Steroid Receptors in Breast Cancer: Understanding of Molecular Function as a Basis for Effective Therapy Development
Abstract
Simple Summary
Abstract
1. Introduction
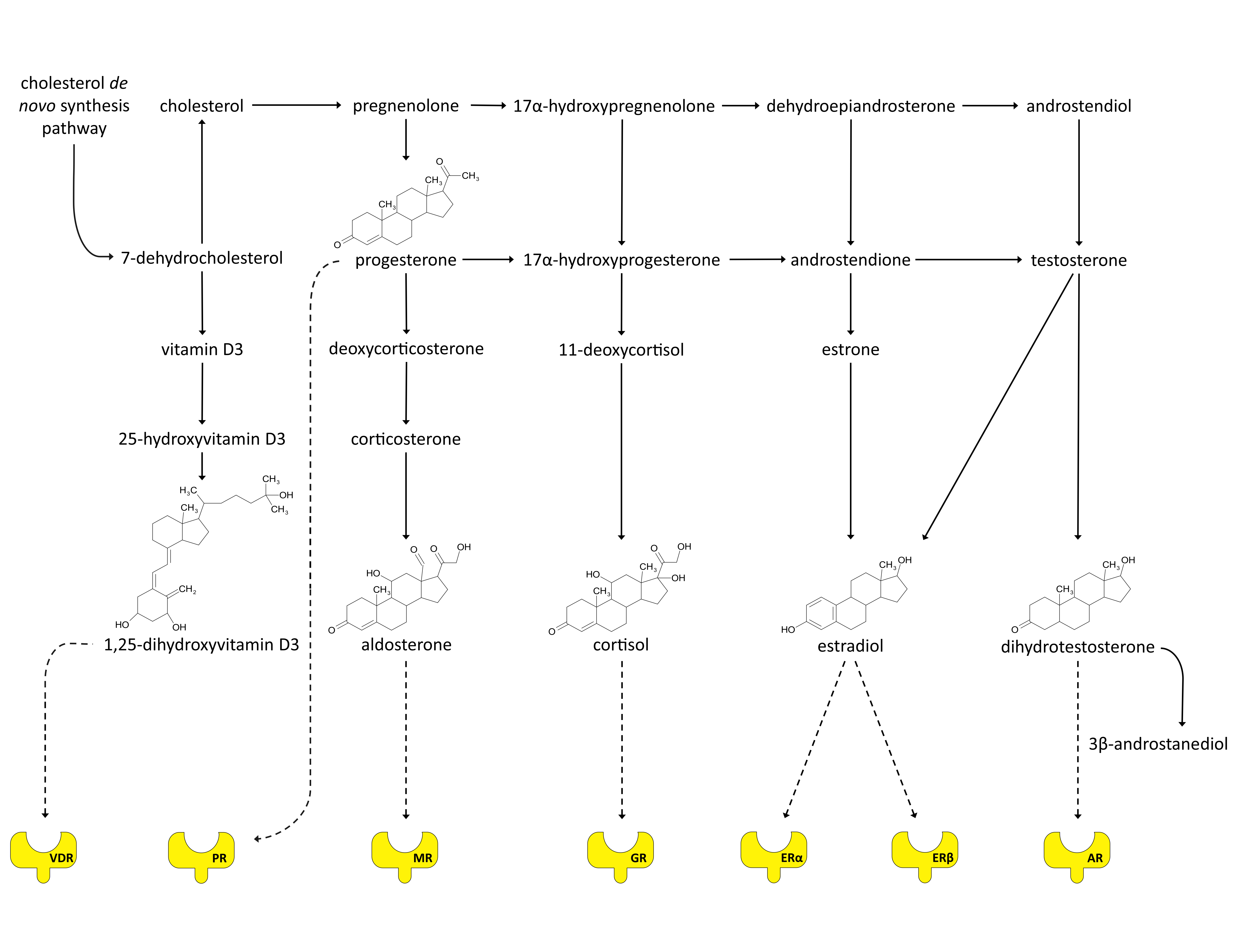
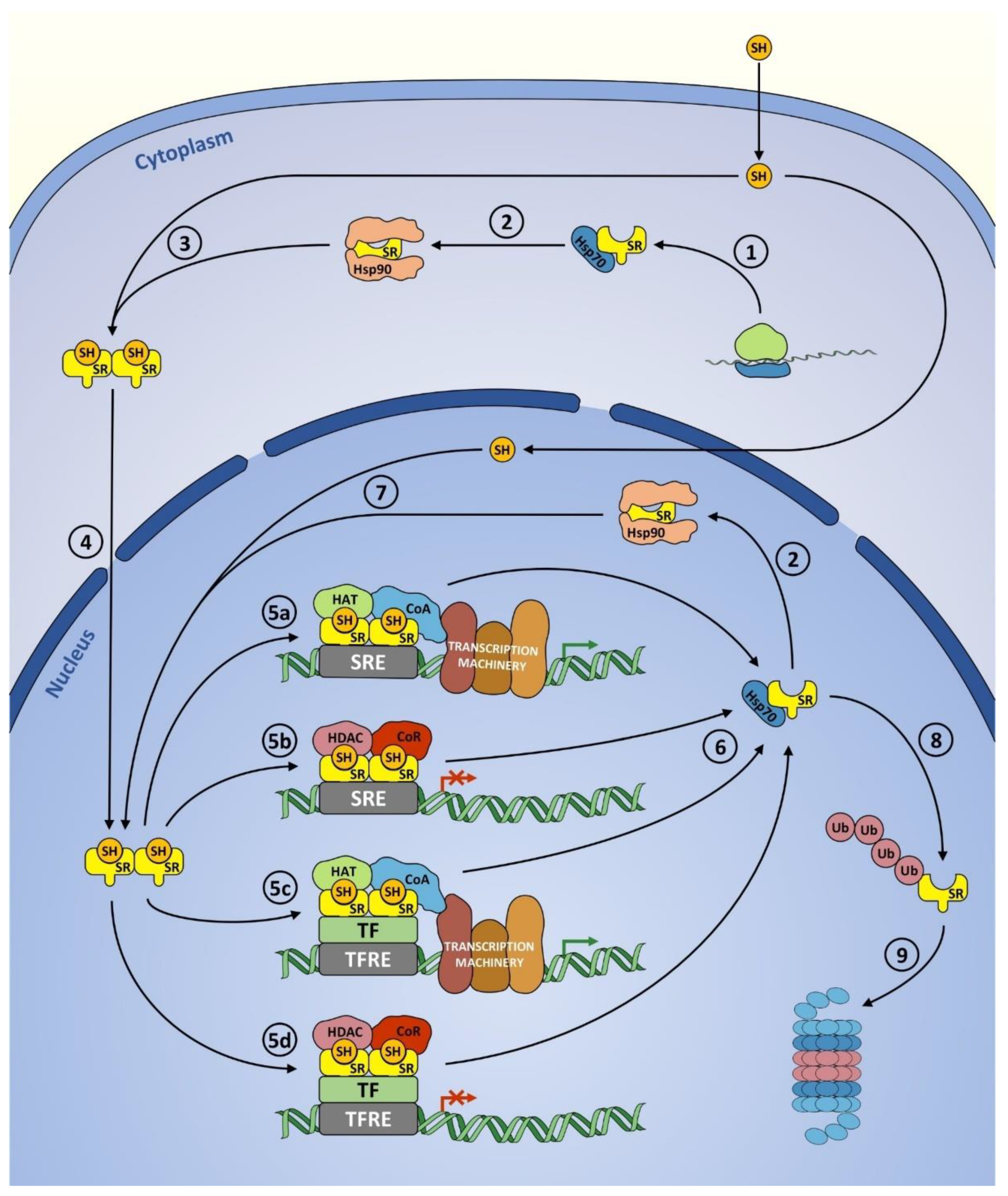
2. Molecular Function of Steroid Receptors—Common Features
3. Estrogen Receptor
4. Progesterone Receptor
4.1. Mechanism of Action
4.2. Post-Translational Modifications
4.3. Role in Healthy Breast
4.4. Role in Breast Cancer
5. Androgen Receptor
5.1. Metabolism of Androgens in Females
5.2. Androgen Receptor Structure and Signaling Pathways
6. Glucocorticoid Receptor
7. Mineralocorticoid Receptor
8. Vitamin D Receptor
9. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Busillo, J.M.; Rhen, T.; Cidlowski, J.A. Steroid Hormone Action. In Yen & Jaffe’s Reproductive Endocrinology; Strauss, J.F., Barbieri, R.L., Seventh, E., Eds.; Elsevier: Philadelphia, PA, USA, 2014; pp. 93–107.e3. ISBN 978-1-4557-2758-2. [Google Scholar]
- Papatheodorou, I.; Moreno, P.; Manning, J.; Fuentes, A.M.-P.; George, N.; Fexova, S.; Fonseca, N.A.; Füllgrabe, A.; Green, M.; Huang, N.; et al. Expression Atlas Update: From Tissues to Single Cells. Nucleic Acids Res. 2020, 48, D77–D83. [Google Scholar] [CrossRef]
- Strauss, J.F. The Synthesis and Metabolism of Steroid Hormones. In Yen & Jaffe’s Reproductive Endocrinology; Strauss, J.F., Barbieri, R.L., Seventh, E., Eds.; Elsevier: Philadelphia, PA, USA, 2014; pp. 66–92.e3. ISBN 978-1-4557-2758-2. [Google Scholar]
- Huss, L.; Butt, S.T.; Borgquist, S.; Elebro, K.; Sandsveden, M.; Rosendahl, A.; Manjer, J. Vitamin D Receptor Expression in Invasive Breast Tumors and Breast Cancer Survival. Breast Cancer Res. 2019, 21, 1–13. [Google Scholar] [CrossRef]
- Murray, R.K.; Granner, D.K.; Mayes, P.A.; Rodwell, V.W. Harper’s Illustrated Biochemistry, 31st ed.; McGraw Hill: New York, NY, USA, 2018; ISBN 978-1-25-983794-4. [Google Scholar]
- Elia, A.; Vanzulli, S.I.; Gass, H.; Lamb, C.A.; Fabris, V.T.; Vazquez, P.M.; Burruchaga, J.; Spengler, E.; Bois, I.C.; Castets, A.; et al. Mipra, a Window of Opportunity Study Evaluating Mifepristone Treatment for Postmenopausal Breast Cancer Patients with Higher Levels of Progesterone Receptor Isoform a than B. In Proceedings of the 2020 San Antonio Breast Cancer Virtual Symposium, San Antonio, TX, USA, 8–11 December 2020; American Association for Cancer Research: Philadelphia, PA, USA, 2021. Abstract nr PS11-35. [Google Scholar]
- Yardley, D.; Peacock, N.; Young, R.; Silber, A.; Chung, G.; Webb, C.; Jones, S.; Shastry, M.; Midha, R.; DeBusk, L.; et al. A Phase 2 Study Evaluating Orteronel, an Inhibitor of Androgen Biosynthesis, in Patients with Androgen Receptor (AR)-Expressing Metastatic Breast Cancer: Interim Analysis. In Proceedings of the Thirty-Eighth Annual CTRC-AACR San Antonio Breast Cancer Symposium, San Antonio, TX, USA, 8–12 December 2015; American Association for Cancer Research: Philadelphia, PA, USA, 2016. Abstract nr P5-14-04. [Google Scholar]
- Lawrence, J.A.; Akman, S.A.; Melin, S.A.; Case, L.D.; Schwartz, G.G. Oral Paricalcitol (19-nor-1,25- Dihydroxyvitamin D2) in Women Receiving Chemotherapy for Metastatic Breast Cancer: A Feasibility Trial. Cancer Biol. Ther. 2013, 14, 476–480. [Google Scholar] [CrossRef]
- Davis, M.K.; Villa, D.; Tsang, T.S.M.; Starovoytov, A.; Gelmon, K.; Virani, S.A. Effect of Eplerenone on Diastolic Function in Women Receiving Anthracycline-Based Chemotherapy for Breast Cancer. JACC Cardio Oncol. 2019, 1, 295–298. [Google Scholar] [CrossRef]
- Cenciarini, M.E.; Proietti, C.J. Molecular Mechanisms Underlying Progesterone Receptor Action in Breast Cancer: Insights into Cell Proliferation and Stem Cell Regulation. Steroids 2019, 152, 108503. [Google Scholar] [CrossRef]
- Tan, M.E.; Li, J.; Xu, H.E.; Melcher, K.; Yong, E. Androgen Receptor: Structure, Role in Prostate Cancer and Drug Discovery. Acta Pharm. Sin. 2015, 36, 3–23. [Google Scholar] [CrossRef]
- Cordera, F.; Jordan, V.C. Steroid Receptors and Their Role in the Biology and Control of Breast Cancer Growth. Semin. Oncol. 2006, 33, 631–641. [Google Scholar] [CrossRef]
- Oakley, R.H.; Cidlowski, J.A. The Biology of the Glucocorticoid Receptor: New Signaling Mechanisms in Health and Disease. J. Allergy Clin. Immunol. 2013, 132, 1033–1044. [Google Scholar] [CrossRef] [PubMed]
- Pascual-Le Tallec, L.; Lombès, M. The Mineralocorticoid Receptor: A Journey Exploring Its Diversity and Specificity of Action. Mol. Endocrinol. 2005, 19, 2211–2221. [Google Scholar] [CrossRef]
- Pratt, W.B.; Galigniana, M.D.; Morishima, Y.; Murphy, P.J.M. Role of Molecular Chaperones in Steroid Receptor Action. Essays Biochem. 2004, 40, 41–58. [Google Scholar] [CrossRef]
- Picard, D. Chaperoning Steroid Hormone Action. Trends Endocrinol. Metab. 2006, 17, 229–235. [Google Scholar] [CrossRef]
- DeMarzo, A.M.; Beck, C.A.; Onate, S.A.; Edwards, D.P. Dimerization of Mammalian Progesterone Receptors Occurs in the Absence of DNA and Is Related to the Release of the 90-KDa Heat Shock Protein. Proc. Natl. Acad. Sci. USA 1991, 88, 72–76. [Google Scholar] [CrossRef]
- Grimm, S.L.; Hartig, S.M.; Edwards, D.P. Progesterone Receptor Signaling Mechanisms. J. Mol. Biol. 2016, 428, 3831–3849. [Google Scholar] [CrossRef]
- Savory, J.G.A.; Préfontaine, G.G.; Lamprecht, C.; Liao, M.; Walther, R.F.; Lefebvre, Y.A.; Haché, R.J.G. Glucocorticoid Receptor Homodimers and Glucocorticoid-Mineralocorticoid Receptor Heterodimers Form in the Cytoplasm through Alternative Dimerization Interfaces. Mol. Cell Biol. 2001, 21, 781–793. [Google Scholar] [CrossRef]
- Tetel, M.J.; Jung, S.; Carbajo, P.; Ladtkow, T.; Skafar, D.F.; Edwards, D.P. Hinge and Amino-Terminal Sequences Contribute to Solution Dimerization of Human Progesterone Receptor. Mol. Endocrinol. 1997, 11, 1114–1128. [Google Scholar] [CrossRef][Green Version]
- Doan, T.B.; Graham, J.D.; Clarke, C.L. Emerging Functional Roles of Nuclear Receptors in Breast Cancer. J. Mol. Endocrinol. 2017, 58, R169–R190. [Google Scholar] [CrossRef] [PubMed]
- Daniel, A.R.; Gaviglio, A.L.; Czaplicki, L.M.; Hillard, C.J.; Housa, D.; Lange, C.A. The Progesterone Receptor Hinge Region Regulates the Kinetics of Transcriptional Responses through Acetylation, Phosphorylation, and Nuclear Retention. Mol. Endocrinol. 2010, 24, 2126–2138. [Google Scholar] [CrossRef]
- Fuentes, N.; Silveyra, P. Estrogen receptor signaling mechanisms. In Advances in Protein Chemistry and Structural Biology; Elsevier Inc.: Amsterdam, The Netherlands, 2019; Volume 116, pp. 135–170. ISBN 9780128155615. [Google Scholar]
- Clarke, R.B.; Anderson, E.; Howell, A. Steroid Receptors in Human Breast Cancer. Trends Endocrinol. Metab. 2004, 15, 316–323. [Google Scholar] [CrossRef] [PubMed]
- Hudson, W.H.; Youn, C.; Ortlund, E.A. Crystal Structure of the Mineralocorticoid Receptor DNA Binding Domain in Complex with DNA. PLoS ONE 2014, 9, e107000. [Google Scholar] [CrossRef]
- Tang, Q.; Chen, Y.; Meyer, C.; Geistlinger, T.; Lupien, M.; Wang, Q.; Liu, T.; Zhang, Y.; Brown, M.; Liu, X.S. A Comprehensive View of Nuclear Receptor Cancer Cistromes. Cancer Res. 2011, 71, 6940–6947. [Google Scholar] [CrossRef] [PubMed]
- Nelson, C.C.; Hendy, S.C.; Shukin, R.J.; Cheng, H.; Bruchovsky, N.; Koop, B.F.; Rennie, P.S. Determinants of DNA Sequence Specificity of the Androgen, Progesterone, and Glucocorticoid Receptors: Evidence for Differential Steroid Receptor Response Elements. Mol. Endocrinol. 1999, 13, 2090–2107. [Google Scholar] [CrossRef]
- Beato, M.; Wright, R.H.G.; Dily, F. Le 90 YEARS OF PROGESTERONE: Molecular Mechanisms of Progesterone Receptor Action on the Breast Cancer Genome. J. Mol. Endocrinol. 2020, 65, T65–T79. [Google Scholar] [CrossRef]
- Scarpin, K.M.; Graham, J.D.; Mote, P.A.; Clarke, C.L. Progesterone Action in Human Tissues: Regulation by Progesterone Receptor (PR) Isoform Expression, Nuclear Positioning and Coregulator Expression. Nucl. Recept. Signal. 2009, 7, 1–13. [Google Scholar] [CrossRef]
- Wójcik, C.; DeMartino, G.N. Intracellular Localization of Proteasomes. Int. J. Biochem. Cell Biol. 2003, 35, 579–589. [Google Scholar] [CrossRef]
- Cicatiello, L.; Addeo, R.; Sasso, A.; Altucci, L.; Petrizzi, V.B.; Borgo, R.; Cancemi, M.; Caporali, S.; Caristi, S.; Scafoglio, C.; et al. Estrogens and Progesterone Promote Persistent CCND1 Gene Activation during G1 by Inducing Transcriptional Derepression via C-Jun/c-Fos/Estrogen Receptor (Progesterone Receptor) Complex Assembly to a Distal Regulatory Element and Recruitment of Cyclin D1 T. Mol. Cell Biol. 2004, 24, 7260–7274. [Google Scholar] [CrossRef]
- Stavreva, D.A.; Müller, W.G.; Hager, G.L.; Smith, C.L.; McNally, J.G. Rapid Glucocorticoid Receptor Exchange at a Promoter Is Coupled to Transcription and Regulated by Chaperones and Proteasomes. Mol. Cell Biol. 2004, 24, 2682–2697. [Google Scholar] [CrossRef] [PubMed]
- Saladin, K. Anatomy & Physiology: The Unity of Form and Function, 6th ed.; McGraw-Hill: New York, NY, USA, 2012. [Google Scholar]
- Shao, W.; Brown, M. Advances in Estrogen Receptor Biology: Prospects for Improvements in Targeted Breast Cancer Therapy. Breast Cancer Res. 2003, 6, 39. [Google Scholar] [CrossRef] [PubMed]
- Lewis, J.S.; Jordan, V.C. Selective Estrogen Receptor Modulators (SERMs): Mechanisms of Anticarcinogenesis and Drug Resistance. Mutat. Res. Mol. Mech. Mutagen. 2005, 591, 247–263. [Google Scholar] [CrossRef]
- Hughes, Z.; Liu, F.; Marquis, K.; Muniz, L.; Pangalos, M.; Ring, R.; Whiteside, G.; Brandon, N. Estrogen Receptor Neurobiology and Its Potential for Translation into Broad Spectrum Therapeutics for CNS Disorders. Curr. Mol. Pharm. 2009, 2, 215–236. [Google Scholar] [CrossRef]
- Xiao, J.; Wang, N.; Sun, B.; Cai, G. Estrogen Receptor Mediates the Effects of Pseudoprotodiocsin on Adipogenesis in 3T3-L1 Cells. Am. J. Physiol. Physiol. 2010, 299, C128–C138. [Google Scholar] [CrossRef]
- Pagano, M.T.; Ortona, E.; Dupuis, M.L. A Role for Estrogen Receptor Alpha36 in Cancer Progression. Front. Endocrinol. 2020, 11, 506. [Google Scholar] [CrossRef]
- Zhou, Y.; Liu, X. The Role of Estrogen Receptor Beta in Breast Cancer. Biomark Res. 2020, 8, 39. [Google Scholar] [CrossRef]
- Jensen, E.V. On the Mechanism of Estrogen Action. Perspect. Biol. Med. 1962, 6, 47–60. [Google Scholar] [CrossRef]
- Park, Y.R.; Lee, J.; Jung, J.H.; Kim, W.W.; Park, C.S.; Lee, R.K.; Chae, Y.S.; Lee, S.J.; Park, J.-Y.; Park, J.Y.; et al. Absence of Estrogen Receptor Is Associated with Worse Oncologic Outcome in Patients Who Were Received Neoadjuvant Chemotherapy for Breast Cancer. Asian J. Surg. 2020, 43, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Salem, K.; Tevaarwerk, A.J.; Strigel, R.M.; Fowler, A.M. Recent Advances in Imaging Steroid Hormone Receptors in Breast Cancer. J. Nucl. Med. 2020, 61, 172–176. [Google Scholar] [CrossRef]
- van Kruchten, M.; Glaudemans, A.W.J.M.; de Vries, E.F.J.; Beets-Tan, R.G.H.; Schröder, C.P.; Dierckx, R.A.; de Vries, E.G.E.; Hospers, G.A.P. PET Imaging of Estrogen Receptors as a Diagnostic Tool for Breast Cancer Patients Presenting with a Clinical Dilemma. J. Nucl. Med. 2012, 53, 182–190. [Google Scholar] [CrossRef]
- Belachew, E.B.; Sewasew, D.T. Molecular Mechanisms of Endocrine Resistance in Estrogen-Receptor-Positive Breast Cancer. Front. Endocrinol. 2021, 12, 188. [Google Scholar] [CrossRef]
- Kasielska-Trojan, A.; Danilewicz, M.; Strużyna, J.; Bugaj, M.; Antoszewski, B. The Role of Oestrogen and Progesterone Receptors in Gigantomastia. Arch. Med. Sci. 2019, 33, 1–5. [Google Scholar] [CrossRef]
- Haque, M.M.; Desai, K.V. Pathways to Endocrine Therapy Resistance in Breast Cancer. Front. Endocrinol. 2019, 10, 573. [Google Scholar] [CrossRef]
- Bhattacharjee, A.; Hossain, M.U.; Chowdhury, Z.M.; Rahman, S.M.A.; Bhuyan, Z.A.; Salimullah, M.; Keya, C.A. Insight of Druggable Cannabinoids against Estrogen Receptor β in Breast Cancer. J. Biomol. Struct. Dyn. 2021, 39, 1688–1697. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; McEachon, J.D.; Pollock, J.A. Synthesis and Characterization of Hydrogen Peroxide Activated Estrogen Receptor Beta Ligands. Bioorg. Med. Chem. 2019, 27, 2075–2082. [Google Scholar] [CrossRef] [PubMed]
- Crandall, D.L.; Busler, D.E.; Novak, T.J.; Weber, R.V.; Kral, J.G. Identification of Estrogen Receptor β RNA in Human Breast and Abdominal Subcutaneous Adipose Tissue. Biochem. Biophys. Res. Commun. 1998, 248, 523–526. [Google Scholar] [CrossRef]
- Kuiper, G.G.J.M.; Shughrue, P.J.; Merchenthaler, I.; Gustafsson, J.-Å. The Estrogen Receptor β Subtype: A Novel Mediator of Estrogen Action in Neuroendocrine Systems. Front. Neuroendocr. 1998, 19, 253–286. [Google Scholar] [CrossRef]
- Balla, B.; Sárvári, M.; Kósa, J.P.; Kocsis-Deák, B.; Tobiás, B.; Árvai, K.; Takács, I.; Podani, J.; Liposits, Z.; Lakatos, P. Long-Term Selective Estrogen Receptor-Beta Agonist Treatment Modulates Gene Expression in Bone and Bone Marrow of Ovariectomized Rats. J. Steroid Biochem. Mol. Biol. 2019, 188, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.Y.H.; Lin, G.; Fang, M.; Rudd, J.A. Localization of Estrogen Receptor ERα, ERβ and GPR30 on Myenteric Neurons of the Gastrointestinal Tract and Their Role in Motility. Gen. Comp. Endocrinol. 2019, 272, 63–75. [Google Scholar] [CrossRef]
- Tamer, S.A.; Yıldırım, A.; Arabacı, Ş.; Çiftçi, S.; Akın, S.; Sarı, E.; Köroğlu, M.K.; Ercan, F.; Yüksel, M.; Çevik, Ö.; et al. Treatment with Estrogen Receptor Agonist ERβ Improves Torsion-Induced Oxidative Testis Injury in Rats. Life Sci. 2019, 222, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Le Moëne, O.; Stavarache, M.; Ogawa, S.; Musatov, S.; Ågmo, A. Estrogen Receptors α and β in the Central Amygdala and the Ventromedial Nucleus of the Hypothalamus: Sociosexual Behaviors, Fear and Arousal in Female Rats during Emotionally Challenging Events. Behav. Brain Res. 2019, 367, 128–142. [Google Scholar] [CrossRef]
- Kim, H.; Park, J.; Leem, H.; Cho, M.; Yoon, J.-H.; Maeng, H.-J.; Lee, Y. Rhododendrin-Induced RNF146 Expression via Estrogen Receptor β Activation Is Cytoprotective Against 6-OHDA-Induced Oxidative Stress. Int. J. Mol. Sci. 2019, 20, 1772. [Google Scholar] [CrossRef]
- Han, S.J.; Jung, S.Y.; Wu, S.-P.; Hawkins, S.M.; Park, M.J.; Kyo, S.; Qin, J.; Lydon, J.P.; Tsai, S.Y.; Tsai, M.-J.; et al. Estrogen Receptor β Modulates Apoptosis Complexes and the Inflammasome to Drive the Pathogenesis of Endometriosis. Cell 2015, 163, 960–974. [Google Scholar] [CrossRef]
- Ibrahim, A.; Hugerth, L.W.; Hases, L.; Saxena, A.; Seifert, M.; Thomas, Q.; Gustafsson, J.; Engstrand, L.; Williams, C. Colitis-induced Colorectal Cancer and Intestinal Epithelial Estrogen Receptor Beta Impact Gut Microbiota Diversity. Int. J. Cancer 2019, 144, 3086–3098. [Google Scholar] [CrossRef]
- Majumdar, S.; Rinaldi, J.C.; Malhotra, N.R.; Xie, L.; Hu, D.-P.; Gauntner, T.D.; Grewal, H.S.; Hu, W.-Y.; Kim, S.H.; Katzenellenbogen, J.A.; et al. Differential Actions of Estrogen Receptor α and β via Nongenomic Signaling in Human Prostate Stem and Progenitor Cells. Endocrinology 2019, 160, 2692–2708. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Williams, M.D.; Bell, D. Expression of PTEN, Androgen Receptor, HER2/Neu, Cytokeratin 5/6, Estrogen Receptor-Beta, HMGA2, and PLAG1 in Salivary Duct Carcinoma. Head Neck Pathol. 2019, 13, 529–534. [Google Scholar] [CrossRef] [PubMed]
- Rani, A.; Stebbing, J.; Giamas, G.; Murphy, J. Endocrine Resistance in Hormone Receptor Positive Breast Cancer—From Mechanism to Therapy. Front. Endocrinol. 2019, 10, 245. [Google Scholar] [CrossRef]
- Hamilton, K.J.; Hewitt, S.C.; Arao, Y.; Korach, K.S. Estrogen hormone biology. In Nuclear Receptors in Development and Disease; Forrest, D., Tsai, S.B.T.-C.T.D.B., Eds.; Academic Press: Cambridge, MA, USA, 2017; Volume 125, pp. 109–146. ISBN 0070-2153. [Google Scholar]
- Omoto, Y.; Iwase, H. Clinical Significance of Estrogen Receptor β in Breast and Prostate Cancer from Biological Aspects. Cancer Sci. 2015, 106, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Dahlman-Wright, K.; Gustafsson, J.-Å. Estrogen Receptor β: An Overview and Update. Nucl. Recept. Signal. 2008, 6, nrs.06003. [Google Scholar] [CrossRef]
- Castoria, G.; Migliaccio, A.; Giovannelli, P.; Auricchio, F. Cell Proliferation Regulated by Estradiol Receptor: Therapeutic Implications. Steroids 2010, 75, 524–527. [Google Scholar] [CrossRef] [PubMed]
- Giraldi, T.; Giovannelli, P.; Di Donato, M.; Castoria, G.; Migliaccio, A.; Auricchio, F. Steroid Signaling Activation and Intracellular Localization of Sex Steroid Receptors. J. Cell Commun. Signal. 2010, 4, 161–172. [Google Scholar] [CrossRef]
- Green, K.A.; Carroll, J.S. Oestrogen-Receptor-Mediated Transcription and the Influence of Co-Factors and Chromatin State. Nat. Rev. Cancer 2007, 7, 713–722. [Google Scholar] [CrossRef]
- Carroll, J.S.; Liu, X.S.; Brodsky, A.S.; Li, W.; Meyer, C.A.; Szary, A.J.; Eeckhoute, J.; Shao, W.; Hestermann, E.V.; Geistlinger, T.R.; et al. Chromosome-Wide Mapping of Estrogen Receptor Binding Reveals Long-Range Regulation Requiring the Forkhead Protein FoxA1. Cell 2005, 122, 33–43. [Google Scholar] [CrossRef]
- Hewitt, S.C.; Li, L.; Grimm, S.A.; Chen, Y.; Liu, L.; Li, Y.; Bushel, P.R.; Fargo, D.; Korach, K.S. Research Resource: Whole-Genome Estrogen Receptor α Binding in Mouse Uterine Tissue Revealed by ChIP-Seq. Mol. Endocrinol. 2012, 26, 887–898. [Google Scholar] [CrossRef]
- Carroll, J.S.; Brown, M. Estrogen Receptor Target Gene: An Evolving Concept. Mol. Endocrinol. 2006, 20, 1707–1714. [Google Scholar] [CrossRef] [PubMed]
- Maselli, A.; Pierdominici, M.; Vitale, C.; Ortona, E. Membrane Lipid Rafts and Estrogenic Signalling: A Functional Role in the Modulation of Cell Homeostasis. Apoptosis 2015, 20, 671–678. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.L. Cross-Talk between Peptide Growth Factor and Estrogen Receptor Signaling Pathways. Biol. Reprod. 1998, 58, 627–632. [Google Scholar] [CrossRef]
- Simpson, E. Sources of Estrogen and Their Importance. J. Steroid Biochem. Mol. Biol. 2003, 86, 225–230. [Google Scholar] [CrossRef]
- Longcope, C. Endocrine Function of the Postmenopausal Ovary. J. Soc. Gynecol. Investig. 2001, 8, S67–S68. [Google Scholar] [CrossRef]
- Kelsey, J.L.; Gammon, M.D.; John, E.M. Reproductive Factors and Breast Cancer. Epidemiol. Rev. 1993, 15, 36–47. [Google Scholar] [CrossRef] [PubMed]
- Titus-Ernstoff, L.; Longnecker, M.P.; Newcomb, P.A.; Dain, B.; Greenberg, E.R.; Mittendorf, R.; Stampfer, M.; Willett, W. Menstrual Factors in Relation to Breast Cancer Risk. Cancer Epidemiol. Prev. Biomark. 1998, 7, 783–789. [Google Scholar]
- Rees, M. The Age of Menarche. ORGYN 1995, 4, 2–4. [Google Scholar]
- Zhao, L.; Huang, S.; Mei, S.; Yang, Z.; Xu, L.; Zhou, N.; Yang, Q.; Shen, Q.; Wang, W.; Le, X.; et al. Pharmacological Activation of Estrogen Receptor Beta Augments Innate Immunity to Suppress Cancer Metastasis. Proc. Natl. Acad. Sci. USA 2018, 115, E3673–E3681. [Google Scholar] [CrossRef]
- Hinsche, O.; Girgert, R.; Emons, G.; Gründker, C. Estrogen Receptor β Selective Agonists Reduce Invasiveness of Triple-Negative Breast Cancer Cells. Int. J. Oncol. 2015, 46, 878–884. [Google Scholar] [CrossRef] [PubMed]
- Song, P.; Li, Y.; Dong, Y.; Liang, Y.; Qu, H.; Qi, D.; Lu, Y.; Jin, X.; Guo, Y.; Jia, Y.; et al. Estrogen Receptor β Inhibits Breast Cancer Cells Migration and Invasion through CLDN6-Mediated Autophagy. J. Exp. Clin. Cancer Res. 2019, 38, 354. [Google Scholar] [CrossRef]
- Braschi, B.; Denny, P.; Gray, K.; Jones, T.; Seal, R.; Tweedie, S.; Yates, B.; Bruford, E. Genenames.Org: The HGNC and VGNC Resources in 2019. Nucleic Acids Res. 2019, 47, D786–D792. [Google Scholar] [CrossRef] [PubMed]
- Singhal, H.; Greene, M.E.; Zarnke, A.L.; Laine, M.; Al Abosy, R.; Chang, Y.F.; Dembo, A.G.; Schoenfelt, K.; Vadhi, R.; Qiu, X.; et al. Progesterone Receptor Isoforms, Agonists and Antagonists Differentially Reprogram Estrogen Signaling. Oncotarget 2018, 9, 4282–4300. [Google Scholar] [CrossRef]
- Ballaré, C.; Castellano, G.; Gaveglia, L.; Althammer, S.; González-Vallinas, J.; Eyras, E.; Le Dily, F.; Zaurin, R.; Soronellas, D.; Vicent, G.P.; et al. Nucleosome-Driven Transcription Factor Binding and Gene Regulation. Mol. Cell 2013, 49, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Proietti, C.J.; Cenciarini, M.E.; Elizalde, P.V. Revisiting Progesterone Receptor (PR) Actions in Breast Cancer: Insights into PR Repressive Functions. Steroids 2018, 133, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Clarke, C.L.; Graham, J.D. Non-Overlapping Progesterone Receptor Cistromes Contribute to Cell-Specific Transcriptional Outcomes. PLoS ONE 2012, 7, e35859. [Google Scholar] [CrossRef]
- Richer, J.K.; Jacobsen, B.M.; Manning, N.G.; Abel, M.G.; Wolf, D.M.; Horwitz, K.B. Differential Gene Regulation by the Two Progesterone Receptor Isoforms in Human Breast Cancer Cells. J. Biol. Chem. 2002, 277, 5209–5218. [Google Scholar] [CrossRef]
- Khan, J.A.; Bellance, C.; Guiochon-Mantel, A.; Lombès, M.; Loosfelt, H. Differential Regulation of Breast Cancer-Associated Genes by Progesterone Receptor Isoforms PRA and PRB in a New Bi-Inducible Breast Cancer Cell Line. PLoS ONE 2012, 7, e45993. [Google Scholar] [CrossRef]
- Diep, C.H.; Knutson, T.P.; Lange, C.A. Active FOXO1 Is a Key Determinant of Isoform-Specific Progesterone Receptor Transactivation and Senescence Programming. Mol. Cancer Res. 2016, 14, 141–162. [Google Scholar] [CrossRef]
- McFall, T.; McKnight, B.; Rosati, R.; Kim, S.; Huang, Y.; Viola-Villegas, N.; Ratnam, M. Progesterone Receptor a Promotes Invasiveness and Metastasis of Luminal Breast Cancer by Suppressing Regulation of Critical MicroRNAs by Estrogen. J. Biol. Chem. 2018, 293, 1163–1177. [Google Scholar] [CrossRef]
- Giangrande, P.H.; Kimbrel, E.A.; Edwards, D.P. The Opposing Transcriptional Activities of the Two Isoforms of the Human Progesterone Receptor Are Due to Differential Cofactor Binding. Mol. Cell. Biol. 2000, 20, 3102–3115. [Google Scholar] [CrossRef] [PubMed]
- Bateman, A.; Martin, M.-J.; Orchard, S.; Magrane, M.; Agivetova, R.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; Bursteinas, B.; et al. UniProt: The Universal Protein Knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Piasecka, D.; Składanowski, A.C.; Kordek, R.; Romańska, H.M.; Sądej, R. Aspekty Regulacji Aktywności Receptora Progesteronu (PR)—Znaczenie w Progresji Raka Gruczołu Piersiowego. Postepy Biochem. 2015, 61, 198–206. [Google Scholar]
- Cohen-Solal, K.; Bailly, A.; Rauch, C.; Quesne, M.; Milgrom, E. Specific Binding of Progesterone Receptor to Progesterone-responsive Elements Does Not Require Prior Dimerization. Eur. J. Biochem. 1993, 214, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Hafiz, H.A.; Horwitz, K.B. Post-Translational Modifications of the Progesterone Receptors. J. Steroid Biochem. Mol. Biol. 2014, 140, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Hagan, C.R.; Lange, C.A. Molecular Determinants of Context-Dependent Progesterone Receptor Action in Breast Cancer. Cancer Cell Signal. Target. Signal. Pathw. Towar Ther. Approaches Cancer 2014, 12, 231–252. [Google Scholar] [CrossRef]
- Qiu, M.; Lange, C.A. MAP Kinases Couple Multiple Functions of Human Progesterone Receptors: Degradation, Transcriptional Synergy, and Nuclear Association. J. Steroid Biochem. Mol. Biol. 2003, 85, 147–157. [Google Scholar] [CrossRef]
- Chung, H.H.; Sze, S.K.; En Woo, A.R.; Sun, Y.; Sim, K.H.; Dong, X.M.; Lin, V.C.L. Lysine Methylation of Progesterone Receptor at Activation Function 1 Regulates Both Ligand-Independent Activity and Ligand Sensitivity of the Receptor. J. Biol. Chem. 2014, 289, 5704–5722. [Google Scholar] [CrossRef]
- Analysis of the Forkhead Box A1 (FOXA1)-Dependent and ORG2058 (ORG058)-Regulated Transcriptome in Human MCF-10A AB32 Mammary Epithelial Cells. Available online: https://www.signalingpathways.org/datasets/dataset.jsf?doi=10.1621/n43gIWhFJc (accessed on 26 August 2021).
- Analysis of the Progesterone (P4) and R5020-Dependent ESR1, PGR and EP300 Cistromes in T47D and MCF-7 Human Breast Cancer Cells. Available online: https://128.249.193.43/datasets/dataset.jsf?doi=10.1621/9pIjse2rDj (accessed on 26 August 2021).
- Analysis of the MEK1/2-Regulated Transcriptome in R5020-Treated Human T47D Cells. Available online: https://www.signalingpathways.org/datasets/dataset.jsf?doi=10.1621/XGSeVZH1LR (accessed on 26 August 2021).
- Hilton, H.N.; Graham, J.D.; Clarke, C.L. Minireview: Progesterone Regulation of Proliferation in the Normal Human Breast and in Breast Cancer: A Tale of Two Scenarios? Mol. Endocrinol. 2015, 29, 1230–1242. [Google Scholar] [CrossRef]
- Lyytinen, H.; Dyba, T.; Ylikorkala, O.; Pukkala, E. A case-control study on hormone therapy as a risk factor for breast cancer in finland. Maturitas 2009, 63, S50. [Google Scholar] [CrossRef]
- Li, C.I.; Beaber, E.F.; Tang, M.T.C.; Porter, P.L.; Daling, J.R.; Malone, K.E. Effect of Depo-Medroxyprogesterone Acetate on Breast Cancer Risk among Women 20 to 44 Years of Age. Cancer Res. 2012, 72, 2028–2035. [Google Scholar] [CrossRef]
- Banks, E.; Beral, V.; Bull, D.; Reeves, G.; Austoker, J.; English, R.; Patnick, J.; Peto, R.; Vessey, M.; Wallis, M.; et al. Breast Cancer and Hormone-Replacement Therapy in the Million Women Study. Lancet 2003, 362, 419–427. [Google Scholar] [CrossRef]
- Hunter, D.J.; Colditz, G.A.; Hankinson, S.E.; Malspeis, S.; Spiegelman, D.; Chen, W.; Stampfer, M.J.; Willett, W.C. Oral Contraceptive Use and Breast Cancer: A Prospective Study of Young Women. Cancer Epidemiol. Biomark. Prev. 2010, 19, 2496–2502. [Google Scholar] [CrossRef]
- Soini, T.; Hurskainen, R.; Grénman, S.; Mäenpää, J.; Paavonen, J.; Pukkala, E. Cancer Risk in Women Using the Levonorgestrel-Releasing Intrauterine System in Finland. Obs. Gynecol. 2014, 124, 292–299. [Google Scholar] [CrossRef]
- Chlebowski, R.T.; Kuller, L.H.; Prentice, R.L.; Stefanick, M.L.; Manson, J.E.; Gass, M.; Aragaki, A.K.; Ockene, J.K.; Lane, D.S.; Sarto, G.E.; et al. Breast Cancer after Use of Estrogen plus Progestin in Postmenopausal Women. N. Engl. J. Med. 2009, 360, 573–587. [Google Scholar] [CrossRef] [PubMed]
- Faivre, E.J.; Lange, C.A. Progesterone Receptors Upregulate Wnt-1 to Induce Epidermal Growth Factor Receptor Transactivation and c-Src-Dependent Sustained Activation of Erk1/2 Mitogen-Activated Protein Kinase in Breast Cancer Cells. Mol. Cell Biol. 2007, 27, 466–480. [Google Scholar] [CrossRef]
- Hu, T.; Li, C. Convergence between Wnt-β-Catenin and EGFR Signaling in Cancer. Mol. Cancer 2010, 9, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Ohtsu, H.; Dempsey, P.J.; Eguchi, S. ADAMs as Mediators of EGF Receptor Transactivation by G Protein-Coupled Receptors. Am. J. Physiol. Cell Physiol. 2006, 291, C1–C10. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z. Transactivation of Epidermal Growth Factor Receptor by g Protein-Coupled Receptors: Recent Progress, Challenges and Future Research. Int. J. Mol. Sci. 2016, 17, 95. [Google Scholar] [CrossRef]
- Schlange, T.; Matsuda, Y.; Lienhard, S.; Huber, A.; Hynes, N.E. Autocrine WNT Signaling Contributes to Breast Cancer Cell Proliferation via the Canonical WNT Pathway and EGFR Transactivation. Breast Cancer Res. 2007, 9, 1–15. [Google Scholar] [CrossRef]
- Cattaneo, F.; Guerra, G.; Parisi, M.; De Marinis, M.; Tafuri, D.; Cinelli, M.; Ammendola, R. Cell-Surface Receptors Transactivation Mediated by G Protein-Coupled Receptors. Int. J. Mol. Sci. 2014, 15, 19700–19728. [Google Scholar] [CrossRef] [PubMed]
- Hagan, C.R.; Knutson, T.P.; Lange, C.A. A Common Docking Domain in Progesterone Receptor-B Links DUSP6 and CK2 Signaling to Proliferative Transcriptional Programs in Breast Cancer Cells. Nucleic Acids Res. 2013, 41, 8926–8942. [Google Scholar] [CrossRef] [PubMed]
- Izzo, F.; Mercogliano, F.; Venturutti, L.; Tkach, M.; Inurrigarro, G.; Schillaci, R.; Cerchietti, L.; Elizalde, P.V.; Proietti, C.J. Progesterone Receptor Activation Downregulates GATA3 by Transcriptional Repression and Increased Protein Turnover Promoting Breast Tumor Growth. Breast Cancer Res. 2014, 16, 491. [Google Scholar] [CrossRef]
- Truong, T.H.; Lange, C.A. Deciphering Steroid Receptor Crosstalk in Hormone-Driven Cancers. Endocrinology 2018, 159, 3897–3907. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, H.; Russell, I.A.; Stark, R.; Rueda, O.M.; Hickey, T.E.; Tarulli, G.A.; Serandour, A.A.A.; Birrell, S.N.; Bruna, A.; Saadi, A.; et al. Progesterone Receptor Modulates ERα Action in Breast Cancer. Nature 2015, 523, 313–317. [Google Scholar] [CrossRef]
- Singhal, H.; Greene, M.E.; Tarulli, G.; Zarnke, A.L.; Bourgo, R.J.; Laine, M.; Chang, Y.F.; Ma, S.; Dembo, A.G.; Raj, G.V.; et al. Genomic Agonism and Phenotypic Antagonism between Estrogen and Progesterone Receptors in Breast Cancer. Sci. Adv. 2016, 2, e1501924. [Google Scholar] [CrossRef] [PubMed]
- Kraus, W.L.; Weis, K.E.; Katzenellenbogen, B.S. Inhibitory Cross-Talk between Steroid Hormone Receptors: Differential Targeting of Estrogen Receptor in the Repression of Its Transcriptional Activity by Agonist- and Antagonist-Occupied Progestin Receptors. Mol. Cell Biol. 1995, 15, 1847–1857. [Google Scholar] [CrossRef]
- Graham, J.D.; Yeates, C.; Balleine, R.L.; Clarke, C.L.; Milliken, J.S.; Bilous, A.M.; Harvey, S.S. Characterization of Progesterone Receptor A and B Expression in Human Breast Cancer. Cancer Res. 1995, 55, 5063–5068. [Google Scholar]
- Mote, P.A.; Bartow, S.; Tran, N.; Clarke, C.L. Loss of Co-Ordinate Expression of Progesterone Receptors A and B Is an Early Event in Breast Carcinogenesis. Breast Cancer Res. Treat. 2002, 72, 163–172. [Google Scholar] [CrossRef]
- Sartorius, C.A.; Shen, T.; Horwitz, K.B. Progesterone Receptors A and B Differentially Affect the Growth of Estrogen-Dependent Human Breast Tumor Xenografts. Breast Cancer Res. Treat. 2003, 79, 287–299. [Google Scholar] [CrossRef]
- Hopp, T.A.; Weiss, H.L.; Hilsenbeck, S.G.; Cui, Y.; Allred, D.C.; Horwitz, K.B.; Fuqua, S.A.W. Breast Cancer Patients with Progesterone Receptor PR-A-Rich Tumors Have Poorer Disease-Free Survival Rates. Clin. Cancer Res. 2004, 10, 2751–2760. [Google Scholar] [CrossRef]
- Mote, P.A.; Gompel, A.; Howe, C.; Hilton, H.N.; Sestak, I.; Cuzick, J.; Dowsett, M.; Hugol, D.; Forgez, P.; Byth, K.; et al. Progesterone Receptor A Predominance Is a Discriminator of Benefit from Endocrine Therapy in the ATAC Trial. Breast Cancer Res. Treat. 2015, 151, 309–318. [Google Scholar] [CrossRef]
- Rojas, P.A.; May, M.; Sequeira, G.R.; Elia, A.; Alvarez, M.; Martínez, P.; Gonzalez, P.; Hewitt, S.; He, X.; Perou, C.M.; et al. Progesterone Receptor Isoform Ratio: A Breast Cancer Prognostic and Predictive Factor for Antiprogestin Responsiveness. J. Natl. Cancer Inst. 2017, 109. [Google Scholar] [CrossRef] [PubMed]
- Wargon, V.; Riggio, M.; Giulianelli, S.; Sequeira, G.R.; Rojas, P.; May, M.; Polo, M.L.; Gorostiaga, M.A.; Jacobsen, B.; Molinolo, A.; et al. Progestin and Antiprogestin Responsiveness in Breast Cancer Is Driven by the PRA/PRB Ratio via AIB1 or SMRT Recruitment to the CCND1 and MYC Promoters. Int. J. Cancer 2015, 136, 2680–2692. [Google Scholar] [CrossRef] [PubMed]
- Burger, H.G. Androgen Production in Women. Fertil. Steril. 2002, 77, 3–5. [Google Scholar] [CrossRef]
- Van der Steen, T.; Tindall, D.; Huang, H. Posttranslational Modification of the Androgen Receptor in Prostate Cancer. Int. J. Mol. Sci. 2013, 14, 14833–14859. [Google Scholar] [CrossRef]
- Ruizeveld De Winter, J.A.; Trapman, J.; Vermey, M.; Mulder, E.; Zegers, N.D.; Van der Kwast, T.H. Androgen Receptor Expression in Human Tissues: An Immunohistochemical Study. J. Histochem. Cytochem. 1991, 39, 927–936. [Google Scholar] [CrossRef]
- Kensler, K.H.; Beca, F.; Baker, G.M.; Heng, Y.J.; Beck, A.H.; Schnitt, S.J.; Hazra, A.; Rosner, B.A.; Eliassen, A.H.; Hankinson, S.E.; et al. Androgen Receptor Expression in Normal Breast Tissue and Subsequent Breast Cancer Risk. NPJ Breast Cancer 2018, 4, 33. [Google Scholar] [CrossRef] [PubMed]
- Fioretti, F.M.; Sita-Lumsden, A.; Bevan, C.L.; Brooke, G.N. Revising the Role of the Androgen Receptor in Breast Cancer. J. Mol. Endocrinol. 2014, 52, R257–R265. [Google Scholar] [CrossRef]
- Garay, J.P.; Park, B.H. Androgen Receptor as a Targeted Therapy for Breast Cancer. Am. J. Cancer Res. 2012, 2, 434–445. [Google Scholar]
- Hickey, T.E.; Irvine, C.M.; Dvinge, H.; Tarulli, G.A.; Hanson, A.R.; Ryan, N.K.; Pickering, M.A.; Birrell, S.N.; Hu, D.G.; Mackenzie, P.I.; et al. Expression of Androgen Receptor Splice Variants in Clinical Breast Cancers. Oncotarget 2015, 6, 44728–44744. [Google Scholar] [CrossRef]
- Thakkar, A.; Wang, B.; Picon-Ruiz, M.; Buchwald, P.; Ince, T.A. Vitamin D and Androgen Receptor-Targeted Therapy for Triple-Negative Breast Cancer. Breast Cancer Res. Treat. 2016, 157, 77–90. [Google Scholar] [CrossRef]
- Nelson, K.A. Androgen Receptor CAG Repeats and Prostate Cancer. Am. J. Epidemiol. 2002, 155, 883–890. [Google Scholar] [CrossRef] [PubMed]
- Palazzolo, I.; Gliozzi, A.; Rusmini, P.; Sau, D.; Crippa, V.; Simonini, F.; Onesto, E.; Bolzoni, E.; Poletti, A. The Role of the Polyglutamine Tract in Androgen Receptor. J. Steroid Biochem. Mol. Biol. 2008, 108, 245–253. [Google Scholar] [CrossRef]
- Lundin, K.B.; Giwercman, A.; Dizeyi, N.; Giwercman, Y.L. Functional in Vitro Characterisation of the Androgen Receptor GGN Polymorphism. Mol. Cell Endocrinol. 2007, 264, 184–187. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Xiao, F.; Zhang, Y.; Lan, A.; Song, Q.; Zhang, R.; Gu, K.; Chen, P.; Li, Z.; Zhang, X.; et al. Shorter GGN Repeats in Androgen Receptor Gene Would Not Increase the Risk of Prostate Cancer. Technol. Cancer Res. Treat. 2017, 16, 159–166. [Google Scholar] [CrossRef]
- Weng, H.; Li, S.; Huang, J.Y.; He, Z.Q.; Meng, X.Y.; Cao, Y.; Fang, C.; Zeng, X.T. Androgen Receptor Gene Polymorphisms and Risk of Prostate Cancer: A Meta-Analysis. Sci. Rep. 2017, 7, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Rebbeck, T.R.; Kantoff, P.W.; Krithivas, K.; Neuhausen, S.; Blackwood, M.A.; Godwin, A.K.; Daly, M.B.; Narod, S.A.; Garber, J.E.; Lynch, H.T.; et al. Modification of BRCA1-Associated Breast Cancer Risk by the Polymorphic Androgen-Receptor CAG Repeat. Am. J. Hum. Genet. 1999, 64, 1371–1377. [Google Scholar] [CrossRef] [PubMed]
- Spurdle, A.B.; Antoniou, A.C.; Duffy, D.L.; Pandeya, N.; Kelemen, L.; Chen, X.; Peock, S.; Cook, M.R.; Smith, P.L.; Purdie, D.M.; et al. The Androgen Receptor CAG Repeat Polymorphism and Modification of Breast Cancer Risk in BRCA1 and BRCA2 Mutation Carriers. Breast Cancer Res. 2005, 7. [Google Scholar] [CrossRef]
- Christopoulos, P.F.; Vlachogiannis, N.I.; Vogkou, C.T.; Koutsilieris, M. The Role of the Androgen Receptor Signaling in Breast Malignancies. Anticancer Res. 2017, 37, 6533–6540. [Google Scholar] [CrossRef]
- Ware, K.E.; Garcia-Blanco, M.A.; Armstrong, A.J.; Dehm, S.M. Biologic and Clinical Significance of Androgen Receptor Variants in Castration Resistant Prostate Cancer. Endocr. Relat Cancer 2014, 21, T87–T103. [Google Scholar] [CrossRef]
- Hsu, F.N.; Chen, M.C.; Chiang, M.C.; Lin, E.; Lee, Y.T.; Huang, P.H.; Lee, G.S.; Lin, H. Regulation of Androgen Receptor and Prostate Cancer Growth by Cyclin-Dependent Kinase 5. J. Biol. Chem. 2011, 286, 33141–33149. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.-S.; Vitto, M.J.; Busby, S.A.; Garcia, B.A.; Kesler, C.T.; Gioeli, D.; Shabanowitz, J.; Hunt, D.F.; Rundell, K.; Brautigan, D.L.; et al. Simian Virus 40 Small t Antigen Mediates Conformation-Dependent Transfer of Protein Phosphatase 2A onto the Androgen Receptor. Mol. Cell Biol. 2005, 25, 1298–1308. [Google Scholar] [CrossRef]
- Lin, H.K.; Yeh, S.; Kang, H.Y.; Chang, C. Akt Suppresses Androgen-Induced Apoptosis by Phosphorylating and Inhibiting Androgen Receptor. Proc. Natl. Acad. Sci. USA 2001, 98, 7200–7205. [Google Scholar] [CrossRef] [PubMed]
- Palazzolo, I.; Burnett, B.G.; Young, J.E.; Brenne, P.L.; La Spada, A.R.; Fischbeck, K.H.; Howell, B.W.; Pennuto, M. Akt Blocks Ligand Binding and Protects against Expanded Polyglutamine Androgen Receptor Toxicity. Hum. Mol. Genet. 2007, 16, 1593–1603. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.K.; Hu, Y.C.; Yang, L.; Altuwaijri, S.; Chen, Y.T.; Kang, H.Y.; Chang, C. Suppression Versus Induction of Androgen Receptor Functions by the Phosphatidylinositol 3-Kinase/Akt Pathway in Prostate Cancer LNCaP Cells with Different Passage Numbers. J. Biol. Chem. 2003, 278, 50902–50907. [Google Scholar] [CrossRef]
- Chen, S.; Gulla, S.; Cai, C.; Balk, S.P. Androgen Receptor Serine 81 Phosphorylation Mediates Chromatin Binding and Transcriptional Activation. J. Biol. Chem. 2012, 287, 8571–8583. [Google Scholar] [CrossRef]
- Mahajan, K.; Challa, S.; Coppola, D.; Lawrence, H.; Luo, Y.; Gevariya, H.; Zhu, W.; Chen, Y.A.; Lawrence, N.J.; Mahajan, N.P. Effect of Ack1 Tyrosine Kinase Inhibitor on Ligand-Independent Androgen Receptor Activity. Prostate 2010, 70, 1274–1285. [Google Scholar] [CrossRef]
- Mahajan, N.P.; Liu, Y.; Majumder, S.; Warren, M.R.; Parker, C.E.; Mohler, J.L.; Earp, H.S.; Whang, Y.E. Activated Cdc42-Associated Kinase Ack1 Promotes Prostate Cancer Progression via Androgen Receptor Tyrosine Phosphorylation. Proc. Natl. Acad. Sci. USA 2007, 104, 8438–8443. [Google Scholar] [CrossRef]
- Mahajan, N.P.; Whang, Y.E.; Mohler, J.L.; Earp, H.S. Activated Tyrosine Kinase Ack1 Promotes Prostate Tumorigenesis: Role of Ack1 in Polyubiquitination of Tumor Suppressor Wwox. Cancer Res. 2005, 65, 10514–10523. [Google Scholar] [CrossRef]
- Liu, Y.; Karaca, M.; Zhang, Z.; Gioeli, D.; Earp, H.S.; Whang, Y.E. Dasatinib Inhibits Site-Specific Tyrosine Phosphorylation of Androgen Receptor by Ack1 and Src Kinases. Oncogene 2010, 29, 3208–3216. [Google Scholar] [CrossRef] [PubMed]
- Shu, S.K.; Liu, Q.; Coppola, D.; Cheng, J.Q. Phosphorylation and Activation of Androgen Receptor by Aurora-A. J. Biol. Chem. 2010, 285, 33045–33053. [Google Scholar] [CrossRef] [PubMed]
- Chymkowitch, P.; Le May, N.; Charneau, P.; Compe, E.; Egly, J.M. The Phosphorylation of the Androgen Receptor by TFIIH Directs the Ubiquitin/Proteasome Process. EMBO J. 2011, 30, 468–479. [Google Scholar] [CrossRef]
- Ponguta, L.A.; Gregory, C.W.; French, F.S.; Wilson, E.M. Site-Specific Androgen Receptor Serine Phosphorylation Linked to Epidermal Growth Factor-Dependent Growth of Castration-Recurrent Prostate Cancer. J. Biol. Chem. 2008, 283, 20989–21001. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Dai, B.; Jiang, T.; Xu, K.; Xie, Y.; Kim, O.; Nesheiwat, I.; Kong, X.; Melamed, J.; Handratta, V.D.; et al. Regulation of Androgen Receptor Activity by Tyrosine Phosphorylation. Cancer Cell 2007, 11, 97. [Google Scholar] [CrossRef]
- Gioeli, D.; Black, B.E.; Gordon, V.; Spencer, A.; Kesler, C.T.; Eblen, S.T.; Paschal, B.M.; Weber, M.J. Stress Kinase Signaling Regulates Androgen Receptor Phosphorylation, Transcription, and Localization. Mol. Endocrinol. 2006, 20, 503–515. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.X.; Kemppainen, J.A.; Wilson, E.M. Identification of Three Proline-Directed Phosphorylation Sites in the Human Androgen Receptor. Mol. Endocrinol. 1995, 9, 605–615. [Google Scholar] [CrossRef]
- Gordon, V.; Bhadel, S.; Wunderlich, W.; Zhang, J.A.; Ficarro, S.B.; Mollah, S.A.; Shabanowitz, J.; Hunt, D.F.; Xenarios, I.; Hahn, W.C.; et al. CDK9 Regulates AR Promoter Selectivity and Cell Growth through Serine 81 Phosphorylation. Mol. Endocrinol. 2010, 24, 2267–2280. [Google Scholar] [CrossRef]
- Linn, D.E.; Yang, X.; Xie, Y.; Alfano, A.; Deshmukh, D.; Wang, X.; Shimelis, H.; Chen, H.; Li, W.; Xu, K.; et al. Differential Regulation of Androgen Receptor by P1M-1 Kinases via Phosphorylation-Dependent Recruitment of Distinct Ubiquitin E3 Ligases. J. Biol. Chem. 2012, 287, 22959–22968. [Google Scholar] [CrossRef]
- Heemers, H.V.; Tindall, D.J. Androgen Receptor (AR) Coregulators: A Diversity of Functions Converging on and Regulating the AR Transcriptional Complex. Endocr. Rev. 2007, 28, 778–808. [Google Scholar] [CrossRef]
- Heinlein, C.A.; Chang, C. Androgen Receptor (AR) Coregulators: An Overview. Endocr. Rev. 2002, 23, 175–200. [Google Scholar] [CrossRef] [PubMed]
- Crown, J.; Fabre, A.; Watson, W.; Gallagher, W.; Prencipe, M. PO-446 Targeting Co-Regulators of Hormone-Receptors as a Novel Therapeutic Approach for Prostate and Breast Cancer. ESMO Open 2018, 3, A406–A407. [Google Scholar] [CrossRef]
- Niemeier, L.A.; Dabbs, D.J.; Beriwal, S.; Striebel, J.M.; Bhargava, R. Androgen Receptor in Breast Cancer: Expression in Estrogen Receptor-Positive Tumors and in Estrogen Receptor-Negative Tumors with Apocrine Differentiation. Mod. Pathol. 2010, 23, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Kensler, K.H.; Poole, E.M.; Heng, Y.J.; Collins, L.C.; Glass, B.; Beck, A.H.; Hazra, A.; Rosner, B.A.; Eliassen, A.H.; Hankinson, S.E.; et al. Androgen Receptor Expression and Breast Cancer Survival: Results from the Nurses’ Health Studies. J. Natl. Cancer Inst. 2019, 111, 700–708. [Google Scholar] [CrossRef]
- Peters, A.A.; Buchanan, G.; Ricciardelli, C.; Bianco-Miotto, T.; Centenera, M.M.; Harris, J.M.; Jindal, S.; Segara, D.; Jia, L.; Moore, N.L.; et al. Androgen Receptor Inhibits Estrogen Receptor-α Activity and Is Prognostic in Breast Cancer. Cancer Res. 2009, 69, 6131–6140. [Google Scholar] [CrossRef]
- Chia, K.M.; Liu, J.; Francis, G.D.; Naderi, A. A Feedback Loop between Androgen Receptor and Erk Signaling in Estrogen Receptor-Negative Breast Cancer. Neoplasia 2011, 13, 154–166. [Google Scholar] [CrossRef]
- Mukherjee, A.; Narayan, K.P.; Pal, K.; Kumar, J.M.; Rangaraj, N.; Kalivendi, S.V.; Banerjee, R. Selective Cancer Targeting via Aberrant Behavior of Cancer Cell-Associated Glucocorticoid Receptor. Mol. Ther. 2009, 17, 623–631. [Google Scholar] [CrossRef]
- Stechschulte, L.A.; Wuescher, L.; Marino, J.S.; Hill, J.W.; Eng, C.; Hinds, T.D. Glucocorticoid Receptor β Stimulates Akt1 Growth Pathway by Attenuation of Pten. J. Biol. Chem. 2014, 289, 17885–17894. [Google Scholar] [CrossRef] [PubMed]
- Vandevyver, S.; Dejager, L.; Libert, C. Comprehensive Overview of the Structure and Regulation of the Glucocorticoid Receptor. Endocr. Rev. 2014, 35, 671–693. [Google Scholar] [CrossRef] [PubMed]
- Præstholm, S.M.; Correia, C.M.; Grøntved, L. Multifaceted Control of GR Signaling and Its Impact on Hepatic Transcriptional Networks and Metabolism. Front. Endocrinol. 2020, 11. [Google Scholar] [CrossRef]
- Wang, Q.; Blackford, J.A.; Song, L.N.; Huang, Y.; Cho, S.; Simons, S.S. Equilibrium Interactions of Corepressors and Coactivators with Agonist and Antagonist Complexes of Glucocorticoid Receptors. Mol. Endocrinol. 2004, 18, 1376–1395. [Google Scholar] [CrossRef]
- Weikum, E.R.; Knuesel, M.T.; Ortlund, E.A.; Yamamoto, K.R. Glucocorticoid Receptor Control of Transcription: Precision and Plasticity via Allostery. Nat. Rev. Mol. Cell Biol. 2017, 18, 159–174. [Google Scholar] [CrossRef] [PubMed]
- Jääskeläinen, A.; Jukkola, A.; Haapasaari, K.M.; Auvinen, P.; Soini, Y.; Karihtala, P. Cytoplasmic Mineralocorticoid Receptor Expression Predicts Dismal Local Relapse-Free Survival in Non-Triple-Negative Breast Cancer. Anticancer Res. 2019, 39, 5879–5890. [Google Scholar] [CrossRef]
- Skor, M.N.; Wonder, E.L.; Kocherginsky, M.; Goyal, A.; Hall, B.A.; Cai, Y.; Conzen, S.D. Glucocorticoid Receptor Antagonism as a Novel Therapy for Triple-Negative Breast Cancer. Clin. Cancer Res. 2013, 19, 6163–6172. [Google Scholar] [CrossRef]
- Kino, T.; Manoli, I.; Kelkar, S.; Wang, Y.; Su, Y.A.; Chrousos, G.P. Glucocorticoid Receptor (GR) β Has Intrinsic, GRα-Independent Transcriptional Activity. Biochem. Biophys. Res. Commun. 2009, 381, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Casey, T.M.; Plaut, K. The Role of Glucocorticoids in Secretory Activation and Milk Secretion, a Historical Perspective. J. Mammary Gland Biol. Neoplasia 2007, 12, 293–304. [Google Scholar] [CrossRef]
- Peng, Y.; Li, C.X.; Chen, F.; Wang, Z.; Ligr, M.; Melamed, J.; Wei, J.; Gerald, W.; Pagano, M.; Garabedian, M.J.; et al. Stimulation of Prostate Cancer Cellular Proliferation and Invasion by the Androgen Receptor Co-Activator ARA70β. Am. J. Pathol. 2008, 172, 225–235. [Google Scholar] [CrossRef]
- Conde, I.; Paniagua, R.; Fraile, B.; Lucio, J.; Arenas, M.I. Glucocorticoid Receptor Changes Its Cellular Location with Breast Cancer Development. Histol. Histopathol. 2008, 23, 77–85. [Google Scholar] [CrossRef] [PubMed]
- West, D.C.; Pan, D.; Tonsing-Carter, E.Y.; Hernandez, K.M.; Pierce, C.F.; Styke, S.C.; Bowie, K.R.; Garcia, T.I.; Kocherginsky, M.; Conzen, S.D. GR and ER Coactivation Alters the Expression of Differentiation Genes and Associates with Improved ER+ Breast Cancer Outcome. Mol. Cancer Res. 2016, 14, 707–719. [Google Scholar] [CrossRef]
- Leo, J.C.L.; Guo, C.; Woon, C.T.; Aw, S.E.; Lin, V.C.L. Glucocorticoid and Mineralocorticoid Cross-Talk with Progesterone Receptor to Induce Focal Adhesion and Growth Inhibition in Breast Cancer Cells. Endocrinology 2004, 145, 1314–1321. [Google Scholar] [CrossRef] [PubMed]
- Abduljabbar, R.; Negm, O.H.; Lai, C.F.; Jerjees, D.A.; Al-Kaabi, M.; Hamed, M.R.; Tighe, P.J.; Buluwela, L.; Mukherjee, A.; Green, A.R.; et al. Clinical and Biological Significance of Glucocorticoid Receptor (GR) Expression in Breast Cancer. Breast Cancer Res. Treat. 2015, 150, 335–346. [Google Scholar] [CrossRef] [PubMed]
- Ritter, H.D.; Antonova, L.; Mueller, C.R. The Unliganded Glucocorticoid Receptor Positively Regulates the Tumor Suppressor Gene BRCA1 through GABP Beta. Mol. Cancer Res. 2012, 10, 558–569. [Google Scholar] [CrossRef]
- Pippal, J.B.; Yao, Y.; Rogerson, F.M.; Fuller, P.J. Structural and Functional Characterization of the Interdomain Interaction in the Mineralocorticoid Receptor. Mol. Endocrinol. 2009, 23, 1360–1370. [Google Scholar] [CrossRef]
- Fuller, P.J.; Yang, J.; Young, M.J. 30 YEARS OF THE MINERALOCORTICOID RECEPTOR: Coregulators as Mediators of Mineralocorticoid Receptor Signalling Diversity. J. Endocrinol. 2017, 234, T23–T34. [Google Scholar] [CrossRef]
- Viengchareun, S.; Le Menuet, D.; Martinerie, L.; Munier, M.; Tallec, L.P.-L.; Lombès, M. The Mineralocorticoid Receptor: Insights into Its Molecular and (Patho)Physiological Biology. Nucl. Recept Signal. 2007, 5, nrs.05012. [Google Scholar] [CrossRef]
- Hultman, M.L.; Krasnoperova, N.V.; Li, S.; Du, S.; Xia, C.; Dietz, J.D.; Lala, D.S.; Welsch, D.J.; Hu, X. The Ligand-Dependent Interaction of Mineralocorticoid Receptor with Coactivator and Corepressor Peptides Suggests Multiple Activation Mechanisms. Mol. Endocrinol. 2005, 19, 1460–1473. [Google Scholar] [CrossRef]
- Yang, J.; Fuller, P.J.; Morgan, J.; Shibata, H.; McDonnell, D.P.; Clyne, C.D.; Young, M.J. Use of Phage Display to Identify Novel Mineralocorticoid Receptor-Interacting Proteins. Mol. Endocrinol. 2014, 28, 1571–1584. [Google Scholar] [CrossRef]
- Pratt, W.B.; Toft, D.O. Steroid Receptor Interactions with Heat Shock Protein and Immunophilin Chaperones. Endocr. Rev. 1997, 18, 306–360. [Google Scholar] [CrossRef]
- Pascual-Le Tallec, L.; Demange, C.; Lombès, M. Human Mineralocorticoid Receptor A and B Protein Forms Produced by Alternative Translation Sites Display Different Transcriptional Activities. Eur. J. Endocrinol. 2004, 150, 585–590. [Google Scholar] [CrossRef] [PubMed]
- Nie, H.; Li, J.; Yang, X.M.; Cao, Q.Z.; Feng, M.X.; Xue, F.; Wei, L.; Qin, W.; Gu, J.; Xia, Q.; et al. Mineralocorticoid Receptor Suppresses Cancer Progression and the Warburg Effect by Modulating the MiR-338-3p-PKLR Axis in Hepatocellular Carcinoma. Hepatology 2015, 62, 1145–1159. [Google Scholar] [CrossRef] [PubMed]
- Rigiracciolo, D.C.; Scarpelli, A.; Lappano, R.; Pisano, A.; Santolla, M.F.; Avino, S.; de Marco, P.; Bussolati, B.; Maggiolini, M.; de Francesco, E.M. GPER Is Involved in the Stimulatory Effects of Aldosterone in Breast Cancer Cells and Breast Tumor-Derived Endothelial Cells. Oncotarget 2015, 7, 94–111. [Google Scholar] [CrossRef] [PubMed]
- Wickert, L.; Watzka, M.; Bolkenius, U.; Bidlingmaier, F.; Ludwig, M. Mineralocorticoid Receptor Splice Variants in Different Human Tissues. Eur. J. Endocrinol. 1998, 138, 702–704. [Google Scholar] [CrossRef]
- Szyniarowski, P.; Corcelle-Termeau, E.; Farkas, T.; Hyøer-Hansen, M.; Nylandsted, J.; Kallunki, T.; Jäättelä, M. A Comprehensive SiRNA Screen for Kinases That Suppress Macroautophagy in Optimal Growth Conditions. Autophagy 2011, 7, 892–903. [Google Scholar] [CrossRef]
- Martin, P.M.; Rolland, P.H.; Raynaud, J.P. Macromolecular Binding of Dexamethasone as Evidence for the Presence of Mineralocorticoid Receptor in Human Breast Cancer. Cancer Res. 1981, 41, 1222–1226. [Google Scholar]
- Ni, H.; Rui, Q.; Zhu, X.; Yu, Z.; Gao, R.; Liu, H. Antihypertensive Drug Use and Breast Cancer Risk: A Metaanalysis of Observational Studies. Oncotarget 2017, 8, 62545–62560. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, R.; Shah, N.P.; Dent, S.F. Strategies to Prevent Cardiovascular Toxicity in Breast Cancer: Is It Ready for Primetime? J. Clin. Med. 2020, 9, 896. [Google Scholar] [CrossRef]
- Sharma, P.; Banerjee, R.; Narayan, K.P. Mineralocorticoid Receptor Mediated Liposomal Delivery System for Targeted Induction of Apoptosis in Cancer Cells. Biochim. Biophys. Acta Biomembr. 2016, 1858, 415–421. [Google Scholar] [CrossRef]
- Welsh, J. Function of the Vitamin D Endocrine System in Mammary Gland and Breast Cancer. Mol. Cell Endocrinol. 2017, 453, 88–95. [Google Scholar] [CrossRef]
- Marik, R.; Fackler, M.J.; Gabrielson, E.; Zeiger, M.A.; Sukumar, S.; Stearns, V.; Umbricht, C.B. DNA Methylation-Related Vitamin D Receptor Insensitivity in Breast Cancer. Cancer Biol. Ther. 2010, 10, 44–53. [Google Scholar] [CrossRef]
- Qi, X.; Pramanik, R.; Wang, J.; Schultz, R.M.; Maitra, R.K.; Han, J.; Deluca, H.F.; Chen, G. The P38 and JNK Pathways Cooperate to Trans-Activate Vitamin D Receptor via c-Jun/AP-1 and Sensitize Human Breast Cancer Cells to Vitamin D3-Induced Growth Inhibition. J. Biol. Chem. 2002, 277, 25884–25892. [Google Scholar] [CrossRef]
- Ditsch, N.; Toth, B.; Mayr, D.; Lenhard, M.; Gallwas, J.; Weissenbacher, T.; Dannecker, C.; Friese, K.; Jeschke, U. The Association between Vitamin D Receptor Expression and Prolonged Overall Survival in Breast Cancer. J. Histochem. Cytochem. 2012, 60, 121–129. [Google Scholar] [CrossRef]
- Tavera-Mendoza, L.E.; Westerling, T.; Libby, E.; Marusyk, A.; Cato, L.; Cassani, R.; Cameron, L.A.; Ficarro, S.B.; Marto, J.A.; Klawitter, J.; et al. Vitamin D Receptor Regulates Autophagy in the Normal Mammary Gland and in Luminal Breast Cancer Cells. Proc. Natl. Acad. Sci. USA 2017, 114, E2186–E2194. [Google Scholar] [CrossRef] [PubMed]
- Pike, J.W.; Meyer, M.B.; Lee, S.-M.; Onal, M.; Benkusky, N.A. The Vitamin D Receptor: Contemporary Genomic Approaches Reveal New Basic and Translational Insights. J. Clin. Investig. 2017, 127, 1146–1154. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, M.U.N.; Khan, T.A. Association between Vitamin D Receptor (Cdx2, Fok1, Bsm1, Apa1, Bgl1, Taq1, and Poly (A)) Gene Polymorphism and Breast Cancer: A Systematic Review and Meta-Analysis. Tumor Biol. 2017, 39, 101042831773128. [Google Scholar] [CrossRef] [PubMed]
- Murray, A.; Madden, S.F.; Synnott, N.C.; Klinger, R.; O’Connor, D.; O’Donovan, N.; Gallagher, W.; Crown, J.; Duffy, M.J. Vitamin D Receptor as a Target for Breast Cancer Therapy. Endocr. Relat. Cancer 2017, 24, 181–195. [Google Scholar] [CrossRef] [PubMed]
- Holm, J.; Eriksson, L.; Ploner, A.; Eriksson, M.; Rantalainen, M.; Li, J.; Hall, P.; Czene, K. Assessment of Breast Cancer Risk Factors Reveals Subtype Heterogeneity. Cancer Res. 2017, 77, 3708–3717. [Google Scholar] [CrossRef]
- Gaugris, S.; Heaney, R.P.; Boonen, S.; Kurth, H.; Bentkover, J.D.; Sen, S.S. Vitamin D Inadequacy among Post-Menopausal Women: A Systematic Review. QJM Mon. J. Assoc. Physicians 2005, 98, 667–676. [Google Scholar] [CrossRef]
- Walsh, J.S.; Bowles, S.; Evans, A.L. Vitamin D in Obesity. Curr. Opin. Endocrinol. Diabetes Obes. 2017, 24, 389–394. [Google Scholar] [CrossRef]
- Zhu, H.; Bhagatwala, J.; Huang, Y.; Pollock, N.K.; Parikh, S.; Raed, A.; Gutin, B.; Harshfield, G.A.; Dong, Y. Race/Ethnicity-Specific Association of Vitamin D and Global DNA Methylation: Cross-Sectional and Interventional Findings. PLoS ONE 2016, 11, e0152849. [Google Scholar] [CrossRef]
- Dibaba, D.T.; Braithwaite, D.; Akinyemiju, T. Metabolic Syndrome and the Risk of Breast Cancer and Subtypes by Race, Menopause and BMI. Cancers 2018, 10, 299. [Google Scholar] [CrossRef] [PubMed]
- Leppert, W.; Strąg-Lemanowicz, A. Rola Leczenia Hormonalnego u Pacjentów z Zaawansowaną Chorobą Nowotworową. Med. Paliatywna W. Prakt. 2015, 9, 30–38. [Google Scholar]

| Coregulator Type | Gene (Protein) Name | Molecular Function |
|---|---|---|
| Coactivator | NCOA1 (Nuclear Receptor Coactivator 1) also known as SRC1 | Acyltransferase, activator |
| NCOA3 (Nuclear Receptor Coactivator 3) also known as SRC3 | ||
| EP300 (Histone acetyltransferase p300) | ||
| CREBBP (CREB-binding protein) | ||
| NCOA2 (Nuclear Receptor Coactivator 2) also known as SRC2, TIF2 or GRIP1 | Activator | |
| Components of the SWI/SNF (Switch/Sucrose Non-Fermentable) complex | Helicase, DNA-binding, hydrolase, chromatin regulator | |
| UBE3A (Ubiquitin-protein ligase E3A) also known as E6AP | Transferase | |
| CARM1 (Histone-arginine methyltransferase CARM1) | Methyltransferase, chromatin regulator | |
| SRA1 (Steroid receptor RNA activator 1) | Receptor, activator | |
| Corepressor | NCOR1 (Nuclear receptor corepressor 1) | DNA-binding, chromatin regulator, repressor |
| NCOR2 (Nuclear receptor corepressor 1) also known as SMRT | DNA-binding, repressor | |
| HDAC1 (Histone deacetylase 1) | Hydrolase, chromatin regulator, repressor | |
| HDAC2 (Histone deacetylase 2) | ||
| RCOR1 (REST corepressor 1) | Chromatin regulator, repressor | |
| CBX3 (Chromobox protein homolog 3) also known as HP1γ | ||
| KDM1A (Lysine-specific histone demethylase 1A) also known as LSD1 | Oxidoreductase, chromatin regulator, repressor | |
| Coactivator/corepressor | SMARCA4 (Transcription activator BRG1) | Helicase, hydrolase, chromatin regulator, activator, repressor, RNA-binding |
| Residue | Kinase/Phosphatase | Function | References |
|---|---|---|---|
| S81 | CDK1, CDK5, CDK9 | Localization, protein stability | [144] |
| PP2 | Cell growth, transcription | [145] | |
| S94 | PP2 | Transcription | [145] |
| S213 | PI3K/AKT1 | Localization | [146,147,148] |
| PIM-1 | Stability | [144,149] | |
| Y267 | Ack | Cell growth, transcription | [150,151,152,153] |
| SRC | |||
| T280/S291 | AurA | Cell growth, transcription | [154] |
| S308 | PP2 | Transcription | [145] |
| Y363 | Ack | Cell growth, transcription | [151] |
| S424 | PP2 | Transcription stability | [145] |
| PP1 | |||
| S515 | MAPK | Transcription, degradation | [155,156] |
| CDK7 | |||
| Y534 | SRC | Localization, cell cycle, transcription | [153,157] |
| S578 | Localization, transcription | [156] | |
| S650 | ERK1/JNK1/p38-alpha | Localization | [158] |
| Transcription | [159] | ||
| PP1 | Localization | [160] | |
| S791 | PI3K/AKT1 | Transcription, apoptosis, localization | [146,147,148] |
| T850 | PIM-1L | Stability | [161] |
| Protein Type | Activation-Associated | Suppression-Associated |
|---|---|---|
| Components of the chromatin remodeling complex | ARIP, BRG, hBRM, BAF57, SRG3/BAF155, SRCAP, hOsa1/BAF250, hOsa2 | |
| Chromatin structure | HMG-1, HMG-2 | |
| Acetyltransferases and deacetylases | NCOA1 (SRC1) [163], NCOA2 (SRC2), NCOA3 (SRC3) (Rac3, ACTR, AIB1, p/CIP, TRAM1) [163], p300, CBP, P/CAF, Tip60 [162,163] | HBO1, SIRT1, HDAC7, other HDAC |
| Methyltransferases and demethylases | CARM1/PRMT5, PRMT1, G9a, NSD1/ARA267α, LSD1, JHDM2A, JMJD2C [162], KDM4A, KDM4D, KDM4C, KDM4B [128] | |
| Ubiquitination/proteasome pathway | E6-AP, PIRH2, SNURF/RNF4, ARA54, USP10, UBCH7 [162], ZIPK [128] | Mdm2, Chip, MKRN1 |
| TSG101 (both groups), ARNIP (no data) | ||
| SUMOylation pathway | SUMO-2, SUMO-3, Ubc9, Zimp7, Zimp10, SENP1 | SUMO-1, PIASy, Uba3 |
| PIAS1, PIAS3, PIASxα/ARIP3, PIASxβ | ||
| Splicing and RNA metabolism | p54nrb, p102 U5snRNP/ANT-1, p44/MEP50 | hnRNPA1 |
| PSF, PSP1, PSP2 (no data) | ||
| DNA repair | Ku70, Ku80, DNA-PKc, BRCA1, BRCA2 | Rad9 |
| Chaperones and cochaperones | Hsp40 (DnaJ, Ydj1p), Hsp90, Hsp70, Cdc37, FKBP52, FKBP51, Bag-1L | DjA1 |
| Cytoskeleton | actin, supervillin, gelsolin, filamin, α-actinin-2 | filamin-A, transgelin, ARA67/PAT1/APPBP |
| α-actinin-4 | ||
| Endocytosis | HIP1, GAK/auxillin2, caveolin-1 | APPL |
| Signal integrators and transducers, scaffolds and adaptors | ARA55 (Hic5) [163], paxillin, FHL2 (DRAL) [163], PELP1/MNAR, vinexin-α, Vav3, Rho GDI, Ack1, PRK1, RanBPM, ARA24/Ran, STAT3, β-catenin [162], calreticulin [163] | PAK6, RACK1, Ebp1, Hey1, Hey2, RNase L, TCF4 |
| Smad3, GSK-3β | ||
| Cell cycle regulators | cyclin E, cdc25B, CDK6, Rb, pp32, RbaK, AATF/Che-1 [162], RAF (IDE) [163] | cyclin D1 |
| Regulators of apoptosis | Par-4 | caspase 8 |
| Viral oncoproteins | E2, Hbx | |
| E6, E7 | ||
| Nuclear receptor coregulators | Asc-1, Asc-2, Trap/Mediator complex proteins, CoCoA, NRIP, PNRC, TIF1-α, MRF1, PDIP1, Zac1, GT198, ARA70 (RFG, ELE1) [163], ART-27, ARA160 (TMF) [163], PGC-1 (LEM6), NCOA2 [163] | Alien, AES, SMRT, NCOR, PATZ, TGIF, TIP110, TZF, ARR19 |
| RIP140 (depending on receptor-coregulator ratio—corepressor in high, coactivator in low) [163] | ||
| Kinases and phosphatases | MAK, ANPK, Dyrk1A, RSK | ERK8, SCP2, PP2A |
| Transcription factors | AML3/CBFα1, EGR1, FOXA1, GATA-2, GATA-3, NF1, PDEF, Sp1, SF1, USF2, SRF [164], FOXO4 | AP-1, ATF2, c-rel, c/EBPα, Dax1, ERα, FKHR, FoxH1, GR, HoxB13, Pod-1, p53, RelA, SRY, SHP, TR2, TR4 |
| Brn-1, c-Jun, Foxa2, Oct-1, RXR, Sox9, Oct-2 (no data) | ||
| Other | DJ-1/PARK7, L-dopa-decarboxylase, MAGEA11, SRA | LATS2/KPM, PTEN, Tob1, Tob2, DJBP |
| Protein Feature | Protein Name | |
|---|---|---|
| GR interactome | Activation-associated | HSP90, p23, FKBP51, FKBP52, Cyp44 [171], PP5 [171], HDAC6, REV-ERBα—protection, activation, translocation |
| MAPKs, CDK, GSK3 (by phosphorylation) | ||
| CBP, P300, PCAF, p160 family: NCOA1, NCOA2, NCOA3 [171,173]—histone acetyl transferases | ||
| Mediator complex—MED1 and MED14 | ||
| C/EBPα, C/EBPβ, COUP-TFII, CREB1, E47, FOXA1, FOXA2, FOXO1, LXRβ, HNF6, PPARα, BMAL1, CLOCK, CRY1/CRY2, PER1/2, REV-ERBα/β, RORα/γ, HNF1α, HNF4α, STAT5, FXR—transcription factors | ||
| AP-1, AP-2, NFB, NF‐κB, ER, CREBP, NF1/CTF1, Yin Yang 1, Sp1, IRF1/2, cMyb, PU.1, EGR1/NGF1-A [171], 14–3–3ζ, 14–3–3η | ||
| CRTC2, SIRT1, PGC-1α, ASCOM complex, SETDB2 | ||
| Suppression-associated | SMRT, HDAC1, CtBP, SMAD6-HDAC3, CRY1, TAZ, NCOR [173] | |
| LXRα—transcription factor | ||
| 14–3–3σ, FLASH, G protein β [171] | ||
| GC responsive factor-1, c-Ets-1/2 [171] | ||
| Deactivated by GR | MAPK, PI3K, TCR complex [171] | |
| GR modifiers [171] | Phosphorylation | CDKs (A-CDK2, A-CDC2, B-CDK2, B-CDC2, E-CDK2, CDK5), p38 MAPKs, AKT, JNKs, GSK-3β, ERK, casein kinase II |
| Dephosphorylation | PP1, PP2a, PP5 | |
| Ubiquitination | E-1, E-2 (UbcH7), E-3 (Hsp70-interacting protein, ET-AP, human homolog of mdm2 (hmdm2) + p53) | |
| SUMOylation | Ubc9, RSUME | |
| Acetylation | CLOCK, BMAL1 | |
| Deacetylation | HDAC2 | |
| Nitrosylation | neuronal NO synthase, NO donors | |
| Oxidation | H2O2 | |
| Reduction | dithiothreitol, N-acetyl-L-cysteine, thioredoxin reductase | |
| Protein Feature | Protein Name | Interaction Site |
|---|---|---|
| Activation-associated | ELL (elongation factor) | NTD [185], AF-1b [186,187] |
| SUMO-1-conjugation enzyme (Ubc9) | NTD | |
| NCOA1 (SRC-1) | AF-2, NTD [187] | |
| SRC-1e isoform | AF-1, NTD [187] | |
| NCOA2 | AF-2 [187] | |
| p300/CBP | AF-1 [186], AF-2 [186,187], NLS1/hinge? [15] | |
| PGC-1α | AF-2 [186,187,188,189,190], LBD [188] | |
| PGC-1β | LBD [188] | |
| ASC2 (NCOA6, RAP250, AIB3, PRIP, TRBP, NRC), ASC2-1 | LBD [188] | |
| CREBP-BP/RNA helicase complex | NTD | |
| FLASH | AF-1, NTD [187] | |
| CIA | LBD [188] | |
| ARA70-1, ARA70-2 | LBD [188] | |
| FAF-1 | AF-1, NTD [187] | |
| TIF1 [187], TIF1α | NTD | |
| NSD1 | LBD [188] | |
| RIP140 | NTD | |
| Tesmin | LBD | |
| RHA (RNA helicase A) [185] | AF-1a [187], NTD [191] | |
| GAL4 response element [185] | DBD [186] | |
| PKA [15] | NTD | |
| p/CAF [15] | NLS1/hinge (probably p300-related) | |
| EEF1A1, XRCC6, other MR dimers [185], EIF5B [189], AGAP002076-PA—similar [189], PKCα [15], Uba3, Ubc12, NEDD [15], HDAC [187]? | ? | |
| Suppression-associated | PIAS1 | NTD/LBD [187] |
| SMRT | LBD | |
| NCOR | LBD | |
| DAXX | NTD | |
| NF-YC | AF-1 | |
| Gemin 4 | LBD? | |
| PIASxβ [187] | NTD? | |
| SSRP1 [186,189], ATRX [189], SAFB [189], AHNAK [189], BUB3 [189], NPIPL3 [189], CCDC55 [189], RPL4 [189], XRCC6 [189], RRBP1 [189], RPL23A [189], SERF2 [189], EEF1A1 [189], ENSA [189], MUC1 [189], Chloride intracellular channel 1 variant [189], GPX3 [189], RRBP1 [189], PCBP2 [189] | ? | |
| MYL2 [189], FRMD4B [189] | ? | |
| Chaperones | hsp90 | LBD |
| hsp70, p23, p48, FKBP-59, CYP40, other immunophilins | indirectly, via Hsp90 | |
| actin [187] | LBD |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kowalczyk, W.; Waliszczak, G.; Jach, R.; Dulińska-Litewka, J. Steroid Receptors in Breast Cancer: Understanding of Molecular Function as a Basis for Effective Therapy Development. Cancers 2021, 13, 4779. https://doi.org/10.3390/cancers13194779
Kowalczyk W, Waliszczak G, Jach R, Dulińska-Litewka J. Steroid Receptors in Breast Cancer: Understanding of Molecular Function as a Basis for Effective Therapy Development. Cancers. 2021; 13(19):4779. https://doi.org/10.3390/cancers13194779
Chicago/Turabian StyleKowalczyk, Wojciech, Grzegorz Waliszczak, Robert Jach, and Joanna Dulińska-Litewka. 2021. "Steroid Receptors in Breast Cancer: Understanding of Molecular Function as a Basis for Effective Therapy Development" Cancers 13, no. 19: 4779. https://doi.org/10.3390/cancers13194779
APA StyleKowalczyk, W., Waliszczak, G., Jach, R., & Dulińska-Litewka, J. (2021). Steroid Receptors in Breast Cancer: Understanding of Molecular Function as a Basis for Effective Therapy Development. Cancers, 13(19), 4779. https://doi.org/10.3390/cancers13194779







