Attenuated Chromosome Oscillation as a Cause of Chromosomal Instability in Cancer Cells
Abstract
:Simple Summary
Abstract
1. Introduction
2. Correction Mechanisms of Erroneous KT-MT Attachments and Its Relevance to CIN
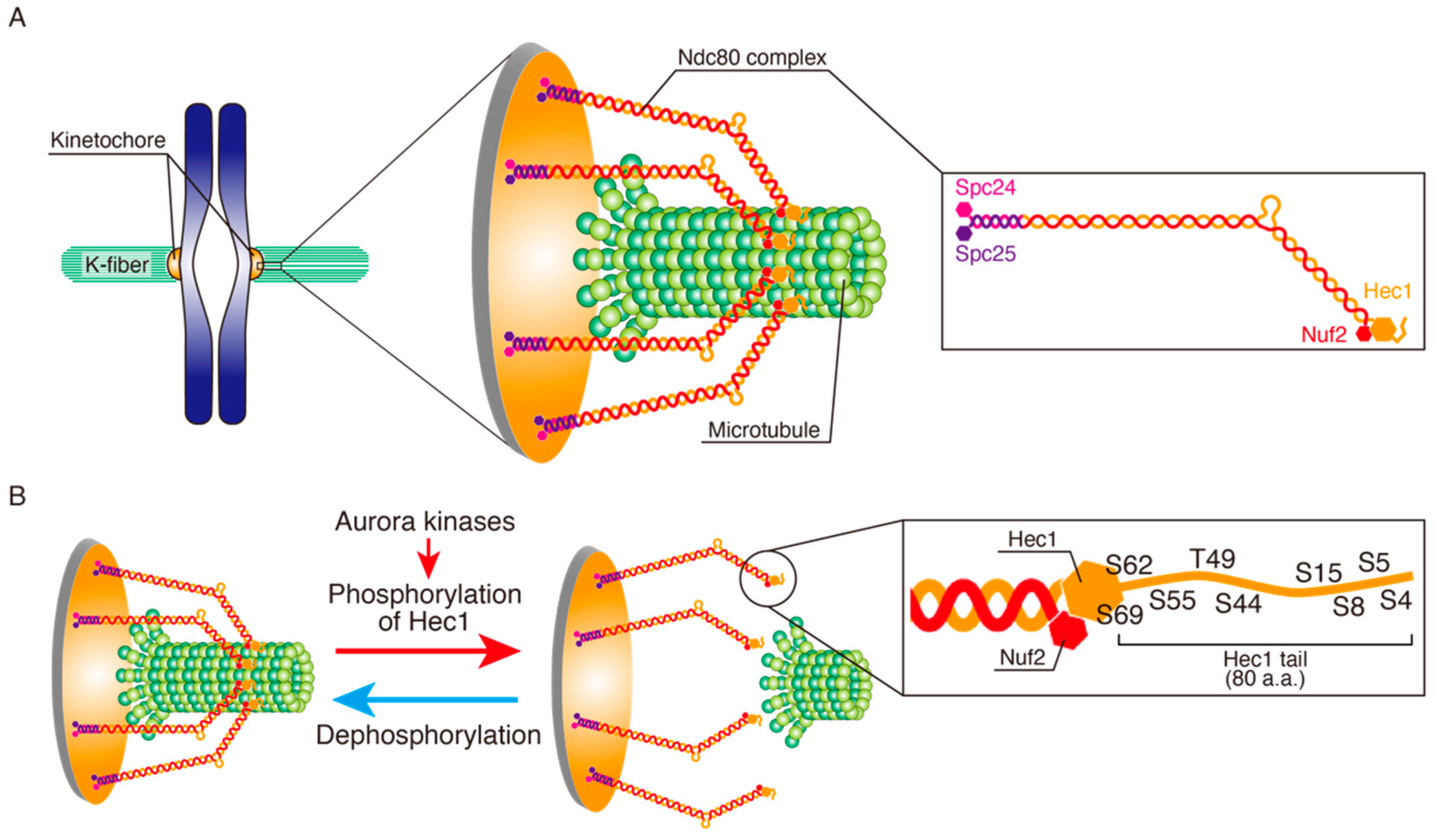
2.1. Overview of Chromosome Dynamics in Mitosis
2.2. Proper and Erroneous KT-MT Attachments
2.3. Error Correction Mechanisms of KT-MT Attachments
2.4. CIN Caused by Insufficient Correction of Erroneous KT-MT Attachments
3. Chromosome Oscillation
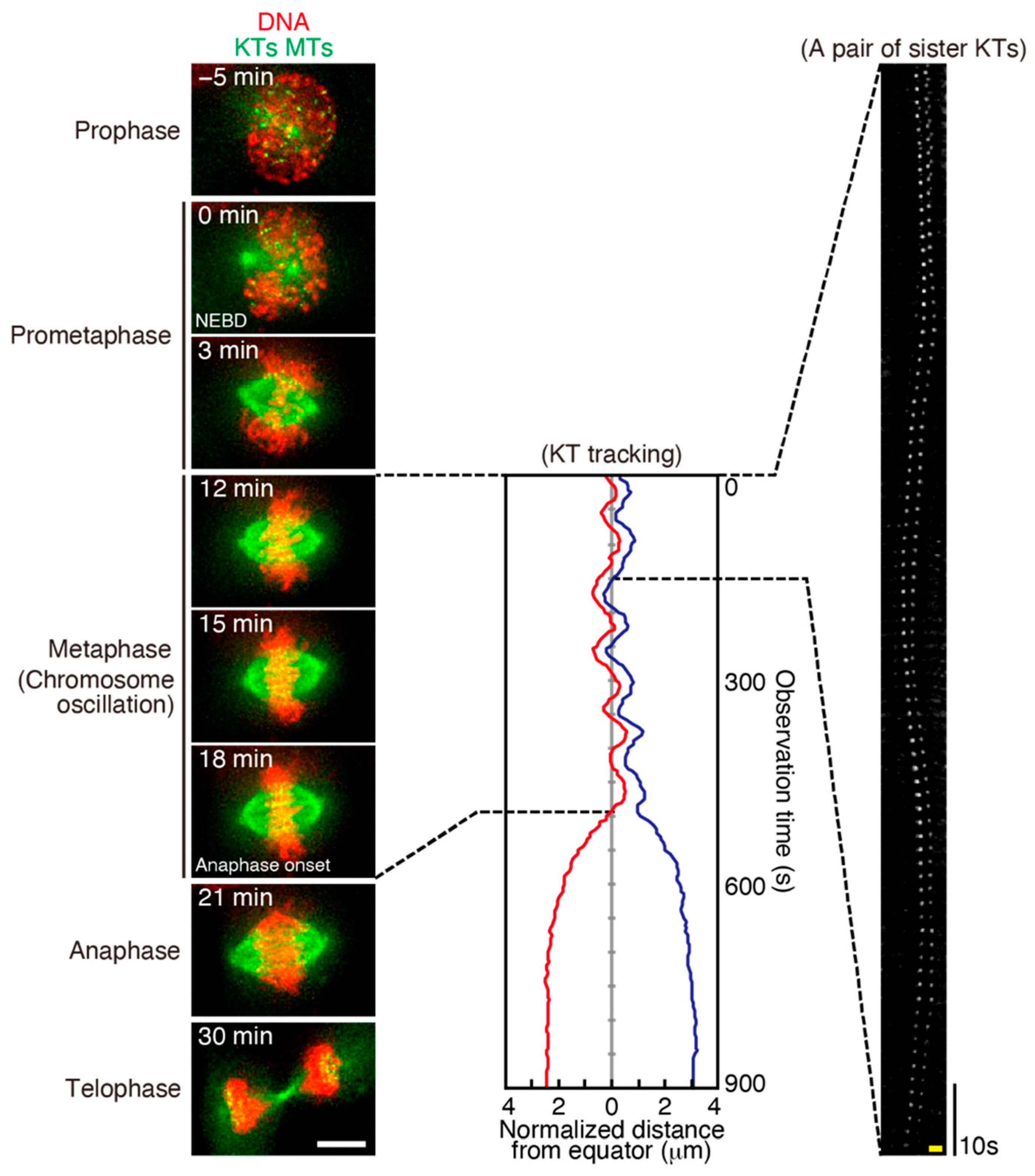
3.1. The Features of Chromosome Oscillation
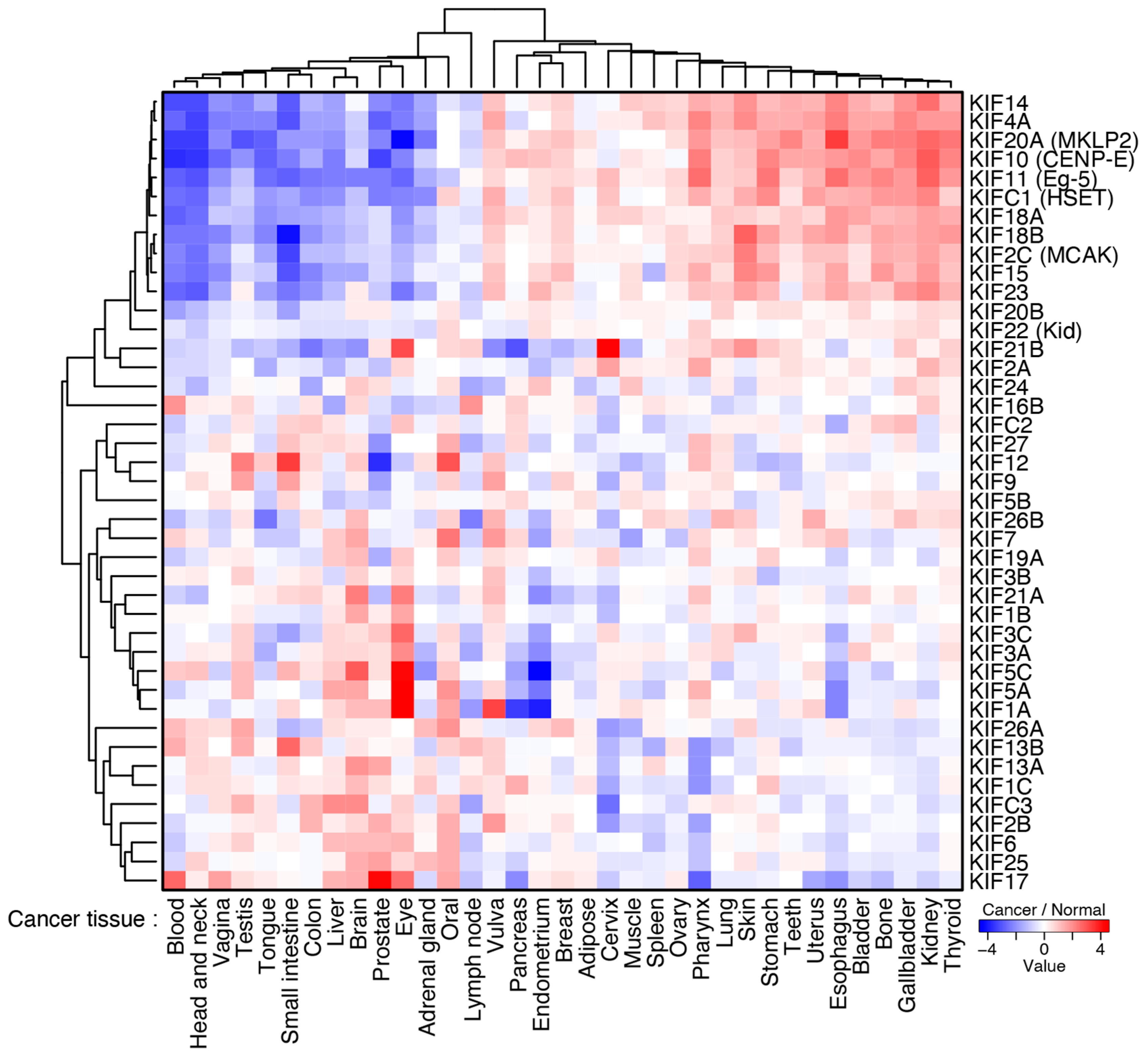
3.2. The Mechanisms of Chromosome Oscillation
4. Chromosome Oscillation Plays a Role in Correction of Erroneous KT-MT Attachments
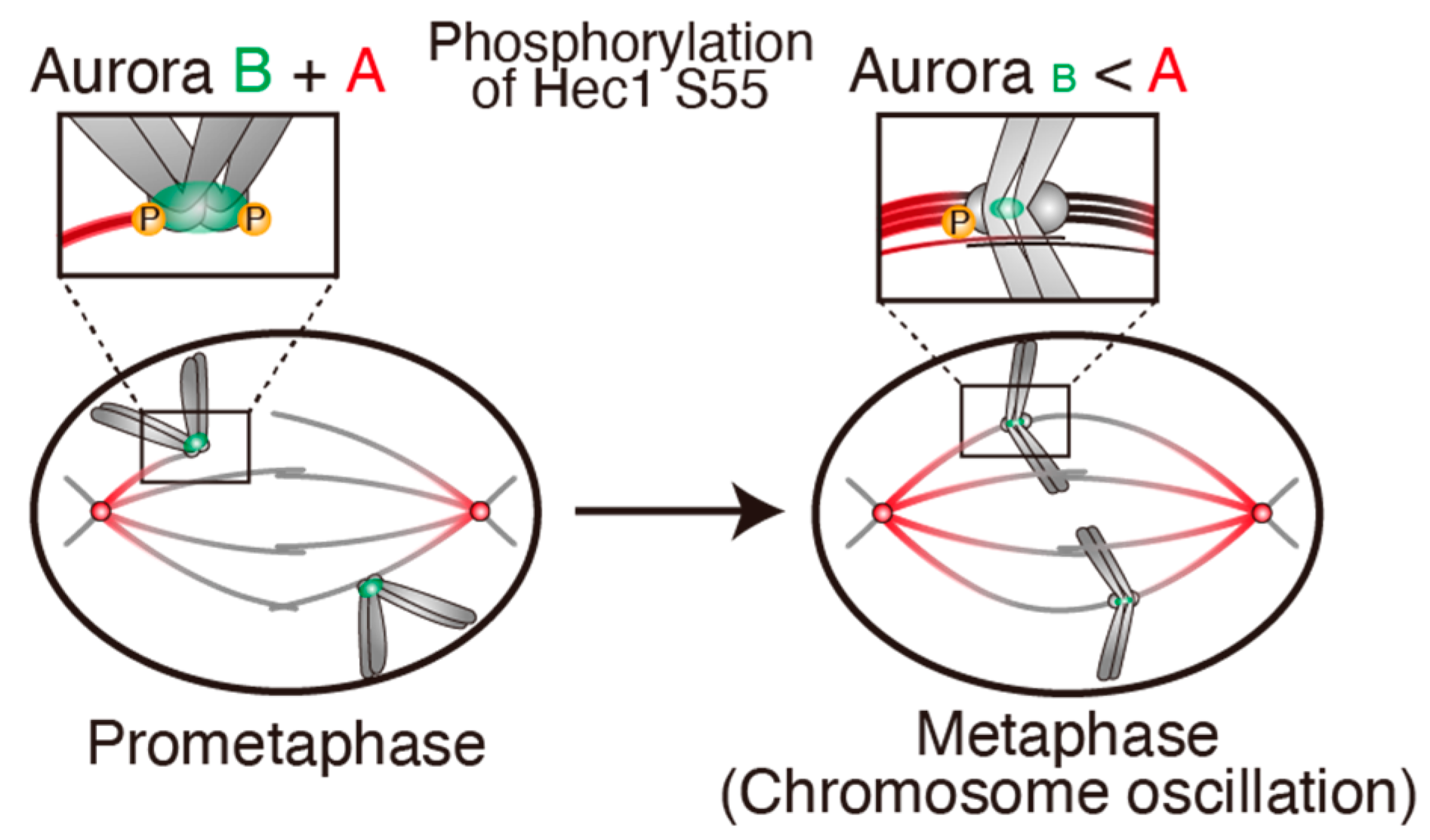
4.1. Hec1 Phosphorylation by Aurora A in Metaphase
4.2. Chromosome Oscillation Promotes Hec1 Phosphorylation by Aurora A
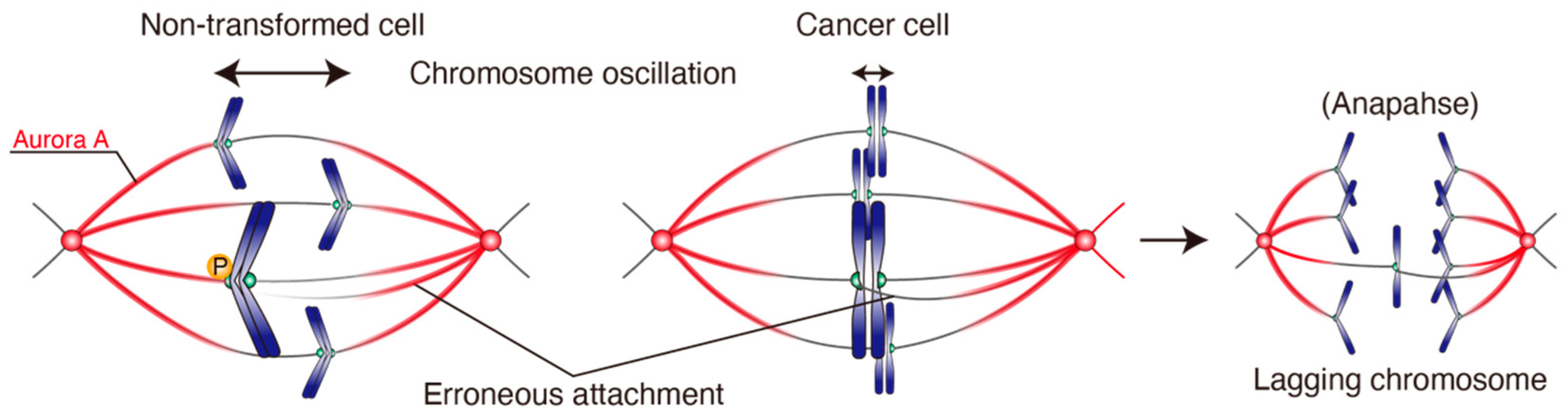
4.3. Chromosome Oscillation Is Attenuated in CIN Cancer Cell Lines
4.4. Chromosome Oscillation Facilitates Correction of Erroneous KT-MT Attachments
5. Conclusions and Outlook
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Weaver, B.A.; Cleveland, D.W. Does aneuploidy cause cancer? Curr. Opin. Cell Biol. 2006, 18, 658–667. [Google Scholar] [CrossRef]
- Taylor, A.M.; Shih, J.; Ha, G.; Gao, G.F.; Zhang, X.; Berger, A.C.; Schumacher, S.E.; Wang, C.; Hu, H.; Liu, J.; et al. Genomic and functional approaches to understanding cancer aneuploidy. Cancer Cell 2018, 33, 676–689.e3. [Google Scholar] [CrossRef] [Green Version]
- Pfau, S.J.; Amon, A. Chromosomal instability and aneuploidy in cancer: From yeast to man. EMBO Rep. 2012, 13, 515–527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orr, B.; Godek, K.M.; Compton, D. Aneuploidy. Curr. Biol. 2015, 25, R538–R542. [Google Scholar] [CrossRef] [Green Version]
- Sansregret, L.; Swanton, C. The role of aneuploidy in cancer evolution. Cold Spring Harb. Perspect. Med. 2017, 7, a028373. [Google Scholar] [CrossRef] [Green Version]
- Burrell, R.A.; McClelland, S.E.; Endesfelder, D.; Groth, P.; Weller, M.C.; Shaikh, N.; Domingo, E.; Kanu, N.; Dewhurst, S.M.; Gronroos, E.; et al. Replication stress links structural and numerical cancer chromosomal instability. Nature 2013, 494, 492–496. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, K.; Hirota, T. Chromosomal instability: A common feature and a therapeutic target of cancer. Biochim. Biophys. Acta 2016, 1866, 64–75. [Google Scholar] [CrossRef]
- Jo, M.; Kusano, Y.; Hirota, T. Unraveling pathologies underlying chromosomal instability in cancers. Cancer Sci. 2021, 112, 2975. [Google Scholar] [CrossRef]
- Bakhoum, S.F.; Compton, D.A. Chromosomal instability and cancer: A complex relationship with therapeutic potential. J. Clin. Investig. 2012, 122, 1138–1143. [Google Scholar] [CrossRef] [Green Version]
- McGranahan, N.; Burrell, R.A.; Endesfelder, D.; Novelli, M.R.; Swanton, C. Cancer chromosomal instability: Therapeutic and diagnostic challenges. EMBO Rep. 2012, 13, 528–538. [Google Scholar] [CrossRef] [Green Version]
- Bakhoum, S.F.; Landau, D.A. Chromosomal instability as a driver of tumor heterogeneity and evolution. Cold Spring Harb. Perspect. Med. 2017, 7, a029611. [Google Scholar] [CrossRef] [PubMed]
- McGranahan, N.; Swanton, C. Clonal heterogeneity and tumor evolution: Past, present, and the future. Cell 2017, 168, 613–628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zasadil, L.M.; Britigan, E.M.; Weaver, B.A. 2n or not 2n: Aneuploidy, polyploidy and chromosomal instability in primary and tumor cells. Semin. Cell Dev. Biol. 2013, 24, 370–379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holland, A.J.; Cleveland, D.W. Boveri revisited: Chromosomal instability, aneuploidy and tumorigenesis. Nat. Rev. Mol. Cell Biol. 2009, 10, 478–487. [Google Scholar] [CrossRef] [Green Version]
- Desai, A.; Mitchison, T.J. Microtubule polymerization dynamics. Annu. Rev. Cell Dev. Biol. 1997, 13, 83–117. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, T.U.; Stark, M.J.; Tanaka, K. Kinetochore capture and bi-orientation on the mitotic spindle. Nat. Rev. Mol. Cell Biol. 2005, 6, 929–942. [Google Scholar] [CrossRef]
- Godek, K.M.; Kabeche, L.; Compton, D.A. Regulation of kinetochore-microtubule attachments through homeostatic control during mitosis. Nat. Rev. Mol. Cell Biol. 2015, 16, 57–64. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, K.; Hirota, T. Chromosome segregation machinery and cancer. Cancer Sci. 2009, 100, 1158–1165. [Google Scholar] [CrossRef]
- Ricke, R.M.; van Deursen, J.M. Correction of microtubule-kinetochore attachment errors: Mechanisms and role in tumor suppression. Semin. Cell Dev. Biol. 2011, 22, 559–565. [Google Scholar] [CrossRef] [Green Version]
- Maiato, H.; Gomes, A.M.; Sousa, F.; Barisic, M. Mechanisms of chromosome congression during mitosis. Biology 2017, 6, 13. [Google Scholar] [CrossRef]
- Auckland, P.; McAinsh, A.D. Building an integrated model of chromosome congression. J. Cell Sci. 2015, 128, 3363–3374. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, K. Dynamic regulation of kinetochore-microtubule interaction during mitosis. J. Biochem. 2012, 152, 415–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Renda, F.; Khodjakov, A. Role of spatial patterns and kinetochore architecture in spindle morphogenesis. Semin. Cell Dev. Biol. 2021, 117, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, L.T.; Maiato, H. Prometaphase. Semin. Cell Dev. Biol. 2021, 117, 52–61. [Google Scholar] [CrossRef]
- Cimini, D. Merotelic kinetochore orientation, aneuploidy, and cancer. Biochim. Biophys. Acta 2008, 1786, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Civelekoglu-Scholey, G.; Cimini, D. Modelling chromosome dynamics in mitosis: A historical perspective on models of metaphase and anaphase in eukaryotic cells. Interface Focus 2014, 4, 20130073. [Google Scholar] [CrossRef] [PubMed]
- Bissonette, S.; Stumpff, J. Quantifying mitotic chromosome dynamics and positioning. J. Cell Physiol. 2014, 229, 1301–1305. [Google Scholar] [CrossRef]
- Winey, M.; Mamay, C.L.; O’Toole, E.T.; Mastronarde, D.N.; Giddings, T.H., Jr.; McDonald, K.L.; McIntosh, J.R. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J. Cell Biol. 1995, 129, 1601–1615. [Google Scholar] [CrossRef] [Green Version]
- O’Toole, E.T.; Winey, M.; McIntosh, J.R. High-voltage electron tomography of spindle pole bodies and early mitotic spindles in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 1999, 10, 2017–2031. [Google Scholar] [CrossRef]
- Ding, R.; McDonald, K.L.; McIntosh, J.R. Three-dimensional reconstruction and analysis of mitotic spindles from the yeast, Schizosaccharomyces pombe. J. Cell Biol. 1993, 120, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Wendell, K.L.; Wilson, L.; Jordan, M.A. Mitotic block in HeLa cells by vinblastine: Ultrastructural changes in kinetochore-microtubule attachment and in centrosomes. J. Cell Sci. 1993, 104 Pt 2, 261–274. [Google Scholar] [CrossRef]
- McEwen, B.F.; Heagle, A.B.; Cassels, G.O.; Buttle, K.F.; Rieder, C.L. Kinetochore fiber maturation in PtK1 cells and its implications for the mechanisms of chromosome congression and anaphase onset. J. Cell Biol. 1997, 137, 1567–1580. [Google Scholar] [CrossRef] [Green Version]
- Rieder, C.L. The formation, structure, and composition of the mammalian kinetochore and kinetochore fiber. Int. Rev. Cytol. 1982, 79, 1–58. [Google Scholar]
- Tanaka, T.U. Kinetochore-microtubule interactions: Steps towards bi-orientation. EMBO J. 2010, 29, 4070–4082. [Google Scholar] [CrossRef]
- Itoh, G.; Ikeda, M.; Iemura, K.; Amin, M.A.; Kuriyama, S.; Tanaka, M.; Mizuno, N.; Osakada, H.; Haraguchi, T.; Tanaka, K. Lateral attachment of kinetochores to microtubules is enriched in prometaphase rosette and facilitates chromosome alignment and bi-orientation establishment. Sci. Rep. 2018, 8, 3888. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, K. Regulatory mechanisms of kinetochore-microtubule interaction in mitosis. Cell Mol. Life Sci. 2013, 70, 559–579. [Google Scholar] [CrossRef]
- Alexander, S.P.; Rieder, C.L. Chromosome motion during attachment to the vertebrate spindle: Initial saltatory-like behavior of chromosomes and quantitative analysis of force production by nascent kinetochore fibers. J. Cell Biol. 1991, 113, 805–815. [Google Scholar] [CrossRef]
- Yang, Z.; Tulu, U.S.; Wadsworth, P.; Rieder, C.L. Kinetochore dynein is required for chromosome motion and congression independent of the spindle checkpoint. Curr. Biol. 2007, 17, 973–980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, K.; Mukae, N.; Dewar, H.; van Breugel, M.; James, E.K.; Prescott, A.R.; Antony, C.; Tanaka, T.U. Molecular mechanisms of kinetochore capture by spindle microtubules. Nature 2005, 434, 987–994. [Google Scholar] [CrossRef] [PubMed]
- Rieder, C.L.; Alexander, S.P. Kinetochores are transported poleward along a single astral microtubule during chromosome attachment to the spindle in newt lung cells. J. Cell Biol. 1990, 110, 81–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharp, D.J.; Rogers, G.C.; Scholey, J.M. Cytoplasmic dynein is required for poleward chromosome movement during mitosis in Drosophila embryos. Nat. Cell Biol. 2000, 2, 922–930. [Google Scholar] [CrossRef]
- Iemura, K.; Tanaka, K. Chromokinesin Kid and kinetochore kinesin CENP-E differentially support chromosome congression without end-on attachment to microtubules. Nat. Commun. 2015, 6, 6447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kapoor, T.M.; Lampson, M.A.; Hergert, P.; Cameron, L.; Cimini, D.; Salmon, E.D.; McEwen, B.F.; Khodjakov, A. Chromosomes can congress to the metaphase plate before biorientation. Science 2006, 311, 388–391. [Google Scholar] [CrossRef] [Green Version]
- Wood, K.W.; Sakowicz, R.; Goldstein, L.S.; Cleveland, D.W. CENP-E is a plus end-directed kinetochore motor required for metaphase chromosome alignment. Cell 1997, 91, 357–366. [Google Scholar] [CrossRef] [Green Version]
- Kops, G.J.; Saurin, A.T.; Meraldi, P. Finding the middle ground: How kinetochores power chromosome congression. Cell Mol. Life Sci. 2010, 67, 2145–2161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, K.; Kitamura, E.; Kitamura, Y.; Tanaka, T.U. Molecular mechanisms of microtubule-dependent kinetochore transport toward spindle poles. J. Cell Biol. 2007, 178, 269–281. [Google Scholar] [CrossRef] [Green Version]
- Shrestha, R.L.; Draviam, V.M. Lateral to end-on conversion of chromosome-microtubule attachment requires kinesins CENP-E and MCAK. Curr. Biol. 2013, 23, 1514–1526. [Google Scholar] [CrossRef] [Green Version]
- Loncarek, J.; Kisurina-Evgenieva, O.; Vinogradova, T.; Hergert, P.; La Terra, S.; Kapoor, T.M.; Khodjakov, A. The centromere geometry essential for keeping mitosis error free is controlled by spindle forces. Nature 2007, 450, 745–749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Indjeian, V.B.; Murray, A.W. Budding yeast mitotic chromosomes have an intrinsic bias to biorient on the spindle. Curr. Biol. 2007, 17, 1837–1846. [Google Scholar] [CrossRef] [Green Version]
- Edelmaier, C.; Lamson, A.R.; Gergely, Z.R.; Ansari, S.; Blackwell, R.; McIntosh, J.R.; Glaser, M.A.; Betterton, M.D. Mechanisms of chromosome biorientation and bipolar spindle assembly analyzed by computational modeling. eLife 2020, 9, e48787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magidson, V.; O’Connell, C.B.; Loncarek, J.; Paul, R.; Mogilner, A.; Khodjakov, A. The spatial arrangement of chromosomes during prometaphase facilitates spindle assembly. Cell 2011, 146, 555–567. [Google Scholar] [CrossRef] [Green Version]
- Silkworth, W.T.; Nardi, I.K.; Paul, R.; Mogilner, A.; Cimini, D. Timing of centrosome separation is important for accurate chromosome segregation. Mol. Biol. Cell 2012, 23, 401–411. [Google Scholar] [CrossRef] [PubMed]
- Kaseda, K.; McAinsh, A.D.; Cross, R.A. Dual pathway spindle assembly increases both the speed and the fidelity of mitosis. Biol. Open 2012, 1, 12–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sacristan, C.; Ahmad, M.U.D.; Keller, J.; Fermie, J.; Groenewold, V.; Tromer, E.; Fish, A.; Melero, R.; Carazo, J.M.; Klumperman, J.; et al. Dynamic kinetochore size regulation promotes microtubule capture and chromosome biorientation in mitosis. Nat. Cell Biol. 2018, 20, 800–810. [Google Scholar] [CrossRef]
- Magidson, V.; Paul, R.; Yang, N.; Ault, J.G.; O’Connell, C.B.; Tikhonenko, I.; McEwen, B.F.; Mogilner, A.; Khodjakov, A. Adaptive changes in the kinetochore architecture facilitate proper spindle assembly. Nat. Cell Biol. 2015, 17, 1134–1144. [Google Scholar] [CrossRef] [Green Version]
- Lampson, M.A.; Grishchuk, E.L. Mechanisms to avoid and correct erroneous kinetochore-microtubule attachments. Biology 2017, 6, 1. [Google Scholar] [CrossRef]
- Bakhoum, S.F.; Compton, D.A. Kinetochores and disease: Keeping microtubule dynamics in check! Curr. Opin. Cell Biol. 2012, 24, 64–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaytsev, A.V.; Grishchuk, E.L. Basic mechanism for biorientation of mitotic chromosomes is provided by the kinetochore geometry and indiscriminate turnover of kinetochore microtubules. Mol. Biol. Cell 2015, 26, 3985–3998. [Google Scholar] [CrossRef] [PubMed]
- Mitchison, T.; Kirschner, M. Dynamic instability of microtubule growth. Nature 1984, 312, 237–242. [Google Scholar] [CrossRef] [PubMed]
- Kirschner, M.; Mitchison, T. Beyond self-assembly: From microtubules to morphogenesis. Cell 1986, 45, 329–342. [Google Scholar] [CrossRef]
- Hayden, J.H.; Bowser, S.S.; Rieder, C.L. Kinetochores capture astral microtubules during chromosome attachment to the mitotic spindle: Direct visualization in live newt lung cells. J. Cell Biol. 1990, 111, 1039–1045. [Google Scholar] [CrossRef]
- Bakhoum, S.F.; Thompson, S.L.; Manning, A.L.; Compton, D.A. Genome stability is ensured by temporal control of kinetochore-microtubule dynamics. Nat. Cell Biol. 2009, 11, 27–35. [Google Scholar] [CrossRef] [Green Version]
- Kabeche, L.; Compton, D.A. Cyclin A regulates kinetochore microtubules to promote faithful chromosome segregation. Nature 2013, 502, 110–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varma, D.; Salmon, E.D. The KMN protein network--chief conductors of the kinetochore orchestra. J. Cell Sci. 2012, 125, 5927–5936. [Google Scholar] [CrossRef] [Green Version]
- Durfee, T.; Becherer, K.; Chen, P.L.; Yeh, S.H.; Yang, Y.; Kilburn, A.E.; Lee, W.H.; Elledge, S.J. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993, 7, 555–569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Riley, D.J.; Chen, P.L.; Lee, W.H. HEC, a novel nuclear protein rich in leucine heptad repeats specifically involved in mitosis. Mol. Cell. Biol. 1997, 17, 6049–6056. [Google Scholar] [CrossRef] [Green Version]
- Tooley, J.; Stukenberg, P.T. The Ndc80 complex: Integrating the kinetochore’s many movements. Chromosome Res. 2011, 19, 377–391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joglekar, A.P.; DeLuca, J.G. Chromosome segregation: Ndc80 can carry the load. Curr. Biol. 2009, 19, R404–R407. [Google Scholar] [CrossRef] [Green Version]
- Wimbish, R.T.; DeLuca, J.G. Hec1/Ndc80 tail domain function at the kinetochore-microtubule interface. Front. Cell Dev. Biol. 2020, 8, 43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeLuca, J.G.; Gall, W.E.; Ciferri, C.; Cimini, D.; Musacchio, A.; Salmon, E.D. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell 2006, 127, 969–982. [Google Scholar] [CrossRef] [Green Version]
- Krenn, V.; Musacchio, A. The Aurora B Kinase in chromosome Bi-orientation and spindle checkpoint signaling. Front. Oncol. 2015, 5, 225. [Google Scholar] [CrossRef] [Green Version]
- Liu, D.; Vleugel, M.; Backer, C.B.; Hori, T.; Fukagawa, T.; Cheeseman, I.M.; Lampson, M.A. Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase. J. Cell Biol. 2010, 188, 809–820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suijkerbuijk, S.J.; Vleugel, M.; Teixeira, A.; Kops, G.J. Integration of kinase and phosphatase activities by BUBR1 ensures formation of stable kinetochore-microtubule attachments. Dev. Cell 2012, 23, 745–755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kruse, T.; Zhang, G.; Larsen, M.S.; Lischetti, T.; Streicher, W.; Kragh Nielsen, T.; Bjorn, S.P.; Nilsson, J. Direct binding between BubR1 and B56-PP2A phosphatase complexes regulate mitotic progression. J. Cell Sci. 2013, 126, 1086–1092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, P.; Raetz, E.A.; Kitagawa, M.; Virshup, D.M.; Lee, S.H. BUBR1 recruits PP2A via the B56 family of targeting subunits to promote chromosome congression. Biol. Open 2013, 2, 479–486. [Google Scholar] [CrossRef] [Green Version]
- Smith, R.J.; Cordeiro, M.H.; Davey, N.E.; Vallardi, G.; Ciliberto, A.; Gross, F.; Saurin, A.T. PP1 and PP2A use opposite phospho-dependencies to control distinct processes at the kinetochore. Cell Rep. 2019, 28, 2206–2219.e8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Welburn, J.P.; Vleugel, M.; Liu, D.; Yates, J.R., III; Lampson, M.A.; Fukagawa, T.; Cheeseman, I.M. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol. Cell 2010, 38, 383–392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keen, N.; Taylor, S. Aurora-kinase inhibitors as anticancer agents. Nat. Rev. Cancer 2004, 4, 927–936. [Google Scholar] [CrossRef] [PubMed]
- Carmena, M.; Wheelock, M.; Funabiki, H.; Earnshaw, W.C. The chromosomal passenger complex (CPC): From easy rider to the godfather of mitosis. Nat. Rev. Mol. Cell Biol. 2012, 13, 789–803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, D.; Vader, G.; Vromans, M.J.; Lampson, M.A.; Lens, S.M. Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science 2009, 323, 1350–1353. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, T.U.; Rachidi, N.; Janke, C.; Pereira, G.; Galova, M.; Schiebel, E.; Stark, M.J.; Nasmyth, K. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell 2002, 108, 317–329. [Google Scholar] [CrossRef]
- Sarangapani, K.K.; Asbury, C.L. Catch and release: How do kinetochores hook the right microtubules during mitosis? Trends Genet. 2014, 30, 150–159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cimini, D.; Wan, X.; Hirel, C.B.; Salmon, E.D. Aurora kinase promotes turnover of kinetochore microtubules to reduce chromosome segregation errors. Curr. Biol. 2006, 16, 1711–1718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knowlton, A.L.; Lan, W.; Stukenberg, P.T. Aurora B is enriched at merotelic attachment sites, where it regulates MCAK. Curr. Biol. 2006, 16, 1705–1710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cimini, D.; Moree, B.; Canman, J.C.; Salmon, E.D. Merotelic kinetochore orientation occurs frequently during early mitosis in mammalian tissue cells and error correction is achieved by two different mechanisms. J. Cell Sci. 2003, 116, 4213–4225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gregan, J.; Polakova, S.; Zhang, L.; Tolic-Norrelykke, I.M.; Cimini, D. Merotelic kinetochore attachment: Causes and effects. Trends Cell Biol. 2011, 21, 374–381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Funabiki, H. Correcting aberrant kinetochore microtubule attachments: A hidden regulation of Aurora B on microtubules. Curr. Opin. Cell Biol. 2019, 58, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, P.; Zaytsev, A.V.; Godzi, M.; Ataullakhanov, F.I.; Grishchuk, E.L.; Stukenberg, P.T. The binding of Borealin to microtubules underlies a tension independent kinetochore-microtubule error correction pathway. Nat. Commun. 2019, 10, 682. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.Y.; Renda, F.; Zhang, H.; Gokden, A.; Wu, D.Z.; Chenoweth, D.M.; Khodjakov, A.; Lampson, M.A. Tension promotes kinetochore-microtubule release by Aurora B kinase. J. Cell Biol. 2021, 220, e202007030. [Google Scholar] [CrossRef] [PubMed]
- Andrews, P.D.; Ovechkina, Y.; Morrice, N.; Wagenbach, M.; Duncan, K.; Wordeman, L.; Swedlow, J.R. Aurora B regulates MCAK at the mitotic centromere. Dev. Cell 2004, 6, 253–268. [Google Scholar] [CrossRef] [Green Version]
- Lan, W.; Zhang, X.; Kline-Smith, S.L.; Rosasco, S.E.; Barrett-Wilt, G.A.; Shabanowitz, J.; Hunt, D.F.; Walczak, C.E.; Stukenberg, P.T. Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr. Biol. 2004, 14, 273–286. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Holland, A.J.; Lan, W.; Cleveland, D.W. Aurora kinases and protein phosphatase 1 mediate chromosome congression through regulation of CENP-E. Cell 2010, 142, 444–455. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, J.C.; Arthanari, H.; Boeszoermenyi, A.; Dashkevich, N.M.; Wilson-Kubalek, E.M.; Monnier, N.; Markus, M.; Oberer, M.; Milligan, R.A.; Bathe, M.; et al. The kinetochore-bound Ska1 complex tracks depolymerizing microtubules and binds to curved protofilaments. Dev. Cell 2012, 23, 968–980. [Google Scholar] [CrossRef] [Green Version]
- Koffa, M.D.; Casanova, C.M.; Santarella, R.; Kocher, T.; Wilm, M.; Mattaj, I.W. HURP is part of a Ran-dependent complex involved in spindle formation. Curr. Biol. 2006, 16, 743–754. [Google Scholar] [CrossRef] [Green Version]
- Wong, J.; Fang, G. HURP controls spindle dynamics to promote proper interkinetochore tension and efficient kinetochore capture. J. Cell Biol. 2006, 173, 879–891. [Google Scholar] [CrossRef] [PubMed]
- Chmatal, L.; Yang, K.; Schultz, R.M.; Lampson, M.A. Spatial regulation of kinetochore microtubule attachments by destabilization at spindle poles in Meiosis, I. Curr. Biol. 2015, 25, 1835–1841. [Google Scholar] [CrossRef] [Green Version]
- Ye, A.A.; Deretic, J.; Hoel, C.M.; Hinman, A.W.; Cimini, D.; Welburn, J.P.; Maresca, T.J. Aurora A Kinase contributes to a pole-based error correction pathway. Curr. Biol. 2015, 25, 1842–1851. [Google Scholar] [CrossRef] [Green Version]
- Hochegger, H.; Hegarat, N.; Pereira-Leal, J.B. Aurora at the pole and equator: Overlapping functions of Aurora kinases in the mitotic spindle. Open Biol. 2013, 3, 120185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maure, J.F.; Kitamura, E.; Tanaka, T.U. Mps1 kinase promotes sister-kinetochore bi-orientation by a tension-dependent mechanism. Curr. Biol. 2007, 17, 2175–2182. [Google Scholar] [CrossRef] [Green Version]
- Jelluma, N.; Brenkman, A.B.; van den Broek, N.J.; Cruijsen, C.W.; van Osch, M.H.; Lens, S.M.; Medema, R.H.; Kops, G.J. Mps1 phosphorylates Borealin to control Aurora B activity and chromosome alignment. Cell 2008, 132, 233–246. [Google Scholar] [CrossRef] [Green Version]
- Santaguida, S.; Tighe, A.; D’Alise, A.M.; Taylor, S.S.; Musacchio, A. Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. J. Cell Biol. 2010, 190, 73–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maciejowski, J.; Drechsler, H.; Grundner-Culemann, K.; Ballister, E.R.; Rodriguez-Rodriguez, J.A.; Rodriguez-Bravo, V.; Jones, M.J.K.; Foley, E.; Lampson, M.A.; Daub, H.; et al. Mps1 regulates kinetochore-microtubule attachment stability via the ska complex to ensure error-free chromosome segregation. Dev. Cell 2017, 41, 143–156.e6. [Google Scholar] [CrossRef] [Green Version]
- Hayward, D.; Bancroft, J.; Mangat, D.; Alfonso-Perez, T.; Dugdale, S.; McCarthy, J.; Barr, F.A.; Gruneberg, U. Checkpoint signaling and error correction require regulation of the MPS1 T-loop by PP2A-B56. J. Cell Biol. 2019, 218, 3188–3199. [Google Scholar] [CrossRef] [Green Version]
- Benzi, G.; Camasses, A.; Atsunori, Y.; Katou, Y.; Shirahige, K.; Piatti, S. A common molecular mechanism underlies the role of Mps1 in chromosome biorientation and the spindle assembly checkpoint. EMBO Rep. 2020, 21, e50257. [Google Scholar] [CrossRef] [PubMed]
- Petsalaki, E.; Akoumianaki, T.; Black, E.J.; Gillespie, D.A.; Zachos, G. Phosphorylation at serine 331 is required for Aurora B activation. J. Cell Biol. 2011, 195, 449–466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petsalaki, E.; Zachos, G. Chk1 and Mps1 jointly regulate correction of merotelic kinetochore attachments. J. Cell Sci. 2013, 126, 1235–1246. [Google Scholar] [CrossRef] [Green Version]
- Miller, M.P.; Asbury, C.L.; Biggins, S. A TOG protein confers tension sensitivity to kinetochore-microtubule attachments. Cell 2016, 165, 1428–1439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herman, J.A.; Miller, M.P.; Biggins, S. chTOG is a conserved mitotic error correction factor. eLife 2020, 9, e61773. [Google Scholar] [CrossRef]
- Musacchio, A. The molecular biology of spindle assembly checkpoint signaling dynamics. Curr. Biol. 2015, 25, R1002–R1018. [Google Scholar] [CrossRef] [Green Version]
- Pinsky, B.A.; Biggins, S. The spindle checkpoint: Tension versus attachment. Trends Cell Biol. 2005, 15, 486–493. [Google Scholar] [CrossRef] [PubMed]
- Nezi, L.; Musacchio, A. Sister chromatid tension and the spindle assembly checkpoint. Curr. Opin. Cell Biol. 2009, 21, 785–795. [Google Scholar] [CrossRef] [PubMed]
- Cimini, D.; Howell, B.; Maddox, P.; Khodjakov, A.; Degrassi, F.; Salmon, E.D. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J. Cell Biol. 2001, 153, 517–527. [Google Scholar] [CrossRef] [PubMed]
- Cimini, D.; Cameron, L.A.; Salmon, E.D. Anaphase spindle mechanics prevent mis-segregation of merotelically oriented chromosomes. Curr. Biol. 2004, 14, 2149–2155. [Google Scholar] [CrossRef] [Green Version]
- Thompson, S.L.; Compton, D.A. Chromosome missegregation in human cells arises through specific types of kinetochore-microtubule attachment errors. Proc. Natl. Acad. Sci. USA 2011, 108, 17974–17978. [Google Scholar] [CrossRef] [Green Version]
- Terradas, M.; Martin, M.; Tusell, L.; Genesca, A. DNA lesions sequestered in micronuclei induce a local defective-damage response. DNA Repair 2009, 8, 1225–1234. [Google Scholar] [CrossRef]
- Terradas, M.; Martin, M.; Tusell, L.; Genesca, A. Genetic activities in micronuclei: Is the DNA entrapped in micronuclei lost for the cell? Mutat. Res. 2010, 705, 60–67. [Google Scholar] [CrossRef]
- Terradas, M.; Martin, M.; Hernandez, L.; Tusell, L.; Genesca, A. Nuclear envelope defects impede a proper response to micronuclear DNA lesions. Mutat. Res. 2012, 729, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Crasta, K.; Ganem, N.J.; Dagher, R.; Lantermann, A.B.; Ivanova, E.V.; Pan, Y.F.; Nezi, L.; Protopopov, A.; Chowdhury, D.; Pellman, D. DNA breaks and chromosome pulverization from errors in mitosis. Nature 2012, 482, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Stephens, P.J.; Greenman, C.D.; Fu, B.; Yang, F.; Bignell, G.R.; Mudie, L.J.; Pleasance, E.D.; Lau, K.W.; Beare, D.; Stebbings, L.A.; et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 2011, 144, 27–40. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.Z.; Spektor, A.; Cornils, H.; Francis, J.M.; Jackson, E.K.; Liu, S.; Meyerson, M.; Pellman, D. Chromothripsis from DNA damage in micronuclei. Nature 2015, 522, 179–184. [Google Scholar] [CrossRef] [Green Version]
- Janssen, A.; van der Burg, M.; Szuhai, K.; Kops, G.J.; Medema, R.H. Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science 2011, 333, 1895–1898. [Google Scholar] [CrossRef] [PubMed]
- Soto, M.; Raaijmakers, J.A.; Medema, R.H. Consequences of genomic diversification induced by segregation errors. Trends Genet. 2019, 35, 279–291. [Google Scholar] [CrossRef] [PubMed]
- Pihan, G.A.; Purohit, A.; Wallace, J.; Knecht, H.; Woda, B.; Quesenberry, P.; Doxsey, S.J. Centrosome defects and genetic instability in malignant tumors. Cancer Res. 1998, 58, 3974–3985. [Google Scholar]
- Chan, J.Y. A clinical overview of centrosome amplification in human cancers. Int. J. Biol. Sci. 2011, 7, 1122–1144. [Google Scholar] [CrossRef] [PubMed]
- Pihan, G.A. Centrosome dysfunction contributes to chromosome instability, chromoanagenesis, and genome reprograming in cancer. Front. Oncol. 2013, 3, 277. [Google Scholar] [CrossRef] [Green Version]
- Quintyne, N.J.; Reing, J.E.; Hoffelder, D.R.; Gollin, S.M.; Saunders, W.S. Spindle multipolarity is prevented by centrosomal clustering. Science 2005, 307, 127–129. [Google Scholar] [CrossRef]
- Kwon, M.; Godinho, S.A.; Chandhok, N.S.; Ganem, N.J.; Azioune, A.; Thery, M.; Pellman, D. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev. 2008, 22, 2189–2203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ganem, N.J.; Godinho, S.A.; Pellman, D. A mechanism linking extra centrosomes to chromosomal instability. Nature 2009, 460, 278–282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silkworth, W.T.; Nardi, I.K.; Scholl, L.M.; Cimini, D. Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome mis-segregation in cancer cells. PLoS ONE 2009, 4, e6564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bakhoum, S.F.; Genovese, G.; Compton, D.A. Deviant kinetochore microtubule dynamics underlie chromosomal instability. Curr. Biol. 2009, 19, 1937–1942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salimian, K.J.; Ballister, E.R.; Smoak, E.M.; Wood, S.; Panchenko, T.; Lampson, M.A.; Black, B.E. Feedback control in sensing chromosome biorientation by the Aurora B kinase. Curr. Biol. 2011, 21, 1158–1165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abe, Y.; Sako, K.; Takagaki, K.; Hirayama, Y.; Uchida, K.S.; Herman, J.A.; DeLuca, J.G.; Hirota, T. HP1-assisted Aurora B Kinase activity prevents chromosome segregation errors. Dev. Cell 2016, 36, 487–497. [Google Scholar] [CrossRef] [Green Version]
- Desai, A.; Maddox, P.S.; Mitchison, T.J.; Salmon, E.D. Anaphase A chromosome movement and poleward spindle microtubule flux occur at similar rates in Xenopus extract spindles. J. Cell Biol. 1998, 141, 703–713. [Google Scholar] [CrossRef] [PubMed]
- LaFountain, J.R., Jr.; Oldenbourg, R.; Cole, R.W.; Rieder, C.L. Microtubule flux mediates poleward motion of acentric chromosome fragments during meiosis in insect spermatocytes. Mol. Biol. Cell 2001, 12, 4054–4065. [Google Scholar] [CrossRef]
- Brust-Mascher, I.; Scholey, J.M. Microtubule flux and sliding in mitotic spindles of Drosophila embryos. Mol. Biol. Cell 2002, 13, 3967–3975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maddox, P.; Desai, A.; Oegema, K.; Mitchison, T.J.; Salmon, E.D. Poleward microtubule flux is a major component of spindle dynamics and anaphase a in mitotic Drosophila embryos. Curr. Biol. 2002, 12, 1670–1674. [Google Scholar] [CrossRef] [Green Version]
- Maddox, P.; Straight, A.; Coughlin, P.; Mitchison, T.J.; Salmon, E.D. Direct observation of microtubule dynamics at kinetochores in Xenopus extract spindles: Implications for spindle mechanics. J. Cell Biol. 2003, 162, 377–382. [Google Scholar] [CrossRef] [Green Version]
- de Lartigue, J.; Brust-Mascher, I.; Scholey, J.M. Anaphase B spindle dynamics in Drosophila S2 cells: Comparison with embryo spindles. Cell Div. 2011, 6, 8. [Google Scholar] [CrossRef] [Green Version]
- Skibbens, R.V.; Skeen, V.P.; Salmon, E.D. Directional instability of kinetochore motility during chromosome congression and segregation in mitotic newt lung cells: A push-pull mechanism. J. Cell Biol. 1993, 122, 859–875. [Google Scholar] [CrossRef] [Green Version]
- Inoue, S.; Salmon, E.D. Force generation by microtubule assembly/disassembly in mitosis and related movements. Mol. Biol. Cell 1995, 6, 1619–1640. [Google Scholar] [CrossRef] [PubMed]
- Banigan, E.J.; Chiou, K.K.; Ballister, E.R.; Mayo, A.M.; Lampson, M.A.; Liu, A.J. Minimal model for collective kinetochore-microtubule dynamics. Proc. Natl. Acad. Sci. USA 2015, 112, 12699–12704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bajer, A.S. Functional autonomy of monopolar spindle and evidence for oscillatory movement in mitosis. J. Cell Biol. 1982, 93, 33–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hafner, J.; Mayr, M.I.; Mockel, M.M.; Mayer, T.U. Pre-anaphase chromosome oscillations are regulated by the antagonistic activities of Cdk1 and PP1 on Kif18A. Nat. Commun. 2014, 5, 4397. [Google Scholar] [CrossRef] [Green Version]
- Su, K.C.; Barry, Z.; Schweizer, N.; Maiato, H.; Bathe, M.; Cheeseman, I.M. A regulatory switch alters chromosome motions at the metaphase-to-anaphase transition. Cell Rep. 2016, 17, 1728–1738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shindo, N.; Otsuki, M.; Uchida, K.S.K.; Hirota, T. Prolonged mitosis causes separase deregulation and chromosome nondisjunction. Cell Rep. 2021, 34, 108652. [Google Scholar] [CrossRef]
- Amaro, A.C.; Samora, C.P.; Holtackers, R.; Wang, E.; Kingston, I.J.; Alonso, M.; Lampson, M.; McAinsh, A.D.; Meraldi, P. Molecular control of kinetochore-microtubule dynamics and chromosome oscillations. Nat. Cell Biol. 2010, 12, 319–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iemura, K.; Natsume, T.; Maehara, K.; Kanemaki, M.T.; Tanaka, K. Chromosome oscillation promotes Aurora A-dependent Hec1 phosphorylation and mitotic fidelity. J. Cell Biol. 2021, 220, e202006116. [Google Scholar] [CrossRef]
- Wan, X.; O’Quinn, R.P.; Pierce, H.L.; Joglekar, A.P.; Gall, W.E.; DeLuca, J.G.; Carroll, C.W.; Liu, S.T.; Yen, T.J.; McEwen, B.F.; et al. Protein architecture of the human kinetochore microtubule attachment site. Cell 2009, 137, 672–684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaqaman, K.; King, E.M.; Amaro, A.C.; Winter, J.R.; Dorn, J.F.; Elliott, H.L.; McHedlishvili, N.; McClelland, S.E.; Porter, I.M.; Posch, M.; et al. Kinetochore alignment within the metaphase plate is regulated by centromere stiffness and microtubule depolymerases. J. Cell Biol. 2010, 188, 665–679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyazaki, W.Y.; Orr-Weaver, T.L. Sister-chromatid cohesion in mitosis and meiosis. Annu. Rev. Genet. 1994, 28, 167–187. [Google Scholar] [CrossRef]
- Wan, X.; Cimini, D.; Cameron, L.A.; Salmon, E.D. The coupling between sister kinetochore directional instability and oscillations in centromere stretch in metaphase PtK1 cells. Mol. Biol. Cell 2012, 23, 1035–1046. [Google Scholar] [CrossRef]
- Stumpff, J.; von Dassow, G.; Wagenbach, M.; Asbury, C.; Wordeman, L. The kinesin-8 motor Kif18A suppresses kinetochore movements to control mitotic chromosome alignment. Dev. Cell 2008, 14, 252–262. [Google Scholar] [CrossRef] [Green Version]
- Stumpff, J.; Wagenbach, M.; Franck, A.; Asbury, C.L.; Wordeman, L. Kif18A and chromokinesins confine centromere movements via microtubule growth suppression and spatial control of kinetochore tension. Dev. Cell 2012, 22, 1017–1029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stumpff, J.; Du, Y.; English, C.A.; Maliga, Z.; Wagenbach, M.; Asbury, C.L.; Wordeman, L.; Ohi, R. A tethering mechanism controls the processivity and kinetochore-microtubule plus-end enrichment of the kinesin-8 Kif18A. Mol. Cell 2011, 43, 764–775. [Google Scholar] [CrossRef] [Green Version]
- Almeida, A.C.; Maiato, H. Chromokinesins. Curr. Biol. 2018, 28, R1131–R1135. [Google Scholar] [CrossRef] [Green Version]
- Rieder, C.L.; Salmon, E.D. Motile kinetochores and polar ejection forces dictate chromosome position on the vertebrate mitotic spindle. J. Cell Biol. 1994, 124, 223–233. [Google Scholar] [CrossRef] [Green Version]
- Wandke, C.; Barisic, M.; Sigl, R.; Rauch, V.; Wolf, F.; Amaro, A.C.; Tan, C.H.; Pereira, A.J.; Kutay, U.; Maiato, H.; et al. Human chromokinesins promote chromosome congression and spindle microtubule dynamics during mitosis. J. Cell Biol. 2012, 198, 847–863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ke, K.; Cheng, J.; Hunt, A.J. The distribution of polar ejection forces determines the amplitude of chromosome directional instability. Curr. Biol. 2009, 19, 807–815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rieder, C.L.; Davison, E.A.; Jensen, L.C.; Cassimeris, L.; Salmon, E.D. Oscillatory movements of monooriented chromosomes and their position relative to the spindle pole result from the ejection properties of the aster and half-spindle. J. Cell Biol. 1986, 103, 581–591. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Xue, C.; Yang, Q.; Low, B.C.; Liou, Y.C. NuSAP governs chromosome oscillation by facilitating the Kid-generated polar ejection force. Nat. Commun. 2016, 7, 10597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitchison, T.J. Polewards microtubule flux in the mitotic spindle: Evidence from photoactivation of fluorescence. J. Cell Biol. 1989, 109, 637–652. [Google Scholar] [CrossRef] [PubMed]
- Ganem, N.J.; Upton, K.; Compton, D.A. Efficient mitosis in human cells lacking poleward microtubule flux. Curr. Biol. 2005, 15, 1827–1832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steblyanko, Y.; Rajendraprasad, G.; Osswald, M.; Eibes, S.; Jacome, A.; Geley, S.; Pereira, A.J.; Maiato, H.; Barisic, M. Microtubule poleward flux in human cells is driven by the coordinated action of four kinesins. EMBO J. 2020, 39, e105432. [Google Scholar] [CrossRef] [PubMed]
- Cassimeris, L.; Morabito, J. TOGp, the human homolog of XMAP215/Dis1, is required for centrosome integrity, spindle pole organization, and bipolar spindle assembly. Mol. Biol. Cell 2004, 15, 1580–1590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Zhuang, X.; Cao, D.; Chu, Y.; Yao, P.; Liu, W.; Liu, L.; Adams, G.; Fang, G.; Dou, Z.; et al. Mitotic regulator SKAP forms a link between kinetochore core complex KMN and dynamic spindle microtubules. J. Biol. Chem. 2012, 287, 39380–39390. [Google Scholar] [CrossRef] [Green Version]
- Kajtez, J.; Solomatina, A.; Novak, M.; Polak, B.; Vukusic, K.; Rudiger, J.; Cojoc, G.; Milas, A.; Sumanovac Sestak, I.; Risteski, P.; et al. Overlap microtubules link sister k-fibres and balance the forces on bi-oriented kinetochores. Nat. Commun. 2016, 7, 10298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Risteski, P.; Jagric, M.; Pavin, N.; Tolic, I.M. Biomechanics of chromosome alignment at the spindle midplane. Curr. Biol. 2021, 31, R574–R585. [Google Scholar] [CrossRef]
- Polak, B.; Risteski, P.; Lesjak, S.; Tolic, I.M. PRC1-labeled microtubule bundles and kinetochore pairs show one-to-one association in metaphase. EMBO Rep. 2017, 18, 217–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jagric, M.; Risteski, P.; Martincic, J.; Milas, A.; Tolic, I.M. Optogenetic control of PRC1 reveals its role in chromosome alignment on the spindle by overlap length-dependent forces. eLife 2021, 10, e61170. [Google Scholar] [CrossRef]
- Gardner, M.K.; Odde, D.J. Modeling of chromosome motility during mitosis. Curr. Opin. Cell Biol. 2006, 18, 639–647. [Google Scholar] [CrossRef] [PubMed]
- Joglekar, A.P.; Hunt, A.J. A simple, mechanistic model for directional instability during mitotic chromosome movements. Biophys. J. 2002, 83, 42–58. [Google Scholar] [CrossRef] [Green Version]
- Armond, J.W.; Harry, E.F.; McAinsh, A.D.; Burroughs, N.J. Inferring the forces controlling metaphase kinetochore oscillations by reverse engineering system dynamics. PLoS Comput. Biol. 2015, 11, e1004607. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Desai, A.; Onuchic, J.N.; Hwa, T. An integrated mechanobiochemical feedback mechanism describes chromosome motility from prometaphase to anaphase in mitosis. Proc. Natl. Acad. Sci. USA 2008, 105, 13752–13757. [Google Scholar] [CrossRef] [Green Version]
- Civelekoglu-Scholey, G.; Sharp, D.J.; Mogilner, A.; Scholey, J.M. Model of chromosome motility in Drosophila embryos: Adaptation of a general mechanism for rapid mitosis. Biophys. J. 2006, 90, 3966–3982. [Google Scholar] [CrossRef] [Green Version]
- Gay, G.; Courtheoux, T.; Reyes, C.; Tournier, S.; Gachet, Y. A stochastic model of kinetochore-microtubule attachment accurately describes fission yeast chromosome segregation. J. Cell Biol. 2012, 196, 757–774. [Google Scholar] [CrossRef] [Green Version]
- Vladimirou, E.; Harry, E.; Burroughs, N.; McAinsh, A.D. Springs, clutches and motors: Driving forward kinetochore mechanism by modelling. Chromosome Res. 2011, 19, 409–421. [Google Scholar] [CrossRef] [Green Version]
- Civelekoglu-Scholey, G.; He, B.; Shen, M.; Wan, X.; Roscioli, E.; Bowden, B.; Cimini, D. Dynamic bonds and polar ejection force distribution explain kinetochore oscillations in PtK1 cells. J. Cell Biol. 2013, 201, 577–593. [Google Scholar] [CrossRef]
- Campos Medina, M.A.; Iemura, K.; Kimura, A.; Tanaka, K. A mathematical model of kinetochore-microtubule attachment regulated by Aurora A activity gradient describes chromosome oscillation and correction of erroneous attachments. Biomed. Res. 2021, in press. [Google Scholar]
- Nishimura, K.; Fukagawa, T.; Takisawa, H.; Kakimoto, T.; Kanemaki, M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods 2009, 6, 917–922. [Google Scholar] [CrossRef] [PubMed]
- Natsume, T.; Kiyomitsu, T.; Saga, Y.; Kanemaki, M.T. Rapid protein depletion in human cells by Auxin-inducible degron tagging with short homology donors. Cell Rep. 2016, 15, 210–218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yesbolatova, A.; Natsume, T.; Hayashi, K.I.; Kanemaki, M.T. Generation of conditional auxin-inducible degron (AID) cells and tight control of degron-fused proteins using the degradation inhibitor auxinole. Methods 2019, 164–165, 73–80. [Google Scholar] [CrossRef]
- DeLuca, K.F.; Meppelink, A.; Broad, A.J.; Mick, J.E.; Peersen, O.B.; Pektas, S.; Lens, S.M.A.; DeLuca, J.G. Aurora A kinase phosphorylates Hec1 to regulate metaphase kinetochore-microtubule dynamics. J. Cell Biol. 2018, 217, 163–177. [Google Scholar] [CrossRef]
- Barisic, M.; Aguiar, P.; Geley, S.; Maiato, H. Kinetochore motors drive congression of peripheral polar chromosomes by overcoming random arm-ejection forces. Nat. Cell Biol. 2014, 16, 1249–1256. [Google Scholar] [CrossRef]
- DeLuca, K.F.; Lens, S.M.; DeLuca, J.G. Temporal changes in Hec1 phosphorylation control kinetochore-microtubule attachment stability during mitosis. J. Cell Sci. 2011, 124, 622–634. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Zhu, C.; Chen, H.; Li, L.; Guo, L.; Jiang, W.; Lu, S.H. Kif18A is involved in human breast carcinogenesis. Carcinogenesis 2010, 31, 1676–1684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagahara, M.; Nishida, N.; Iwatsuki, M.; Ishimaru, S.; Mimori, K.; Tanaka, F.; Nakagawa, T.; Sato, T.; Sugihara, K.; Hoon, D.S.; et al. Kinesin 18A expression: Clinical relevance to colorectal cancer progression. Int. J. Cancer 2011, 129, 2543–2552. [Google Scholar] [CrossRef] [PubMed]
- Rath, O.; Kozielski, F. Kinesins and cancer. Nat. Rev. Cancer 2012, 12, 527–539. [Google Scholar] [CrossRef]
- Cohen-Sharir, Y.; McFarland, J.M.; Abdusamad, M.; Marquis, C.; Bernhard, S.V.; Kazachkova, M.; Tang, H.; Ippolito, M.R.; Laue, K.; Zerbib, J.; et al. Aneuploidy renders cancer cells vulnerable to mitotic checkpoint inhibition. Nature 2021, 590, 486–491. [Google Scholar] [CrossRef]
- Quinton, R.J.; DiDomizio, A.; Vittoria, M.A.; Kotynkova, K.; Ticas, C.J.; Patel, S.; Koga, Y.; Vakhshoorzadeh, J.; Hermance, N.; Kuroda, T.S.; et al. Whole-genome doubling confers unique genetic vulnerabilities on tumour cells. Nature 2021, 590, 492–497. [Google Scholar] [CrossRef] [PubMed]
- Marquis, C.; Fonseca, C.L.; Queen, K.A.; Wood, L.; Vandal, S.E.; Malaby, H.L.H.; Clayton, J.E.; Stumpff, J. Chromosomally unstable tumor cells specifically require KIF18A for proliferation. Nat. Commun. 2021, 12, 1213. [Google Scholar] [CrossRef] [PubMed]
- Ertych, N.; Stolz, A.; Stenzinger, A.; Weichert, W.; Kaulfuss, S.; Burfeind, P.; Aigner, A.; Wordeman, L.; Bastians, H. Increased microtubule assembly rates influence chromosomal instability in colorectal cancer cells. Nat. Cell Biol. 2014, 16, 779–791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Long, A.F.; Udy, D.B.; Dumont, S. Hec1 tail phosphorylation differentially regulates mammalian kinetochore coupling to polymerizing and depolymerizing microtubules. Curr. Biol. 2017, 27, 1692–1699.e3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farag, S.S. The potential role of Aurora kinase inhibitors in haematological malignancies. Br. J. Haematol. 2011, 155, 561–579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wakiya, M.; Nishi, E.; Kawai, S.; Yamada, K.; Katsumata, K.; Hirayasu, A.; Itabashi, Y.; Yamamoto, A. Chiasmata and the kinetochore component Dam1 are crucial for elimination of erroneous chromosome attachments and centromere oscillation at meiosis I. Open Biol. 2021, 11, 200308. [Google Scholar] [CrossRef]
- Yamamoto, A. Shake it off: The elimination of erroneous kinetochore-microtubule attachments and chromosome oscillation. Int. J. Mol. Sci. 2021, 22, 3174. [Google Scholar] [CrossRef]
- Kuniyasu, K.; Iemura, K.; Tanaka, K. Delayed chromosome alignment to the spindle equator increases the rate of chromosome missegregation in cancer cell lines. Biomolecules 2019, 9, 10. [Google Scholar] [CrossRef] [Green Version]
- Orth, J.D.; Kohler, R.H.; Foijer, F.; Sorger, P.K.; Weissleder, R.; Mitchison, T.J. Analysis of mitosis and antimitotic drug responses in tumors by in vivo microscopy and single-cell pharmacodynamics. Cancer Res. 2011, 71, 4608–4616. [Google Scholar] [CrossRef] [Green Version]
- Knouse, K.A.; Lopez, K.E.; Bachofner, M.; Amon, A. Chromosome segregation fidelity in epithelia requires tissue architecture. Cell 2018, 175, 200–211.e13. [Google Scholar] [CrossRef] [Green Version]
- Bolhaqueiro, A.C.F.; Ponsioen, B.; Bakker, B.; Klaasen, S.J.; Kucukkose, E.; van Jaarsveld, R.H.; Vivie, J.; Verlaan-Klink, I.; Hami, N.; Spierings, D.C.J.; et al. Ongoing chromosomal instability and karyotype evolution in human colorectal cancer organoids. Nat. Genet. 2019, 51, 824–834. [Google Scholar] [CrossRef]
- Nelson, L.; Tighe, A.; Golder, A.; Littler, S.; Bakker, B.; Moralli, D.; Murtuza Baker, S.; Donaldson, I.J.; Spierings, D.C.J.; Wardenaar, R.; et al. A living biobank of ovarian cancer ex vivo models reveals profound mitotic heterogeneity. Nat. Commun. 2020, 11, 822. [Google Scholar] [CrossRef]
- Du, Y.; English, C.A.; Ohi, R. The kinesin-8 Kif18A dampens microtubule plus-end dynamics. Curr. Biol. 2010, 20, 374–380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fonseca, C.L.; Malaby, H.L.H.; Sepaniac, L.A.; Martin, W.; Byers, C.; Czechanski, A.; Messinger, D.; Tang, M.; Ohi, R.; Reinholdt, L.G.; et al. Mitotic chromosome alignment ensures mitotic fidelity by promoting interchromosomal compaction during anaphase. J. Cell Biol. 2019, 218, 1148–1163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, J.; Mitchison, T.J. Cell death response to anti-mitotic drug treatment in cell culture, mouse tumor model and the clinic. Endocr. Relat. Cancer 2017, 24, T83–T96. [Google Scholar] [CrossRef] [Green Version]
- Ohashi, A.; Ohori, M.; Iwai, K.; Nakayama, Y.; Nambu, T.; Morishita, D.; Kawamoto, T.; Miyamoto, M.; Hirayama, T.; Okaniwa, M.; et al. Aneuploidy generates proteotoxic stress and DNA damage concurrently with p53-mediated post-mitotic apoptosis in SAC-impaired cells. Nat. Commun. 2015, 6, 7668. [Google Scholar] [CrossRef] [Green Version]
- Braun, J.; Mockel, M.M.; Strittmatter, T.; Marx, A.; Groth, U.; Mayer, T.U. Synthesis and biological evaluation of optimized inhibitors of the mitotic kinesin Kif18A. ACS Chem. Biol. 2015, 10, 554–560. [Google Scholar] [CrossRef] [PubMed]
- Kollareddy, M.; Zheleva, D.; Dzubak, P.; Brahmkshatriya, P.S.; Lepsik, M.; Hajduch, M. Aurora kinase inhibitors: Progress towards the clinic. Investig. New Drugs 2012, 30, 2411–2432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hilton, J.F.; Shapiro, G.I. Aurora kinase inhibition as an anticancer strategy. J. Clin. Oncol. 2014, 32, 57–59. [Google Scholar] [CrossRef]
- Huang, L.Y.; Lee, Y.S.; Huang, J.J.; Chang, C.C.; Chang, J.M.; Chuang, S.H.; Kao, K.J.; Tsai, Y.J.; Tsai, P.Y.; Liu, C.W.; et al. Characterization of the biological activity of a potent small molecule Hec1 inhibitor TAI-1. J. Exp. Clin. Cancer Res. 2014, 33, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iemura, K.; Yoshizaki, Y.; Kuniyasu, K.; Tanaka, K. Attenuated Chromosome Oscillation as a Cause of Chromosomal Instability in Cancer Cells. Cancers 2021, 13, 4531. https://doi.org/10.3390/cancers13184531
Iemura K, Yoshizaki Y, Kuniyasu K, Tanaka K. Attenuated Chromosome Oscillation as a Cause of Chromosomal Instability in Cancer Cells. Cancers. 2021; 13(18):4531. https://doi.org/10.3390/cancers13184531
Chicago/Turabian StyleIemura, Kenji, Yujiro Yoshizaki, Kinue Kuniyasu, and Kozo Tanaka. 2021. "Attenuated Chromosome Oscillation as a Cause of Chromosomal Instability in Cancer Cells" Cancers 13, no. 18: 4531. https://doi.org/10.3390/cancers13184531
APA StyleIemura, K., Yoshizaki, Y., Kuniyasu, K., & Tanaka, K. (2021). Attenuated Chromosome Oscillation as a Cause of Chromosomal Instability in Cancer Cells. Cancers, 13(18), 4531. https://doi.org/10.3390/cancers13184531






