Musings from the Tribbles Research and Innovation Network
Abstract
Simple Summary
Abstract
1. Introduction
2. Establishment of TRAIN
3. Tissue- and Cell-Specific Expression of TRIB Family Members
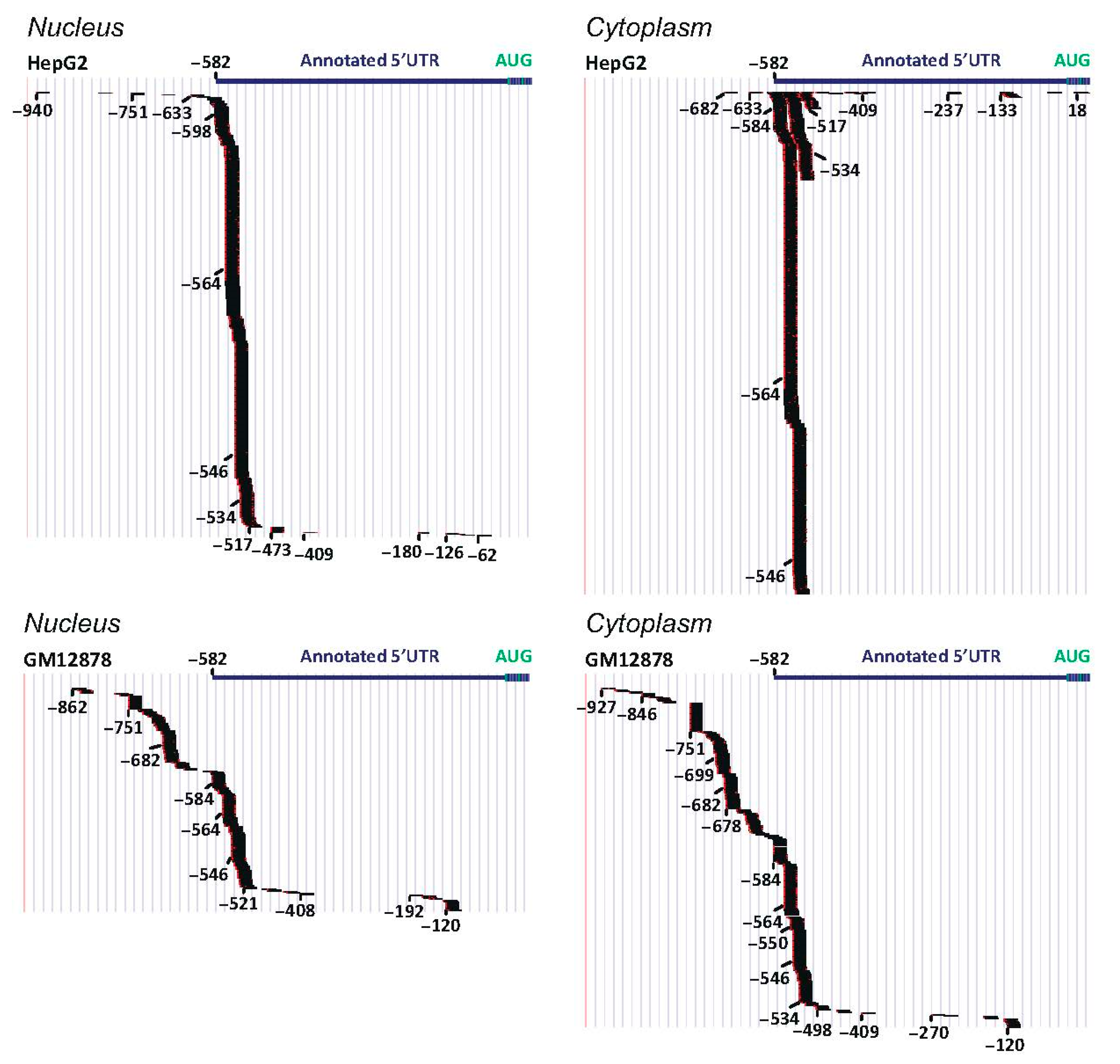
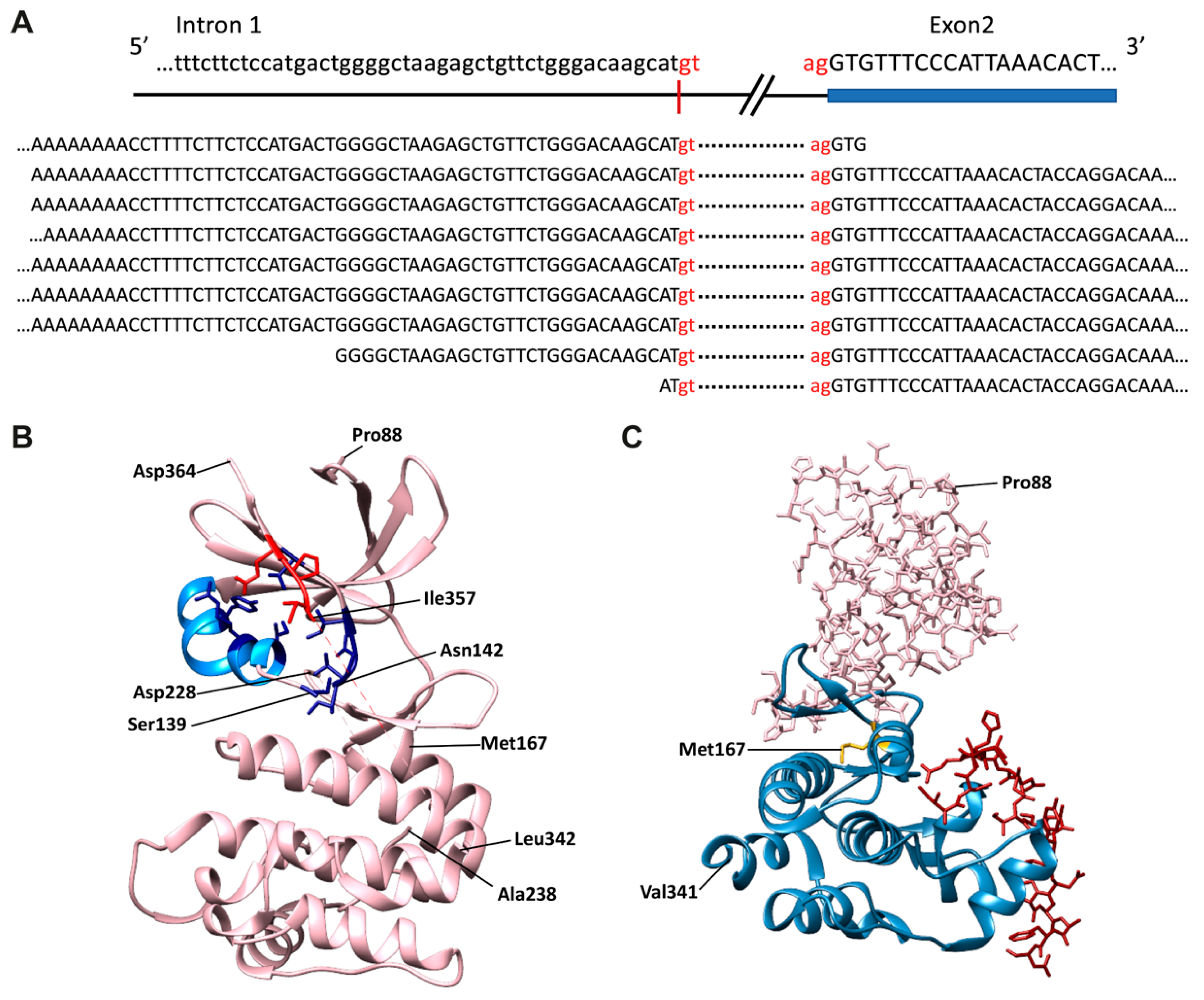
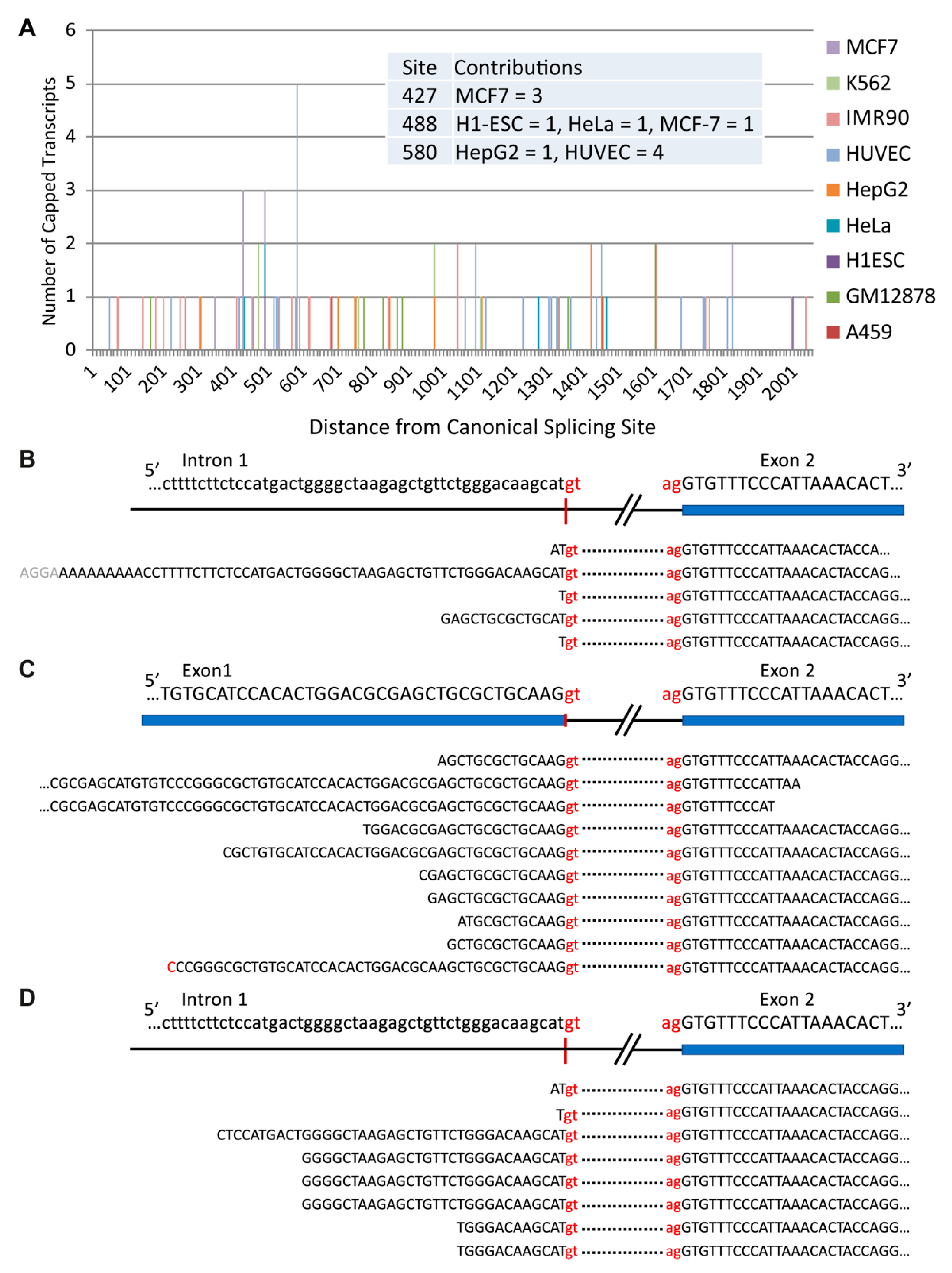
4. Genomic and Structural Data: Insights into TRIB Expression
5. Growth of TRIB (and COP1) Research
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Calandra, S.; Tarugi, P.; Speedy, H.E.; Dean, A.F.; Bertolini, S.; Shoulders, C.C. Mechanisms and genetic determinants regulating sterol absorption, circulating LDL levels, and sterol elimination: Implications for classification and disease risk. J. Lipid Res. 2011, 52, 1885–1926. [Google Scholar] [CrossRef]
- Örd, T.; Örd, T. Mammalian Pseudokinase TRIB3 in Normal Physiology and Disease: Charting the Progress in Old and New Avenues. Curr. Protein Pept. Sci. 2017, 18, 819–842. [Google Scholar] [CrossRef]
- Dobens, L.; Nauman, C.; Fischer, Z.; Yao, X. Control of Cell Growth and Proliferation by the Tribbles Pseudokinase: Lessons from Drosophila. Cancers 2021, 13, 883. [Google Scholar] [CrossRef]
- Jadhav, K.S.; Bauer, R.C. Trouble with Tribbles-1. Arter. Thromb. Vasc. Biol. 2019, 39, 998–1005. [Google Scholar] [CrossRef]
- Mayoral-Varo, V.; Jiménez, L.; Link, W. The Critical Role of TRIB2 in Cancer and Therapy Resistance. Cancers 2021, 13, 2701. [Google Scholar] [CrossRef]
- Fang, Y.; Zekiy, A.O.; Ghaedrahmati, F.; Timoshin, A.; Farzaneh, M.; Anbiyaiee, A.; Khoshnam, S.E. Tribbles homolog 2 (Trib2), a pseudo serine/threonine kinase in tumorigenesis and stem cell fate decisions. Cell Commun. Signal. 2021, 19, 41. [Google Scholar] [CrossRef]
- Pernar, C.H.; Ebot, E.M.; Wilson, K.M.; Mucci, L.A. The Epidemiology of Prostate Cancer. Cold Spring Harb. Perspect. Med. 2018, 8, a030361. [Google Scholar] [CrossRef]
- Blücher, C.; Stadler, S.C. Obesity and Breast Cancer: Current Insights on the Role of Fatty Acids and Lipid Metabolism in Promoting Breast Cancer Growth and Progression. Front. Endocrinol. 2017, 8, 293. [Google Scholar] [CrossRef] [PubMed]
- Dobens, L.L.; Bouyain, S. Developmental roles of tribbles protein family members. Dev. Dyn. 2012, 241, 1239–1248. [Google Scholar] [CrossRef] [PubMed]
- Großhans, J.; Wieschaus, E. A Genetic Link between Morphogenesis and Cell Division during Formation of the Ventral Furrow in Drosophila. Cell 2000, 101, 523–531. [Google Scholar] [CrossRef]
- Saka, Y.; Smith, J. A Xenopus tribbles orthologue is required for the progression of mitosis and for development of the nervous system. Dev. Biol. 2004, 273, 210–225. [Google Scholar] [CrossRef]
- Abdelilah-Seyfried, S.; Chan, Y.M.; Zeng, C.; Justice, N.J.; Younger-Shepherd, S.; Sharp, L.E.; Barbel, S.; Meadows, S.A.; Jan, L.Y.; Jan, Y.N. A gain-of-function screen for genes that affect the development of the Drosophila adult external sensory or-gan. Genetics 2000, 155, 733–752. [Google Scholar] [CrossRef]
- Mata, J.; Curado, S.; Ephrussi, A.; Rørth, P. Tribbles Coordinates Mitosis and Morphogenesis in Drosophila by Regulating String/CDC25 Proteolysis. Cell 2000, 101, 511–522. [Google Scholar] [CrossRef]
- Masoner, V.; Das, R.; Pence, L.; Anand, G.; LaFerriere, H.; Zars, T.; Bouyain, S.; Dobens, L.L. The kinase domain of Drosophila Tribbles is required for turnover of fly C/EBP during cellmigration. Dev. Biol. 2013, 375, 33–44. [Google Scholar] [CrossRef][Green Version]
- Levine, B.; Hackney, J.F.; Bergen, A.; Dobens, L.; Truesdale, A.; Dobens, L. Opposing interactions between Drosophila Cut and the C/EBP encoded by Slow Border Cells direct apical constriction and epithelial invagination. Dev. Biol. 2010, 344, 196–209. [Google Scholar] [CrossRef][Green Version]
- Ogienko, A.A.; Yarinich, L.A.; Fedorova, E.V.; Dorogova, N.V.; Bayborodin, S.I.; Baricheva, E.M.; Pindyurin, A.V. GAGA Regulates Border Cell Migration in Drosophila. Int. J. Mol. Sci. 2020, 21, 7468. [Google Scholar] [CrossRef]
- Carracedo, A.; Lorente, M.; Egia, A.; Blázquez, C.; García, S.; Giroux, V.; Malicet, C.; Villuendas, R.; Gironella, M.; González-Feria, L.; et al. The stress-regulated protein p8 mediates cannabinoid-induced apoptosis of tumor cells. Cancer Cell 2006, 9, 301–312. [Google Scholar] [CrossRef]
- Salazar, M.; Lorente, M.; García-Taboada, E.; Hernández-Tiedra, S.; Davila, D.; Francis, S.; Guzmán, M.; Kiss-Toth, E.; Velasco, G. The pseudokinase tribbles homologue-3 plays a crucial role in cannabinoid anticancer action. Biochim. et Biophys. Acta (BBA) Mol. Cell Biol. Lipids 2013, 1831, 1573–1578. [Google Scholar] [CrossRef]
- Klingenspor, M.; Xu, P.; Cohen, R.D.; Welch, C.; Reue, K. Altered Gene Expression Pattern in the Fatty Liver Dystrophy Mouse Reveals Impaired Insulin-mediated Cytoskeleton Dynamics. J. Biol. Chem. 1999, 274, 23078–23084. [Google Scholar] [CrossRef]
- Péterfy, M.; Phan, J.; Xu, P.; Reue, K. Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nat. Genet. 2001, 27, 121–124. [Google Scholar] [CrossRef]
- Tseng, Y.-H.; Butte, A.J.; Kokkotou, E.; Yechoor, V.K.; Taniguchi, C.M.; Kriauciunas, K.M.; Cypess, A.M.; Niinobe, M.; Yoshikawa, K.; Patti, M.E.; et al. Prediction of preadipocyte differentiation by gene expression reveals role of insulin receptor substrates and necdin. Nat. Cell Biol. 2005, 7, 601–611. [Google Scholar] [CrossRef]
- Qi, L.; Heredia, J.E.; Altarejos, J.Y.; Screaton, R.; Goebel, N.; Niessen, S.; MacLeod, I.X.; Liew, C.W.; Kulkarni, R.N.; Bain, J.R.; et al. TRB3 Links the E3 Ubiquitin Ligase COP1 to Lipid Metabolism. Science 2006, 312, 1763–1766. [Google Scholar] [CrossRef]
- Dornan, D.; Wertz, I.; Shimizu, H.; Arnott, D.; Frantz, G.D.; Dowd, P.; Rourke, K.O.; Koeppen, H.; Dixit, V.M. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature 2004, 429, 86–92. [Google Scholar] [CrossRef]
- Keeshan, K.; Bailis, W.; Dedhia, P.H.; Vega, M.E.; Shestova, O.; Xu, L.; Toscano, K.; Uljon, S.N.; Blacklow, S.C.; Pear, W.S. Transformation by Tribbles homolog 2 (Trib2) requires both the Trib2 kinase domain and COP1 binding. Blood 2010, 116, 4948–4957. [Google Scholar] [CrossRef]
- Kiss-Toth, E.; Bagstaff, S.M.; Sung, H.Y.; Jozsa, V.; Dempsey, C.; Caunt, J.C.; Oxley, K.M.; Wyllie, D.H.; Polgar, T.; Harte, M.; et al. Human Tribbles, a Protein Family Controlling Mitogen-activated Protein Kinase Cascades. J. Biol. Chem. 2004, 279, 42703–42708. [Google Scholar] [CrossRef]
- Sung, H.Y.; Guan, H.; Czibula, A.; King, A.R.; Eder, K.; Heath, E.; Suvarna, S.K.; Dower, S.K.; Wilson, A.G.; Francis, S.; et al. Human Tribbles-1 Controls Proliferation and Chemotaxis of Smooth Muscle Cells via MAPK Signaling Pathways. J. Biol. Chem. 2007, 282, 18379–18387. [Google Scholar] [CrossRef]
- Ashton-Chess, J.; Giral, M.; Mengel, M.; Renaudin, K.; Foucher, Y.; Gwinner, W.; Braud, C.; Dugast, E.; Quillard, T.; Thebault, P.; et al. Tribbles-1 as a Novel Biomarker of Chronic Antibody-Mediated Rejection. J. Am. Soc. Nephrol. 2008, 19, 1116–1127. [Google Scholar] [CrossRef]
- Alvarez, C.M.; Opelz, G.; Garcia, L.F.; Süsal, C. Expression of Regulatory T–Cell-Related Molecule Genes and Clinical Outcome in Kidney Transplant Recipients. Transplantation 2009, 87, 857–863. [Google Scholar] [CrossRef]
- Dugast, E.; Kiss-Toth, E.; Docherty, L.; Danger, R.; Chesneau, M.; Pichard, V.; Judor, J.-P.; Pettré, S.; Conchon, S.; Soulillou, J.-P.; et al. Identification of Tribbles-1 as a Novel Binding Partner of Foxp3 in Regulatory T Cells. J. Biol. Chem. 2013, 288, 10051–10060. [Google Scholar] [CrossRef]
- Mockler, M.B.; Conroy, M.J.; Lysaght, J. Targeting T Cell Immunometabolism for Cancer Immunotherapy; Understanding the Impact of the Tumor Microenvironment. Front. Oncol. 2014, 4, 107. [Google Scholar] [CrossRef]
- Dedhia, P.H.; Keeshan, K.; Uljon, S.; Xu, L.; Vega, M.E.; Shestova, O.; Zaks-Zilberman, M.; Romany, C.; Blacklow, S.C.; Pear, W.S. Differential ability of Tribbles family members to promote degradation of C/EBPα and induce acute myelogenous leukemia. Blood 2010, 116, 1321–1328. [Google Scholar] [CrossRef]
- Miyajima, C.; Itoh, Y.; Inoue, Y.; Hayashi, H. Positive Regulation of Interleukin-2 Expression by a Pseudokinase, Tribbles 1, in Activated T Cells. Biol. Pharm. Bull. 2015, 38, 1126–1133. [Google Scholar] [CrossRef]
- Wilkin, F.; Savonet, V.; Radulescu, A.; Petermans, J.; Dumont, J.E.; Maenhaut, C. Identification and Characterization of Novel Genes Modulated in the Thyroid of Dogs Treated with Methimazole and Propylthiouracil. J. Biol. Chem. 1996, 271, 28451–28457. [Google Scholar] [CrossRef] [PubMed]
- Wilkin, F.; Suarez-Huerta, N.; Robaye, B.; Peetermans, J.; Libert, F.; Dumont, J.E.; Maenhaut, C. Characterization of a Phosphoprotein whose mRNA is Regulated by the Mitogenic Pathways in Dog Thyroid Cells. Eur. J. Biochem. 1997, 248, 660–668. [Google Scholar] [CrossRef] [PubMed]
- Bowers, A.; Scully, S.; Boylan, J.F. SKIP3, a novel Drosophila tribbles ortholog, is overexpressed in human tumors and is regulated by hypoxia. Oncogene 2003, 22, 2823–2835. [Google Scholar] [CrossRef]
- Simonovsky, E.; Schuster, R.; Yeger-Lotem, E. Large-scale analysis of human gene expression variability associates highly variable drug targets with lower drug effectiveness and safety. Bioinformatics 2019, 35, 3028–3037. [Google Scholar] [CrossRef] [PubMed]
- GTEx Portal. Available online: https://gtexportal.org/ (accessed on 12 February 2021).
- McCall, M.N.; Illei, P.B.; Halushka, M.K. Complex Sources of Variation in Tissue Expression Data: Analysis of the GTEx Lung Transcriptome. Am. J. Hum. Genet. 2016, 99, 624–635. [Google Scholar] [CrossRef]
- Farahbod, M.; Pavlidis, P. Untangling the effects of cellular composition on coexpression analysis. Genome Res. 2020, 30, 849–859. [Google Scholar] [CrossRef]
- Kim-Hellmuth, S.; Aguet, F.; Oliva, M.; Muñoz-Aguirre, M.; Kasela, S.; Wucher, V.; Castel, S.E.; Hamel, A.R.; Viñuela, A.; Roberts, A.L.; et al. Cell type–specific genetic regulation of gene expression across human tissues. Science 2020, 369, eaaz8528. [Google Scholar] [CrossRef]
- The GTEx Consortium The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 2020, 369, 1318–1330. [CrossRef] [PubMed]
- Salomé, M.; Hopcroft, L.; Keeshan, K. Inverse and correlative relationships between TRIBBLES genes indicate non-redundant functions during normal and malignant hemopoiesis. Exp. Hematol. 2018, 66, 63–78.e13. [Google Scholar] [CrossRef]
- Yoshino, S.; Yokoyama, T.; Sunami, Y.; Takahara, T.; Nakamura, A.; Yamazaki, Y.; Tsutsumi, S.; Aburatani, H.; Nakamura, T. Trib1 promotes acute myeloid leukemia progression by modulating the transcriptional programs of Hoxa9. Blood 2021, 137, 75–88. [Google Scholar] [CrossRef]
- Aznarez, I.; Nomakuchi, T.T.; Tetenbaum-Novatt, J.; Rahman, M.A.; Fregoso, O.; Rees, H.; Krainer, A.R. Mechanism of Nonsense-Mediated mRNA Decay Stimulation by Splicing Factor SRSF1. Cell Rep. 2018, 23, 2186–2198. [Google Scholar] [CrossRef] [PubMed]
- Calviello, L.; Hirsekorn, A.; Ohler, U. Quantification of translation uncovers the functions of the alternative transcriptome. Nat. Struct. Mol. Biol. 2020, 27, 717–725. [Google Scholar] [CrossRef]
- Imamachi, N.; Salam, K.A.; Suzuki, Y.; Akimitsu, N. A GC-rich sequence feature in the 3′ UTR directs UPF1-dependent mRNA decay in mammalian cells. Genome Res. 2017, 27, 407–418. [Google Scholar] [CrossRef]
- Won, J.-I.; Shin, J.; Park, S.Y.; Yoon, J.; Jeong, D.-H. Global Analysis of the Human RNA Degradome Reveals Widespread Decapped and Endonucleolytic Cleaved Transcripts. Int. J. Mol. Sci. 2020, 21, 6452. [Google Scholar] [CrossRef]
- Wu, G.; Schmid, M.; Rib, L.; Polak, P.; Meola, N.; Sandelin, A.; Jensen, T.H. A Two-Layered Targeting Mechanism Underlies Nuclear RNA Sorting by the Human Exosome. Cell Rep. 2020, 30, 2387–2401.e5. [Google Scholar] [CrossRef]
- Noguchi, S.; Arakawa, T.; Fukuda, S.; Furuno, M.; Hasegawa, A.; Hori, F.; Ishikawa-Kato, S.; Kaida, K.; Kaiho, A.; Kanamori-Katayama, M.; et al. FANTOM5 CAGE profiles of human and mouse samples. Sci. Data 2017, 4, 170112. [Google Scholar] [CrossRef]
- Wade, J.; Grainger, D.C. Spurious transcription and its impact on cell function. Transcription 2018, 9, 182–189. [Google Scholar] [CrossRef]
- Cui, B.; Eyers, P.; Dobens, L.; Tan, N.; Mace, P.; Link, W.; Kiss-Toth, E.; Keeshan, K.; Nakamura, T.; Pear, W.; et al. High-lights of the 2nd International Symposium on Tribbles and Diseases: Tribbles tremble in therapeutics for immunity, metabo-lism, fundamental cell biology and cancer. Acta Pharm. Sinica B 2019, 9, 443–454. [Google Scholar] [CrossRef]
- Murphy, J.; Nakatani, Y.; Jamieson, S.A.; Dai, W.; Lucet, I.S.; Mace, P.D. Molecular Mechanism of CCAAT-Enhancer Binding Protein Recruitment by the TRIB1 Pseudokinase. Structure 2015, 23, 2111–2121. [Google Scholar] [CrossRef]
- Jamieson, S.A.; Ruan, Z.; Burgess, A.E.; Curry, J.R.; McMillan, H.D.; Brewster, J.L.; Dunbier, A.K.; Axtman, A.D.; Kannan, N.; Mace, P.D. Substrate binding allosterically relieves autoinhibition of the pseudokinase TRIB1. Sci. Signal. 2018, 11, eaau0597. [Google Scholar] [CrossRef]
- Nagy, E.; Maquat, L. A rule for termination-codon position within intron-containing genes: When nonsense affects RNA abundance. Trends Biochem. Sci. 1998, 23, 198–199. [Google Scholar] [CrossRef]
- Kurosaki, T.; Popp, M.W.; Maquat, L.E. Quality and quantity control of gene expression by nonsense-mediated mRNA decay. Nat. Rev. Mol. Cell Biol. 2019, 20, 406–420. [Google Scholar] [CrossRef]
- Niespolo, C.; Johnston, J.M.; Deshmukh, S.R.; Satam, S.; Shologu, Z.; Villacanas, O.; Sudbery, I.M.; Wilson, H.L.; Kiss-Toth, E. Tribbles-1 Expression and Its Function to Control Inflammatory Cytokines, Including Interleukin-8 Levels are Regulated by miRNAs in Macrophages and Prostate Cancer Cells. Front. Immunol. 2020, 11, 574046. [Google Scholar] [CrossRef]
- Briskin, D.; Wang, P.Y.; Bartel, D.P. The biochemical basis for the cooperative action of microRNAs. Proc. Natl. Acad. Sci. USA 2020, 117, 17764–17774. [Google Scholar] [CrossRef]
- Mashima, T.; Soma-Nagae, T.; Migita, T.; Kinoshita, R.; Iwamoto, A.; Yuasa, T.; Yonese, J.; Ishikawa, Y.; Seimiya, H. TRIB1 Supports Prostate Tumorigenesis and Tumor-Propagating Cell Survival by Regulation of Endoplasmic Reticulum Chaperone Expression. Cancer Res. 2014, 74, 4888–4897. [Google Scholar] [CrossRef]
- Shahrouzi, P.; Astobiza, I.; Cortazar, A.; Torrano, V.; Macchia, A.; Flores, J.; Niespolo, C.; Mendizabal, I.; Caloto, R.; Ercilla, A.; et al. Genomic and Functional Regulation of TRIB1 Contributes to Prostate Cancer Pathogenesis. Cancers 2020, 12, 2593. [Google Scholar] [CrossRef]
- E Kung, J.; Jura, N. The pseudokinase TRIB 1 toggles an intramolecular switch to regulate COP 1 nuclear export. EMBO J. 2019, 38, e99708. [Google Scholar] [CrossRef]
- Vlietstra, R.J.; van Alewijk, D.C.; Hermans, K.G.; van Steenbrugge, G.J.; Trapman, J. Frequent inactivation of PTEN in pros-tate cancer cell lines and xenografts. Cancer Res. 1998, 58, 2720–2723. [Google Scholar]
- Kim, H.-J.; Park, Y.I.; Dong, M.-S. Comparison of prostate cancer cell lines for androgen receptor-mediated reporter gene assays. Toxicol. In Vitro 2006, 20, 1159–1167. [Google Scholar] [CrossRef]
- Migliorini, D.; Bogaerts, S.; Defever, D.; Vyas, R.; Denecker, G.; Radaelli, E.; Zwolinska, A.; Depaepe, V.; Hochepied, T.; Skarnes, W.C.; et al. Cop1 constitutively regulates c-Jun protein stability and functions as a tumor suppressor in mice. J. Clin. Investig. 2011, 121, 1329–1343. [Google Scholar] [CrossRef] [PubMed]
- Vitari, A.C.; Leong, K.G.; Newton, K.; Yee, C.; O’Rourke, K.; Liu, J.; Phu, L.; Vij, R.; Ferrando, R.; Couto, S.S.; et al. COP1 is a tumour suppressor that causes degradation of ETS transcription factors. Nature 2011, 474, 403–406. [Google Scholar] [CrossRef]
- Bastola, D.R.; Pahwa, G.S.; Lin, M.-F.; Cheng, P.-W. Downregulation of PTEN/MMAC/TEP1 expression in human prostate cancer cell line DU145 by growth stimuli. Mol. Cell. Biochem. 2002, 236, 75–81. [Google Scholar] [CrossRef]
- Minamiguchi, K.; Kawada, M.; Someno, T.; Ishizuka, M. Androgen-independent prostate cancer DU145 cells suppress androgen-dependent growth of prostate stromal cells through production of inhibitory factors for androgen responsiveness. Biochem. Biophys. Res. Commun. 2003, 306, 629–636. [Google Scholar] [CrossRef]
- Dallavalle, C.; Albino, D.; Civenni, G.; Merulla, J.; Ostano, P.; Mello-Grand, M.; Rossi, S.; Losa, M.; D’Ambrosio, G.; Sessa, F.; et al. MicroRNA-424 impairs ubiquitination to activate STAT3 and promote prostate tumor progression. J. Clin. Investig. 2016, 126, 4585–4602. [Google Scholar] [CrossRef]
- Skjøth, I.; Issinger, O.-G. Profiling of signaling molecules in four different human prostate carcinoma cell lines before and after induction of apoptosis. Int. J. Oncol. 2006, 28, 217–229. [Google Scholar] [CrossRef]
- Fraser, M.; Zhao, H.; Luoto, K.R.; Lundin, C.; Coackley, C.; Chan, N.; Joshua, A.; Bismar, T.A.; Evans, A.; Helleday, T.; et al. PTEN Deletion in Prostate Cancer Cells Does Not Associate with Loss of RAD51 Function: Implications for Radiotherapy and Chemotherapy. Clin. Cancer Res. 2011, 18, 1015–1027. [Google Scholar] [CrossRef]
- Johnston, J.M.; Angyal, A.; Bauer, R.C.; Hamby, S.; Suvarna, S.K.; Baidžajevas, K.; Hegedus, Z.; Dear, T.N.; Turner, M.; Wilson, H.L.; et al. Myeloid Tribbles 1 induces early atherosclerosis via enhanced foam cell expansion. Sci. Adv. 2019, 5, eaax9183. [Google Scholar] [CrossRef]



| Cell Line | Characteristics | TRIB1 Protein Levels 1 | COP1 Protein | Publications |
|---|---|---|---|---|
| PC3 | PTEN– [61] | Genome rearrangement | Shahrouzi et al., 2020 [59] | |
| Androgen-independent [62] | High | undetectable protein [63,64] | ||
| Bone metastasis | Niespolo et al., 2020 [56] | |||
| DU145 | PTEN+ [65] | |||
| Androgen-independent [66] | NA | Yes [63,67] | Shahrouzi et al., 2020 [59] | |
| Brain metastasis | ||||
| LNCaP | PTEN– [61] | |||
| Androgen-sensitive [62] | Low | Yes [63,64,67] | Niespolo et al., 2020 [56] | |
| Lymph node metastasis | ||||
| 22Rv1 | PTEN+ [68,69] | |||
| Androgen-sensitive [62] | Low | Yes, based on RNA 2 | ||
| Prostate |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruiz-Cantos, M.; Hutchison, C.E.; Shoulders, C.C. Musings from the Tribbles Research and Innovation Network. Cancers 2021, 13, 4517. https://doi.org/10.3390/cancers13184517
Ruiz-Cantos M, Hutchison CE, Shoulders CC. Musings from the Tribbles Research and Innovation Network. Cancers. 2021; 13(18):4517. https://doi.org/10.3390/cancers13184517
Chicago/Turabian StyleRuiz-Cantos, Miriam, Claire E. Hutchison, and Carol C. Shoulders. 2021. "Musings from the Tribbles Research and Innovation Network" Cancers 13, no. 18: 4517. https://doi.org/10.3390/cancers13184517
APA StyleRuiz-Cantos, M., Hutchison, C. E., & Shoulders, C. C. (2021). Musings from the Tribbles Research and Innovation Network. Cancers, 13(18), 4517. https://doi.org/10.3390/cancers13184517






