukemias. Cancer Res. 2009, 69, 3986–3994. [Google Scholar] [CrossRef]
- Akkoç, Y.; Berrak, Ö.; Arısan, E.D.; Obakan, P.; Çoker-Gürkan, A.; Palavan-Ünsal, N. Inhibition of PI3K signaling triggered apoptotic potential of curcumin which is hindered by Bcl-2 through activation of autophagy in MCF-7 cells. Biomed. Pharmacother. 2015, 71, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; Liu, S.; Sams, G.H.; Curphey, D.P.; Santhanam, R.; Rush, L.J.; Schaefer, D.; Falkenberg, L.G.; Sullivan, L.; Jaroncyk, L.; et al. Aberrant overexpression of IL-15 initiates large granular lymphocyte leukemia through chromosomal instability and DNA hypermethylation. Cancer Cell 2012, 22, 645–655. [Google Scholar] [CrossRef] [PubMed]
- Langerak, A.W.; Assmann, J.L.J.C. Large granular lymphocyte cells and immune dysregulation diseases—The chicken or the egg? Haematologica 2018, 103, 193–194. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.H.; Wei, S.; Lamy, T.; Li, Y.; Epling-Burnette, P.K.; Djeu, J.Y.; Loughran, T.P., Jr. Blockade of Fas-dependent apoptosis by soluble Fas in LGL leukemia. Blood 2002, 100, 1449–1453. [Google Scholar] [CrossRef]
- Schade, A.E.; Wlodarski, M.W.; Maciejewski, J.P. Pathophysiology defined by altered signal transduction pathways: The role of JAK-STAT and PI3K signaling in leukemic large granular lymphocytes. Cell Cycle 2006, 5, 2571–2574. [Google Scholar] [CrossRef]
- Teramo, A.; Gattazzo, C.; Passeri, F.; Lico, A.; Tasca, G.; Cabrelle, A.; Martini, V.; Frezzato, F.; Trimarco, V.; Ave, E.; et al. Intrinsic and extrinsic mechanisms contribute to maintain the JAK/STAT pathway aberrantly activated in T-type large granular lymphocyte leukemia. Blood 2013, 121, 3843–3854. [Google Scholar] [CrossRef]
- Rajala, H.L.; Eldfors, S.; Kuusanmäki, H.; van Adrichem, A.J.; Olson, T.; Lagström, S.; Andersson, E.I.; Jerez, A.; Clemente, M.J.; Yan, Y.; et al. Discovery of somatic STAT5b mutations in large granular lymphocytic leukemia. Blood 2013, 121, 4541–4550. [Google Scholar] [CrossRef]
- Andersson, E.I.; Tanahashi, T.; Sekiguchi, N.; Gasparini, V.R.; Bortoluzzi, S.; Kawakami, T.; Matsuda, K.; Mitsui, T.; Eldfors, S.; Bortoluzzi, S.; et al. High incidence of activating STAT5B mutations in CD4-positive T-cell large granular lymphocyte leukemia. Blood 2016, 128, 2465–2468. [Google Scholar] [CrossRef] [PubMed]
- Moosic, K.B.; Paila, U.; Olson, K.C.; Dziewulska, K.; Wang, T.T.; Xing, J.C.; Ratan, A.; Feith, D.J.; Loughran, T.P., Jr.; Olson, T.L. Genomics of LGL leukemia and select other rare leukemia/lymphomas. Best Pract. Res. Clin. Haematol. 2019, 32, 196–206. [Google Scholar] [CrossRef] [PubMed]
- Lamy, T.; Liu, J.H.; Landowski, T.H.; Dalton, W.S.; Loughran, T.P., Jr. Dysregulation of CD95/CD95 ligand-apoptotic pathway in CD3(1) large granular lymphocyte leukemia. Blood 1998, 92, 4771–4777. [Google Scholar] [CrossRef] [PubMed]
- Teramo, A.; Barilà, G.; Calabretto, G.; Ercolin, C.; Lamy, T.; Moignet, A.; Roussel, M.; Pastoret, C.; Leoncin, M.; Gattazzo, C.; et al. STAT3 mutation impacts biological and clinical features of T-LGL leukemia. Oncotarget 2017, 8, 61876–61889. [Google Scholar] [CrossRef] [PubMed]
- Barilà, G.; Teramo, A.; Calabretto, G.; Vicenzetto, C.; Gasparini, V.R.; Pavan, L.; Leoncin, M.; Vedovato, S.; Frigo, A.C.; Facco, M.; et al. Stat3 mutations impact on overall survival in large granular lymphocyte leukemia: A single-center experience of 205 patients. Leukemia 2020, 34, 1116–1124. [Google Scholar] [CrossRef]
- Mariotti, B.; Calabretto, G.; Rossato, M.; Teramo, A.; Castellucci, M.; Barilà, G.; Leoncin, M.; Vicenzetto, C.; Facco, M.; Semenzato, G.; et al. Identification of a miR-146b-Fas ligand axis in the development of neutropenia in T large granular lymphocyte leukemia. Haematologica 2020, 105, 1351–1360. [Google Scholar] [CrossRef] [PubMed]
- Hosokawa, K.; Kajigaya, S.; Feng, X.; Desierto, M.J.; Fernandez Ibanez, M.D.; Rios, O.; Weinstein, B.; Scheinberg, P.; Townsley, D.M.; Young, N.S. A plasma microRNA signature as a biomarker for acquired aplastic anemia. Haematologica 2017, 102, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; LeBlanc, F.R.; Dighe, S.A.; Hamele, C.E.; Olson, T.L.; Feith, D.J.; Loughran, T.P., Jr. TRAIL mediates and sustains constitutive NF-κB activation in LGL leukemia. Blood 2018, 131, 2803–2815. [Google Scholar] [CrossRef]
- Pontikoglou, C.; Kalpadakis, C.; Papadaki, H.A. Pathophysiologic mechanisms and management of neutropenia associated with large granular lymphocytic leukemia. Expert Rev. Hematol. 2011, 4, 317–328. [Google Scholar] [CrossRef] [PubMed]
- Langerak, A.W.; Brüggemann, M.; Davi, F.; Darzentas, N.; van Dongen, J.J.M.; Gonzalez, D.; Cazzaniga, G.; Giudicelli, V.; Lefranc, M.P.; Giraud, M.; et al. High-Throughput Immunogenetics for Clinical and Research Applications in Immunohematology: Potential and Challenges. J. Immunol. 2017, 198, 3765–3774. [Google Scholar] [CrossRef] [PubMed]
- Minervina, A.; Pogorelyy, M.; Mamedov, I. T-cell receptor and B-cell receptor repertoire profiling in adaptive immunity. Transpl. Int. 2019, 32, 1111–1123. [Google Scholar] [CrossRef] [PubMed]
- Giudice, V.; Feng, X.; Lin, Z.; Hu, W.; Zhang, F.; Qiao, W.; Ibanez, M.D.P.F.; Rios, O.; Young, N.S. Deep sequencing and flow cytometric characterization of expanded effector memory CD8+CD57+ T cells frequently reveals T-cell receptor Vβ oligoclonality and CDR3 homology in acquired aplastic anemia. Haematologica 2018, 103, 759–769. [Google Scholar] [CrossRef] [PubMed]
- Calis, J.J.; Rosenberg, B.R. Characterizing immune repertoires by high throughput sequencing: Strategies and applications. Trends Immunol. 2014, 35, 581–590. [Google Scholar] [CrossRef] [PubMed]
- Schuster, F.R.; Hubner, B.; Führer, M.; Eckermann, O.; Gombert, M.; Dornmair, K.; Binder, V.; Reuther, S.; Krell, P.; Keller, T.; et al. Highly skewed T-cell receptor V-beta chain repertoire in the bone marrow is associated with response to immunosuppressive drug therapy in children with very severe aplastic anemia. Blood Cancer J. 2011, 1, e8. [Google Scholar] [CrossRef]
- Ma, L.; Yang, L.; Shi, B.; He, X.; Peng, A.; Li, Y.; Zhang, T.; Sun, S.; Ma, R.; Yao, X. Analyzing the CDR3 Repertoire with respect to TCR-Beta Chain V-D-J and V-J Rearrangements in Peripheral T Cells using HTS. Sci. Rep. 2016, 6, 29544. [Google Scholar] [CrossRef]
- Bretschneider, I.; Clemente, M.J.; Meisel, C.; Guerreiro, M.; Streitz, M.; Hopfenmüller, W.; Maciejewski, J.P.; Wlodarski, M.W.; Volk, H.D. Discrimination of T-cell subsets and T-cell receptor repertoire distribution. Immunol. Res. 2014, 58, 20–27. [Google Scholar] [CrossRef]
- Teng, G.; Papavasiliou, F.N. Immunoglobulin somatic hypermutation. Annu. Rev Genet. 2007, 41, 107–120. [Google Scholar] [CrossRef]
- Schatz, D.G.; Ji, Y. Recombination centres and the orchestration of V(D)J recombination. Nat. Rev. Immunol. 2011, 11, 251–263. [Google Scholar] [CrossRef]
- Gauss, G.H.; Lieber, M.R. Mechanistic constraints on diversity in human V(D)J recombination. Mol. Cell Biol. 1996, 16, 258–269. [Google Scholar] [CrossRef]
- Matthews, A.J.; Zheng, S.; DiMenna, L.J.; Chaudhuri, J. Regulation of immunoglobulin class-switch recombination: Choreography of noncoding transcription; targeted DNA deamination; and long-range DNA repair. Adv. Immunol. 2014, 122, 1–57. [Google Scholar]
- Arstila, T.P.; Casrouge, A.; Baron, V.; Even, J.; Kanellopoulos, J.; Kourilsky, P. A direct estimate of the human alphabeta T cell receptor diversity. Science 1999, 286, 958–961. [Google Scholar] [CrossRef] [PubMed]
- Murray, J.M.; Messier, T.; Rivers, J.; O’Neill, J.P.; Walker, V.E.; Vacek, P.M.; Finette, B.A. VDJ recombinase-mediated TCR β locus gene usage and coding joint processing in peripheral T cells during perinatal and pediatric development. J. Immunol. 2012, 189, 2356–2364. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.S.; Bassing, C.H. Inefficient V(D)J recombination underlies monogenic T cell receptor β expression. Proc. Natl. Acad. Sci. USA 2020, 117, 18172–18174. [Google Scholar] [CrossRef] [PubMed]
- Janeway, C.A., Jr.; Travers, P.; Walport, M. T-cell receptor gene rearrangement. In Immunobiology: The Immune System in Health and Disease; Garland Science: New York, NY, USA, 2001. [Google Scholar]
- Appay, V.; van Lier, R.A.; Sallusto, F.; Roederer, M. Phenotype and function of human T lymphocyte subsets: Consensus and issues. Cytom. A 2008, 73, 975–983. [Google Scholar] [CrossRef] [PubMed]
- Farber, D.L.; Yudanin, N.A.; Restifo, N.P. Human memory T cells: Generation; compartmentalization and homeostasis. Nat. Rev. Immunol. 2014, 14, 24–35. [Google Scholar] [CrossRef]
- Filaci, G.; Fenoglio, D.; Fravega, M.; Ansaldo, G.; Borgonovo, G.; Traverso, P.; Villaggio, B.; Ferrera, A.; Kunkl, A.; Rizzi, M.; et al. CD8+ CD28- T regulatory lymphocytes inhibiting T cell proliferative and cytotoxic functions infiltrate human cancers. J. Immunol. 2007, 179, 4323–4334. [Google Scholar] [CrossRef] [PubMed]
- Characiejus, D.; Pasukoniene, V.; Jonusauskaite, R.; Azlauskaite, N.; Aleknavicius, E.; Mauricas, M.; Otter, W.D. Peripheral blood CD8highCD57+ lymphocyte levels may predict outcome in melanoma patients treated with adjuvant interferon-alpha. Anticancer Res. 2008, 28, 1139–1142. [Google Scholar]
- Risitano, A.M.; Maciejewski, J.P.; Green, S.; Plasilova, M.; Zeng, W.; Young, N.S. In-vivo dominant immune responses in aplastic anaemia: Molecular tracking of putatively pathogenetic T-cell clones by TCR beta-CDR3 sequencing. Lancet 2004, 364, 355–364. [Google Scholar] [CrossRef]
- Risitano, A.M.; Kook, H.; Zeng, W.; Chen, G.; Young, N.S.; Maciejewski, J.P. Oligoclonal and polyclonal CD4 and CD8 lymphocytes in aplastic anemia and paroxysmal nocturnal hemoglobinuria measured by V beta CDR3 spectratyping and flow cytometry. Blood 2002, 100, 178–183. [Google Scholar] [CrossRef]
- Kook, H.; Zeng, W.; Guibin, C.; Kirby, M.; Young, N.S.; Maciejewski, J.P. Increased cytotoxic T cells with effector phenotype in aplastic anemia and myelodysplasia. Exp. Hematol. 2001, 29, 1270–1277. [Google Scholar] [CrossRef]
- Wlodarski, M.W.; Nearman, Z.; Jankowska, A.; Babel, N.; Powers, J.; Leahy, P.; Volk, H.D.; Maciejewski, J.P. Phenotypic differences between healthy effector CTL and leukemic LGL cells support the notion of antigen-triggered clonal transformation in T-LGL leukemia. J. Leukoc. Biol. 2008, 83, 589–601. [Google Scholar] [CrossRef] [PubMed]
- Clemente, M.J.; Wlodarski, M.W.; Makishima, H.; Viny, A.D.; Bretschneider, I.; Shaik, M.; Bejanyan, N.; Lichtin, A.E.; His, E.D.; Paquette, R.L.; et al. Clonal drift demonstrates unexpected dynamics of the T-cell repertoire in T-large granular lymphocyte leukemia. Blood 2011, 118, 4384–4393. [Google Scholar] [CrossRef] [PubMed]
- Clemente, M.J.; Przychodzen, B.; Jerez, A.; Dienes, B.E.; Afable, M.G.; Husseinzadeh, H.; Rajala, H.L.; Wlodarski, M.W.; Mustjoki, S.; Maciejewski, J.P. Deep sequencing of the T-cell receptor repertoire in CD8+ T-large granular lymphocyte leukemia identifies signature landscapes. Blood 2013, 122, 4077–4085. [Google Scholar] [CrossRef]
- Langerak, A.W.; van Den Beemd, R.; Wolvers-Tettero, I.L.; Boor, P.P.; van Lochem, E.G.; Hooijkaas, H.; van Dongen, J.J. Molecular and flow cytometric analysis of the Vbeta repertoire for clonality assessment in mature TCRalphabeta T-cell proliferations. Blood 2001, 98, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Langerak, A.W.; Wolvers-Tettero, I.L.; van Dongen, J.J. Detection of T cell receptor beta (TCRB) gene rearrangement patterns in T cell malignancies by Southern blot analysis. Leukemia 1999, 13, 965–974. [Google Scholar] [CrossRef][Green Version]
- Moreau, E.J.; Langerak, A.W.; van Gastel-Mol, E.J.; Wolvers-Tettero, I.L.; Zhan, M.; Zhou, Q.; Koop, B.F.; van Dongen, J.J. Easy detection of all T cell receptor gamma (TCRG) gene rearrangements by Southern blot analysis: Recommendations for optimal results. Leukemia 1999, 13, 1620–1626. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gorski, J.; Yassai, M.; Zhu, X.; Kissela, B.; Keever, C.; Flomenberg, N. Circulating T cell repertoire complexity in normal individuals and bone marrow recipients analyzed by CDR3 size spectratyping. Correlation with immune status. J. Immunol. 1994, 152, 5109–5119. [Google Scholar]
- Langerak, A.W.; Szczepański, T.; van der Burg, M.; Wolvers-Tettero, I.L.; van Dongen, J.J. Heteroduplex PCR analysis of rearranged T cell receptor genes for clonality assessment in suspect T cell proliferations. Leukemia 1997, 11, 2192–2199. [Google Scholar] [CrossRef]
- Wu, D.; Sherwood, A.; Fromm, J.R.; Winter, S.S.; Dunsmore, K.P.; Loh, M.L.; Greisman, H.A.; Sabath, D.E.; Wood, B.L.; Robins, H. High-throughput sequencing detects minimal residual disease in acute T lymphoblastic leukemia. Sci. Transl. Med. 2012, 4, 134ra63. [Google Scholar] [CrossRef] [PubMed]
- Robins, H.S.; Campregher, P.V.; Srivastava, S.K.; Wacher, A.; Turtle, C.J.; Kahsai, O.; Riddell, S.R.; Warren, E.H.; Carlson, C.S. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood 2009, 114, 4099–4107. [Google Scholar] [CrossRef]
- Sherwood, A.M.; Desmarais, C.; Livingston, R.J.; Andriesen, J.; Haussler, M.; Carlson, C.S.; Robins, H. Deep sequencing of the human TCRγ and TCRβ repertoires suggests that TCRβ rearranges after αβ and γδ T cell commitment. Sci. Transl. Med. 2011, 3, 90ra61. [Google Scholar] [CrossRef]
- Boylston, A.W.; Lancaster, F.C. Recognition of malignant cells by antibodies to the T-cell antigen receptor: Potential for diagnosis. Cancer Cells 1991, 3, 236–238. [Google Scholar]
- van den Beemd, R.; Boor, P.P.; van Lochem, E.G.; Hop, W.C.; Langerak, A.W.; Wolvers-Tettero, I.L.; Hooijkaas, H.; van Dongen, J.J. Flow cytometric analysis of the Vbeta repertoire in healthy controls. Cytometry 2000, 40, 336–345. [Google Scholar] [CrossRef]
- Lefranc, M.P. Immunoglobulin and T Cell Receptor Genes: IMGT(®) and the Birth and Rise of Immunoinformatics. Front. Immunol. 2014, 5, 22. [Google Scholar] [CrossRef] [PubMed]
- WHO-IUIS Nomenclature Subcommittee for immunoglobulins and Tcell Receptors; Lefranc, M.P. WHO-IUIS Nomenclature Subcommittee for immunoglobulins and T cell receptors report August 2007, 13th International Congress of Immunology, Rio de Janeiro; Brazil. Dev. Comp. Immunol. 2008, 32, 461–463. [Google Scholar] [CrossRef] [PubMed]
- Andersson, E.; Kuusanmäki, H.; Bortoluzzi, S.; Lagström, S.; Parsons, A.; Rajala, H.; van Adrichem, A.; Eldfors, S.; Olson, T.; Clemente, M.J.; et al. Activating somatic mutations outside the SH2-domain of STAT3 in LGL leukemia. Leukemia 2016, 30, 1204–1208. [Google Scholar] [CrossRef]
- Hsieh, Y.C.; Chang, S.T.; Huang, W.T.; Kuo, S.Y.; Chiang, T.A.; Chuang, S.S. A comparative study of flow cytometric T cell receptor Vβ repertoire and T cell receptor gene rearrangement in the diagnosis of large granular lymphocytic lymphoproliferation. Int. J. Lab. Hematol. 2013, 35, 501–509. [Google Scholar] [CrossRef] [PubMed]
- Starkie, D.O.; Compson, J.E.; Rapecki, S.; Lightwood, D.J. Generation of Recombinant Monoclonal Antibodies from Immunised Mice and Rabbits via Flow Cytometry and Sorting of Antigen-Specific IgG+ Memory B Cells. PLoS ONE 2016, 11, e0152282. [Google Scholar] [CrossRef]
- Morice, W.G.; Kimlinger, T.; Katzmann, J.A.; Lust, J.A.; Heimgartner, P.J.; Halling, K.C.; Hanson, C.A. Flow cytometric assessment of TCR-Vbeta expression in the evaluation of peripheral blood involvement by T-cell lymphoproliferative disorders: A comparison with conventional T-cell immunophenotyping and molecular genetic techniques. Am. J. Clin. Pathol. 2004, 121, 373–383. [Google Scholar] [CrossRef]
- Posnett, D.N.; Sinha, R.; Kabak, S.; Russo, C. Clonal populations of T cells in normal elderly humans: The T cell equivalent to “benign monoclonal gammapathy”. J. Exp. Med. 1994, 179, 609–618. [Google Scholar] [CrossRef] [PubMed]
- Boucher, N.; Dufeu-Duchesne, T.; Vicaut, E.; Farge, D.; Effros, R.B.; Schächter, F. CD28 expression in T cell aging and human longevity. Exp. Gerontol. 1998, 33, 267–282. [Google Scholar] [CrossRef]
- Wikby, A.; Ferguson, F.; Forsey, R.; Thompson, J.; Strindhall, J.; Löfgren, S.; Nilsson, B.O.; Ernerudh, J.; Pawelec, G.; Johansson, B. An immune risk phenotype; cognitive impairment; and survival in very late life: Impact of allostatic load in Swedish octogenarian and nonagenarian humans. J. Gerontol. A Biol. Sci. Med. Sci. 2005, 60, 556–565. [Google Scholar] [CrossRef] [PubMed]
- Stalika, E.; Papalexandri, A.; Kannelis, G.; Batsis, I.; Papadaki, T.; Anagnostopoulos, A.; Stamatopoulos, K. Transient monoclonal CD3+ T large granular lymphocyte proliferation in a case of mantle cell lymphoma with Rituximab-associated late onset neutropenia. Hematol. Oncol. 2011, 29, 144–146. [Google Scholar] [CrossRef] [PubMed]
- Garban, F.; Carras, S.; Drillat, P.; Jacob, M.C.; Fabre, B.; Callanan, M.; Courby, S.; Makowski, C.; Cahn, J.Y.; Gressin, R. Extracorporeal photopheresis as a curative treatment strategy in non epidermotropic T-cell lymphoma and large granular lymphocyte leukemia. Ann. Oncol. 2012, 23, 2386–2390. [Google Scholar] [CrossRef]
- Koskela, H.L.; Eldfors, S.; Ellonen, P.; van Adrichem, A.J.; Kuusanmäki, H.; Andersson, E.I.; Lagström, S.; Clemente, M.J.; Olson, T.; Jalkanen, S.E.; et al. Somatic STAT3 mutations in large granular lymphocytic leukemia. N. Engl. J. Med. 2012, 366, 1905–1913. [Google Scholar] [CrossRef]
- Andersson, E.I.; Rajala, H.L.; Eldfors, S.; Ellonen, P.; Olson, T.; Jerez, A.; Clemente, M.J.; Kallioniemi, O.; Porkka, K.; Heckman, C.; et al. Novel somatic mutations in large granular lymphocytic leukemia affecting the STAT-pathway and T-cell activation. Blood Cancer J. 2013, 3, e168. [Google Scholar] [CrossRef] [PubMed]
- Papalexandri, A.; Stalika, E.; Iskas, M.; Karypidou, M.; Zerva, P.; Touloumenidou, T.; Tachynopoulou, V.; Batsis, I.; Papadaki, T.; Sakellari, I.; et al. Molecular evidence for repertoire skewing of T large granular lymphocyte proliferation after allogeneic hematopoietic SCT: Report of two cases. Bone Marrow Transplant. 2013, 48, 1260–1261. [Google Scholar] [CrossRef]
- Stalika, E.; Papalexandri, A.; Iskas, M.; Stavroyianni, N.; Kanellis, G.; Kotta, K.; Pontikoglou, C.; Siorenta, A.; Anagnostopoulos, A.; Papadaki, H.; et al. Familial CD3+ T large granular lymphocyte leukemia: Evidence that genetic predisposition and antigen selection promote clonal cytotoxic T-cell responses. Leuk. Lymphoma 2014, 55, 1781–1787. [Google Scholar] [CrossRef]
- Singleton, T.P.; Yin, B.; Teferra, A.; Mao, J.Z. Spectrum of Clonal Large Granular Lymphocytes (LGLs) of αβ T Cells: T-Cell Clones of Undetermined Significance; T-Cell LGL Leukemias; and T-Cell Immunoclones. Am. J. Clin. Pathol. 2015, 144, 137–144. [Google Scholar] [CrossRef]
- Qiu, Z.Y.; Fan, L.; Wang, R.; Gale, R.P.; Liang, H.J.; Wang, M.; Wang, L.; Wu, Y.J.; Qiao, C.; Chen, Y.Y.; et al. Methotrexate therapy of T-cell large granular lymphocytic leukemia impact of STAT3 mutation. Oncotarget 2016, 7, 61419–61425. [Google Scholar] [CrossRef]
- Peng, G.; Yang, W.; Zhang, L.; Zhou, K.; Li, Y.; Li, Y.; Ye, L.; Li, J.; Fan, H.; Song, L.; et al. Moderate-dose cyclophosphamide in the treatment of relapsed/refractory T-cell large granular lymphocytic leukemia-associated pure red cell aplasia. Hematology 2016, 21, 138–143. [Google Scholar] [CrossRef] [PubMed]
- Awada, H.; Mahfouz, R.Z.; Durrani, J.; Kishtagari, A.; Jagadeesh, D.; Lichtin, A.E.; Hill, B.T.; Hamilton, B.K.; Carraway, H.E.; Nazha, A.; et al. Large granular lymphocytic leukaemia after solid organ and haematopoietic stem cell transplantation. Br. J. Haematol. 2020, 189, 318–322. [Google Scholar] [CrossRef]
- Zhu, Y.; Gao, Q.; Hu, J.; Liu, X.; Guan, D.; Zhang, F. Clinical features and treatment outcomes in patients with T-cell large granular lymphocytic leukemia: A single-institution experience. Leuk. Res. 2020, 90, 106299. [Google Scholar] [CrossRef]
- Minish, J.M.; Hamed, N.; Seifert, R.; Kelkar, A.H. T-Cell Large Granular Lymphocytic Leukemia as a Cause for Severe Neutropenia. Cureus 2020, 12, e10186. [Google Scholar] [CrossRef]
- Durrani, J.; Awada, H.; Kishtagari, A.; Visconte, V.; Kerr, C.; Adema, V.; Nagata, Y.; Kuzmanovic, T.; Hong, S.; Patel, B.; et al. Large granular lymphocytic leukemia coexists with myeloid clones and myelodysplastic syndrome. Leukemia 2020, 34, 957–962. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Arias, R.; López-Vázquez, A.; Diaz-Peña, R.; Sampere, A.; Tricas, L.; Asensi, V.; Rodrigo, L.; López-Larrea, C. CD8dim and NKG2D expression defines related subsets of CD4+ T cells in HIV-infected patients with worse prognostic factors. J. Acquir. Immune Defic. Syndr. 2009, 51, 390–398. [Google Scholar] [CrossRef]
- Rodriguez-Caballero, A.; Garcia-Montero, A.C.; Almeida, J.; Balanzategui, A.; Munoz-Criado, S.; Orfao, A. Association between the HLA haplotype and the TCR-Vbeta repertoire of anti-hCMV specific memory T-cells in immunocompetent healthy adults. Cytom. B Clin. Cytom. 2007, 72, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Garrido, P.; Ruiz-Cabello, F.; Bárcena, P.; Sandberg, Y.; Cantón, J.; Lima, M.; Balanzategui, A.; González, M.; López-Nevot, M.A.; Langerak, A.W.; et al. Monoclonal TCR-Vbeta13.1+/CD4+/NKa+/CD8-/+dim T-LGL lymphocytosis: Evidence for an antigen-driven chronic T-cell stimulation origin. Blood 2007, 109, 4890–4898. [Google Scholar] [CrossRef]
- Groh, V.; Bruhl, A.; El-Gabalawy, H.; Nelson, J.L.; Spies, T. Stimulation of T cell autoreactivity by anomalous expression of NKG2D and its MIC ligands in rheumatoid arthritis. Proc. Natl. Acad. Sci. USA 2003, 100, 9452–9457. [Google Scholar] [CrossRef]
- Groh, V.; Smythe, K.; Dai, Z.; Spies, T. Fas-ligand-mediated paracrine T cell regulation by the receptor NKG2D in tumor immunity. Nat. Immunol. 2006, 7, 755–762. [Google Scholar] [CrossRef] [PubMed]
- Gargiulo, L.; Lastraioli, S.; Cerruti, G.; Serra, M.; Loiacono, F.; Zupo, S.; Luzzatto, L.; Notaro, R. Highly homologous T-cell receptor beta sequences support a common target for autoreactive T cells in most patients with paroxysmal nocturnal hemoglobinuria. Blood 2007, 109, 5036–5042. [Google Scholar] [CrossRef] [PubMed]
- De Simone, M.; Rossetti, G.; Pagani, M. Single Cell T Cell Receptor Sequencing: Techniques and Future Challenges. Front. Immunol. 2018, 9, 1638. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).