Organoid Models for Cancer Research—From Bed to Bench Side and Back
Abstract
:Simple Summary
Abstract
1. Introduction
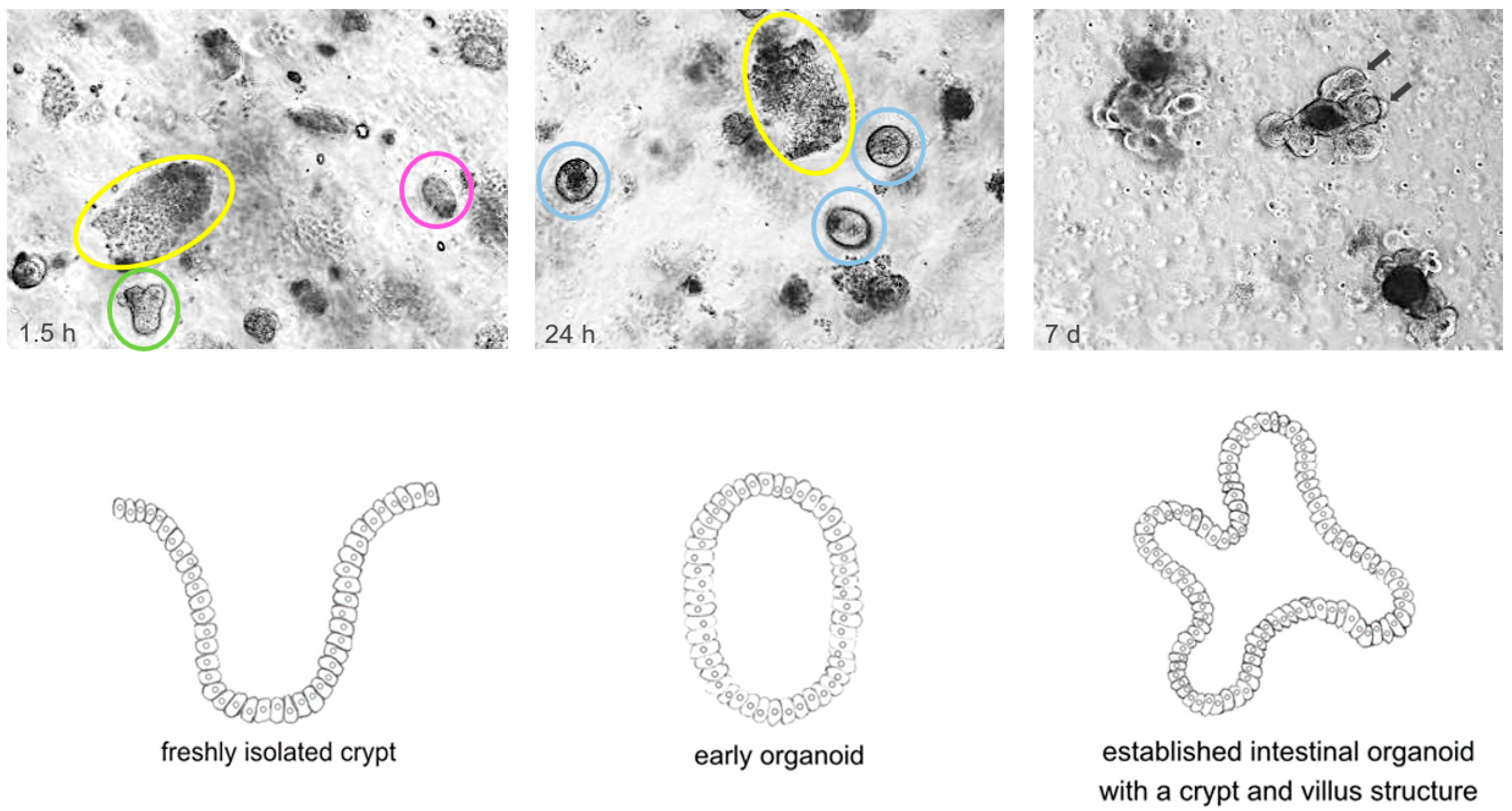
2. Origin and Establishment of Organoid Models
3. Organoids as a Valuable Model for the Investigation of Tumor Development and Progression
4. Organoids as an Enhanced Model for (Personalized) Drug Screening
5. Organoids as a Promising Tool for the Improvement of Therapy Efficiency Prediction
| Tumor Type | Summation | Reference |
|---|---|---|
| Esophageal cancer | only small sample size; organoids derived from esophageal adenocarcinoma resemble the individual patients´ poor clinical response to classic chemotherapeutics such as 5-FU paclitaxel | [69,100] |
| Gastric cancer | correlation of treatment response of tumor organoids from primary tumor to therapeutic response of metastases for exemplary two patients | [70] |
| ambiguous results in correlation of organoid treatment effects to patients´ clinical response | [101,102] | |
| Colorectal cancer | Showing for the first time potential of organoids to predict clinical response; shown for metastatic CRC, gastroesophageal and cholangiocellular cancer | [90] |
| APOLLO trial—first interventional trial; drug screening and next generation sequencing in organoids from peritoneal metastases of CRC; providing organoid-screening stratified therapy for 2 patients | [89] | |
| single-arm, single-center prospective intervention trial in metastatic CRC that missed to show feasibility of optimal therapy selection by organoid based drug screen | [98] | |
| ClinCare study—evaluating the predictive value of PDOs from 80 therapeutically naive locally advanced rectal cancer patients for patients´ clinical response to standard of care chemo(radio)therapy; sensitivity data of 68 organoids matched clinical outcome of the patients´, only 12 did not match | [103] | |
| retrospective correlation of treatment response of 7 rectal PDOs to corresponding patients´ clinical performance regarding 5-FU or FOLFOX treatment | [56] | |
| Pancreatic cancer | prospective trial evaluating and correlating PDOs from primary or metastatic tissue as predictors of clinical drug response, mainly including ductal adenocarcinoma but also less frequent subtypes | [96] |
| correlating treatment efficiency in PDOs to clinical results of the corresponding four patients for gemcitabine treatment demonstrating an overall correlation | [74] | |
| performing therapeutic profiling (pharmacotyping) in 66 PDOs for five mainly used chemotherapeutic agents in PDAC and retrospectively correlating patients´ outcome to their corresponding PDO performance demonstrating good correlation; longitudinal organoid sampling reflected patients´ individual clinical courses | [75] | |
| evaluating individual response of one patient with metastatic pancreatic cancer to PDO selected chemotherapy; PDO insensitivity to initial chemotherapeutic regime was represented in clinical setting as well as good response to PDO sensitive agents | [104] | |
| Liver cancer | no study correlating PDO response to clinical performance | |
| Breast cancer | showing response correlation to tamoxifen of 12 PDOs from needle biopsy of metastatic breast cancer patients to the corresponding patient (their complete large PDO library (95 lines) could not be used for treatment response matching due to the establishment of organoids from surgically in sano resected tumor) | [63] |
| drug identification for one patient by PDO drug screen | [105] | |
| Ovarian cancer | showing statistically significant correlation in response of seven PDOs of five patients with high grade serous ovarian cancer to patients´ clinical history under carboplatin/paclitaxel therapy | [106] |
| HNSCC | matching PDO response to radiotherapy to the effects in the corresponding patients, good correlation | [95] |
| Glioblastom | evaluating glioblastoma organoid reaction to standard-of-care post-surgical treatment (temzolomide and radiation) to patients´ clinical performance with tendency to positive correlation; no prediction of treatment response by MGMT methylation status | [83] |
| Melanoma | establishing PDOs and corresponding immune-enhanced PDOs (iPDO) by usage of matching lymphnodes or WBC, treatment with different kinds of immunotherapeutic drugs (pembrolizumab, nivolumab, ipilimumab, dabrafenib/trametinib); positive correlation of iPDOs to patients´ performance in 85%; in addition longitudinal evaluation of 2 distinct tumors and corresponding organoids | [107] |
6. Organoids as a Valuable Tool of Predicting and Understanding Therapy-Associated Side Effects
7. Conclusions and Perspectives
Funding
Conflicts of Interest
References
- Ferlay, J.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Pineros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today; International Agency for Research on Cancer: Lyon, France, 2018. [Google Scholar]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehár, J.; Kryukov, G.; Sonkin, D.; et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef]
- Ghandi, M.; Huang, F.W.; Jané-Valbuena, J.; Kryukov, G.; Lo, C.C.; Iii, E.R.M.; Barretina, J.; Gelfand, E.T.; Bielski, C.M.; Li, H.; et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 2019, 569, 503–508. [Google Scholar] [CrossRef]
- Drost, J.; Clevers, H. Organoids in cancer research. Nat. Rev. Cancer 2018, 18, 407–418. [Google Scholar] [CrossRef]
- Ben-David, U.; Siranosian, B.; Ha, G.; Tang, H.; Oren, Y.; Hinohara, K.; Strathdee, C.A.; Dempster, J.; Lyons, N.J.; Burns, R.; et al. Genetic and transcriptional evolution alters cancer cell line drug response. Nature 2018, 560, 325–330. [Google Scholar] [CrossRef] [PubMed]
- Schutgens, F.; Clevers, H. Human Organoids: Tools for Understanding Biology and Treating Diseases. Annu. Rev. Pathol. Mech. Dis. 2020, 15, 211–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.; Koo, B.-K.; Knoblich, J.A. Human organoids: Model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 2020, 21, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.H.; Siah, K.W.; Lo, A.W. Estimation of clinical trial success rates and related parameters. Biostatistics 2019, 20, 273–286. [Google Scholar] [CrossRef] [PubMed]
- Hay, M.; Thomas, D.W.; Craighead, J.L.; Economides, C.; Rosenthal, J. Clinical development success rates for investigational drugs. Nat. Biotechnol. 2014, 32, 40–51. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Yu, L.; Li, Z.; Li, W.; Huang, W.R. Patient-Derived Organoid (PDO) Platforms to Facilitate Clinical Decision Making. J. Transl. Med. 2021, 19, 40. [Google Scholar] [CrossRef]
- Holch, J.W.; Metzeler, K.H.; Jung, A.; Riedmann, K.; Jost, P.J.; Weichert, W.; Kirchner, T.; Heinemann, V.; Westphalen, C.B. Universal Genomic Testing: The next step in oncological decision-making or a dead end street? Eur. J. Cancer 2017, 82, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Vries, R.G.; Snippert, H.J.; Van De Wetering, M.; Barker, N.; Stange, D.; Van Es, J.H.; Abo, A.; Kujala, P.; Peters, P.J.; et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009, 459, 262–265. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, M.A.; Huch, M. Disease modelling in human organoids. Dis. Model. Mech. 2019, 12, dmm039347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weiswald, L.-B.; Bellet, D.; Dangles-Marie, V. Spherical Cancer Models in Tumor Biology. Neoplasia 2015, 17, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Farhat, J.; Pandey, I.; AlWahsh, M. Transcending toward Advanced 3D-Cell Culture Modalities: A Review about an Emerging Paradigm in Translational Oncology. Cells 2021, 10, 1657. [Google Scholar] [CrossRef]
- Yamanaka, S. Strategies and New Developments in the Generation of Patient-Specific Pluripotent Stem Cells. Cell Stem Cell 2007, 1, 39–49. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [Green Version]
- Eiraku, M.; Takata, N.; Ishibashi, H.; Kawada, M.; Sakakura, E.; Okuda, S.; Sekiguchi, K.; Adachi, T.; Sasai, Y. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature 2011, 472, 51–56. [Google Scholar] [CrossRef]
- Workman, M.; Mahe, M.M.; Trisno, S.; Poling, H.M.; Watson, C.L.; Sundaram, N.; Chang, C.-F.; Schiesser, J.; Aubert, P.; Stanley, E.G.; et al. Engineered human pluripotent-stem-cell-derived intestinal tissues with a functional enteric nervous system. Nat. Med. 2017, 23, 49–59. [Google Scholar] [CrossRef] [Green Version]
- Freedman, B.S.; Brooks, C.R.; Lam, A.Q.; Fu, H.; Morizane, R.; Agrawal, V.; Saad, A.F.; Li, M.K.; Hughes, M.R.; Werff, R.V.; et al. Modelling kidney disease with CRISPR-mutant kidney organoids derived from human pluripotent epiblast spheroids. Nat. Commun. 2015, 6, 8715. [Google Scholar] [CrossRef] [Green Version]
- Takasato, M.; Er, P.X.; Chiu, H.S.; Maier, B.; Baillie, G.; Ferguson, C.; Parton, R.; Wolvetang, E.; Roost, M.S.; Lopes, S.M.C.D.S.; et al. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature 2015, 526, 564–568. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, M.; Renner, M.; Martin, C.-A.; Wenzel, D.; Bicknell, L.S.; Hurles, M.E.; Homfray, T.; Penninger, J.; Jackson, A.; Knoblich, J.A. Cerebral organoids model human brain development and microcephaly. Nature 2013, 501, 373–379. [Google Scholar] [CrossRef]
- Guan, Y.; Xu, D.; Garfin, P.M.; Ehmer, U.; Hurwitz, M.; Enns, G.; Michie, S.; Wu, M.; Zheng, M.; Nishimura, T.; et al. Human hepatic organoids for the analysis of human genetic diseases. JCI Insight 2017, 2, e94954. [Google Scholar] [CrossRef] [Green Version]
- Völkner, M.; Zschätzsch, M.; Rostovskaya, M.; Overall, R.; Busskamp, V.; Anastassiadis, K.; Karl, M.O. Retinal Organoids from Pluripotent Stem Cells Efficiently Recapitulate Retinogenesis. Stem Cell Rep. 2016, 6, 525–538. [Google Scholar] [CrossRef] [Green Version]
- Sato, T.; Clevers, H. Growing Self-Organizing Mini-Guts from a Single Intestinal Stem Cell: Mechanism and Applications. Science 2013, 340, 1190–1194. [Google Scholar] [CrossRef] [Green Version]
- Clevers, H. Modeling Development and Disease with Organoids. Cell 2016, 165, 1586–1597. [Google Scholar] [CrossRef] [Green Version]
- Georgakopoulos, N.; Prior, N.; Angres, B.; Mastrogiovanni, G.; Cagan, A.; Harrison, D.; Hindley, C.J.; Arnes-Benito, R.; Liau, S.-S.; Curd, A.; et al. Long-term expansion, genomic stability and in vivo safety of adult human pancreas organoids. BMC Dev. Biol. 2020, 20, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Lewis, S.K.; Nachun, D.; Martin, M.G.; Horvath, S.; Coppola, G.; Jones, D.L. DNA Methylation Analysis Validates Organoids as a Viable Model for Studying Human Intestinal Aging. Cell. Mol. Gastroenterol. Hepatol. 2020, 9, 527–541. [Google Scholar] [CrossRef] [Green Version]
- Sato, T.; Stange, D.; Ferrante, M.; Vries, R.G.; van Es, J.H.; Brink, S.V.D.; van Houdt, W.; Pronk, A.; van Gorp, J.; Siersema, P.D.; et al. Long-term Expansion of Epithelial Organoids From Human Colon, Adenoma, Adenocarcinoma, and Barrett’s Epithelium. Gastroenterology 2011, 141, 1762–1772. [Google Scholar] [CrossRef] [PubMed]
- Neal, J.; Li, X.; Zhu, J.; Giangarra, V.; Grzeskowiak, C.L.; Ju, J.; Liu, I.H.; Chiou, S.-H.; Salahudeen, A.A.; Smith, A.R.; et al. Organoid Modeling of the Tumor Immune Microenvironment. Cell 2018, 175, 1972–1988.e16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Ootani, A.; Kuo, C. An Air–Liquid Interface Culture System for 3D Organoid Culture of Diverse Primary Gastroin-testinal Tissues. In Gastrointestinal Physiology and Diseases; Humana Press: New York, NY, USA, 2016. [Google Scholar]
- Kitajima, S.; Ivanova, E.; Guo, S.; Yoshida, R.; Campisi, M.; Sundararaman, S.K.; Tange, S.; Mitsuishi, Y.; Thai, T.C.; Masuda, S.; et al. Suppression of STING Associated with LKB1 Loss in KRAS-Driven Lung Cancer. Cancer Discov. 2019, 9, 34–45. [Google Scholar] [CrossRef] [Green Version]
- Jenkins, R.W.; Aref, A.R.; Lizotte, P.; Ivanova, E.; Stinson, S.; Zhou, C.; Bowden, M.; Deng, J.; Liu, H.; Miao, D.; et al. Ex Vivo Profiling of PD-1 Blockade Using Organotypic Tumor Spheroids. Cancer Discov. 2018, 8, 196–215. [Google Scholar] [CrossRef] [Green Version]
- Tuveson, D.; Clevers, H. Cancer modeling meets human organoid technology. Science 2019, 364, 952–955. [Google Scholar] [CrossRef] [PubMed]
- Vogelstein, B.; Fearon, E.R.; Hamilton, S.R.; Kern, S.E.; Preisinger, A.C.; Leppert, M.; Smits, A.M.; Bos, J.L. Genetic Alterations during Colorectal-Tumor Development. N. Engl. J. Med. 1988, 319, 525–532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Snover, D.C. Update on the serrated pathway to colorectal carcinoma. Hum. Pathol. 2011, 42, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Drost, J.; Van Boxtel, R.; Blokzijl, F.; Mizutani, T.; Sasaki, N.; Sasselli, V.; De Ligt, J.; Behjati, S.; Grolleman, J.E.; Van Wezel, T.; et al. Use of CRISPR-modified human stem cell organoids to study the origin of mutational signatures in cancer. Science 2017, 358, 234–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matano, M.; Date, S.; Shimokawa, M.; Takano, A.; Fujii, M.; Ohta, Y.; Watanabe, T.; Kanai, T.; Sato, T. Modeling colorectal cancer using CRISPR-Cas9–mediated engineering of human intestinal organoids. Nat. Med. 2015, 21, 256–262. [Google Scholar] [CrossRef] [PubMed]
- Fumagalli, A.; Drost, J.; Suijkerbuijk, S.J.E.; van Boxtel, R.; de Ligt, J.; Offerhaus, G.J.; Begthel, H.; Beerling, E.; Tan, E.H.; Sansom, O.J.; et al. Genetic dissection of colorectal cancer progression by orthotopic transplantation of engineered cancer organoids. Proc. Natl. Acad. Sci. USA 2017, 114, E2357–E2364. [Google Scholar] [CrossRef] [Green Version]
- Gao, X.; Bali, A.S.; Randell, S.H.; Hogan, B.L. GRHL2 coordinates regeneration of a polarized mucociliary epithelium from basal stem cells. J. Cell Biol. 2015, 211, 669–682. [Google Scholar] [CrossRef]
- Lannagan, T.; Lee, Y.K.; Wang, T.; Roper, J.; Bettington, M.; Fennell, L.; Vrbanac, L.; Jonavicius, L.; Somashekar, R.; Gieniec, K.; et al. Genetic editing of colonic organoids provides a molecularly distinct and orthotopic preclinical model of serrated carcinogenesis. Gut 2019, 68, 684–692. [Google Scholar] [CrossRef]
- Liu, X.; Cheng, Y.; Abraham, J.M.; Wang, Z.; Wang, Z.; Ke, X.; Yan, R.; Shin, E.J.; Ngamruengphong, S.; Khashab, M.A.; et al. Modeling Wnt signaling by CRISPR-Cas9 genome editing recapitulates neoplasia in human Barrett epithelial organoids. Cancer Lett. 2018, 436, 109–118. [Google Scholar] [CrossRef]
- Fessler, E.; Drost, J.; Van Hooff, S.R.; Linnekamp, J.F.; Wang, X.; Jansen, M.; Melo, F.D.S.E.; Prasetyanti, P.R.; Ijspeert, J.E.; Franitza, M.; et al. TGFβ signaling directs serrated adenomas to the mesenchymal colorectal cancer subtype. EMBO Mol. Med. 2016, 8, 745–760. [Google Scholar] [CrossRef]
- Nadauld, L.D.; Garcia, S.; Natsoulis, G.; Bell, J.M.; Miotke, L.; Hopmans, E.S.; Xu, H.; Pai, R.K.; Palm, C.; Regan, J.F.; et al. Metastatic tumor evolution and organoid modeling implicate TGFBR2as a cancer driver in diffuse gastric cancer. Genome Biol. 2014, 15, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Drost, J.; van Jaarsveld, R.; Ponsioen, B.; Zimberlin, C.D.; van Boxtel, R.; Buijs, A.; Sachs, N.; Overmeer, R.M.; Offerhaus, G.J.; Begthel, H.; et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature 2015, 521, 43–47. [Google Scholar] [CrossRef]
- Dekkers, J.F.; Whittle, J.R.; Vaillant, F.; Chen, H.-R.; Dawson, C.; Liu, K.; Geurts, M.H.; Herold, M.; Clevers, H.; Lindeman, G.J.; et al. Modeling Breast Cancer Using CRISPR-Cas9–Mediated Engineering of Human Breast Organoids. J. Natl. Cancer Inst. 2020, 112, 540–544. [Google Scholar] [CrossRef]
- Kawasaki, K.; Fujii, M.; Sugimoto, S.; Ishikawa, K.; Matano, M.; Ohta, Y.; Toshimitsu, K.; Takahashi, S.; Hosoe, N.; Sekine, S.; et al. Chromosome Engineering of Human Colon-Derived Organoids to Develop a Model of Traditional Serrated Adenoma. Gastroenterology 2020, 158, 638–651.e8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seino, T.; Kawasaki, S.; Shimokawa, M.; Tamagawa, H.; Toshimitsu, K.; Fujii, M.; Ohta, Y.; Matano, M.; Nanki, K.; Kawasaki, K.; et al. Human Pancreatic Tumor Organoids Reveal Loss of Stem Cell Niche Factor Dependence during Disease Progression. Cell Stem Cell 2018, 22, 454–467.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teriyapirom, I.; Batista-Rocha, A.S.; Koo, B.K. Genetic Engineering in Organoids. J. Mol. Med. 2021, 99, 555–568. [Google Scholar] [CrossRef]
- Ogawa, J.; Pao, G.M.; Shokhirev, M.N.; Verma, I.M. Glioblastoma Model Using Human Cerebral Organoids. Cell Rep. 2018, 23, 1220–1229. [Google Scholar] [CrossRef] [Green Version]
- Parisian, A.D.; Koga, T.; Miki, S.; Johann, P.D.; Kool, M.; Crawford, J.R.; Furnari, F.B. SMARCB1 loss interacts with neuronal differentiation state to block maturation and impact cell stability. Genes Dev. 2020, 34, 1316–1329. [Google Scholar] [CrossRef] [PubMed]
- Tejero-Villalba, R.; Huang, Y.; Katsyv, I.; Kluge, M.; Lin, J.-Y.; Garcia, J.T.; Daviaud, N.; Wang, Y.; Zhang, B.; Tsankova, N.M.; et al. Gene signatures of quiescent glioblastoma cells reveal mesenchymal shift and interactions with niche microenvironment. EBioMedicine 2019, 42, 252–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weeber, F.; van de Wetering, M.; Hoogstraat, M.; Dijkstra, K.; Krijgsman, O.; Kuilman, T.; Hooijdonk, C.G.M.G.-V.; van der Velden, D.L.; Peeper, D.S.; Cuppen, E.; et al. Preserved genetic diversity in organoids cultured from biopsies of human colorectal cancer metastases. Proc. Natl. Acad. Sci. USA 2015, 112, 13308–13311. [Google Scholar] [CrossRef] [Green Version]
- Fujii, M.; Shimokawa, M.; Date, S.; Takano, A.; Matano, M.; Nanki, K.; Ohta, Y.; Toshimitsu, K.; Nakazato, Y.; Kawasaki, K.; et al. A Colorectal Tumor Organoid Library Demonstrates Progressive Loss of Niche Factor Requirements during Tumorigenesis. Cell Stem Cell 2016, 18, 827–838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ganesh, K.; Wu, C.; O’Rourke, K.P.; Szeglin, B.C.; Zheng, Y.; Sauvé, C.-E.G.; Adileh, M.; Wasserman, I.; Marco, M.R.; Kim, A.S.; et al. A rectal cancer organoid platform to study individual responses to chemoradiation. Nat. Med. 2019, 25, 1607–1614. [Google Scholar] [CrossRef] [PubMed]
- Pasch, C.A.; Favreau, P.F.; Yueh, A.E.; Babiarz, C.P.; Gillette, A.A.; Sharick, J.T.; Karim, M.R.; Nickel, K.P.; DeZeeuw, A.K.; Sprackling, C.M.; et al. Patient-Derived Cancer Organoid Cultures to Predict Sensitivity to Chemotherapy and Radiation. Clin. Cancer Res. 2019, 25, 5376–5387. [Google Scholar] [CrossRef]
- Broutier, L.; Mastrogiovanni, G.; Verstegen, M.M.; Francies, H.E.; Gavarró, L.M.; Bradshaw, C.R.; Allen, G.E.; Arnes-Benito, R.; Sidorova, O.; Gaspersz, M.P.; et al. Human primary liver cancer–derived organoid cultures for disease modeling and drug screening. Nat. Med. 2017, 23, 1424–1435. [Google Scholar] [CrossRef] [PubMed]
- Maier, C.F.; Zhu, L.; Nanduri, L.K.; Kühn, D.; Kochall, S.; Thepkaysone, M.-L.; William, D.; Grützmann, K.; Klink, B.; Betge, J.; et al. Patient-Derived Organoids of Cholangiocarcinoma. Int. J. Mol. Sci. 2021, 22, 8675. [Google Scholar] [CrossRef]
- Buzzelli, J.N.; Ouaret, D.; Brown, G.; Allen, P.D.; Muschel, R.J. Colorectal cancer liver metastases organoids retain characteristics of original tumor and acquire chemotherapy resistance. Stem Cell Res. 2018, 27, 109–120. [Google Scholar] [CrossRef]
- Gao, D.; Vela, I.; Sboner, A.; Iaquinta, P.J.; Karthaus, W.R.; Gopalan, A.; Dowling, C.; Wanjala, J.N.; Undvall, E.A.; Arora, V.K.; et al. Organoid Cultures Derived from Patients with Advanced Prostate Cancer. Cell 2014, 159, 176–187. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Shiratsuchi, H.; Lin, J.; Chen, G.; Reddy, R.M.; Azizi, E.; Fouladdel, S.; Chang, A.; Lin, L.; Jiang, H.; et al. Expansion of CTCs from early stage lung cancer patients using a microfluidic co-culture model. Oncotarget 2014, 5, 12383–12397. [Google Scholar] [CrossRef] [Green Version]
- Sachs, N.; De Ligt, J.; Kopper, O.; Gogola, E.; Bounova, G.; Weeber, F.; Balgobind, A.V.; Wind, K.; Gracanin, A.; Begthel, H.; et al. A Living Biobank of Breast Cancer Organoids Captures Disease Heterogeneity. Cell 2018, 172, 373–386.e10. [Google Scholar] [CrossRef] [Green Version]
- van de Wetering, M.; Francies, H.E.; Francis, J.M.; Bounova, G.; Iorio, F.; Pronk, A.; van Houdt, W.; van Gorp, J.; Taylor-Weiner, A.; Kester, L.; et al. Prospective Derivation of a Living Organoid Biobank of Colorectal Cancer Patients. Cell 2015, 161, 933–945. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Sun, L.; Liu, M.; Mao, Y. Patient-derived organoids: A promising model for personalized cancer treatment. Gastroenterol. Rep. 2018, 6, 243–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wensink, G.E.; Elias, S.G.; Mullenders, J.; Koopman, M.; Boj, S.F.; Kranenburg, O.W.; Roodhart, J.M.L. Patient-derived organoids as a predictive biomarker for treatment response in cancer patients. NPJ Precis. Oncol. 2021, 5, 1–13. [Google Scholar] [CrossRef]
- Tiriac, H.; Bucobo, J.C.; Tzimas, D.; Grewel, S.; Lacomb, J.F.; Rowehl, L.M.; Nagula, S.; Wu, M.; Kim, J.; Sasson, A.; et al. Successful creation of pancreatic cancer organoids by means of EUS-guided fine-needle biopsy sampling for personalized cancer treatment. Gastrointest. Endosc. 2018, 87, 1474–1480. [Google Scholar] [CrossRef] [PubMed]
- Kijima, T.; Nakagawa, H.; Shimonosono, M.; Chandramouleeswaran, P.M.; Hara, T.; Sahu, V.; Kasagi, Y.; Kikuchi, O.; Tanaka, K.; Giroux, V.; et al. Three-Dimensional Organoids Reveal Therapy Resistance of Esophageal and Oropharyngeal Squamous Cell Carcinoma Cells. Cell. Mol. Gastroenterol. Hepatol. 2019, 7, 73–91. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Francies, H.E.; Secrier, M.; Perner, J.; Miremadi, A.; Galeano-Dalmau, N.; Barendt, W.J.; Letchford, L.; Leyden, G.M.; Goffin, E.K.; et al. Organoid cultures recapitulate esophageal adenocarcinoma heterogeneity providing a model for clonality studies and precision therapeutics. Nat. Commun. 2018, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.H.; Siu, H.C.; Law, S.; Ho, S.L.; Yue, S.S.; Tsui, W.Y.; Chan, D.; Chan, A.S.; Ma, S.K.Y.; Lam, K.O.; et al. A Comprehensive Human Gastric Cancer Organoid Biobank Captures Tumor Subtype Heterogeneity and Enables Therapeutic Screening. Cell Stem Cell 2018, 23, 882–897.e11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mullenders, J.; de Jongh, E.; Brousali, A.; Roosen, M.; Blom, J.P.A.; Begthel, H.; Korving, J.; Jonges, T.; Kranenburg, O.; Meijer, R.; et al. Mouse and human urothelial cancer organoids: A tool for bladder cancer research. Proc. Natl. Acad. Sci. USA 2019, 116, 4567–4574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nuciforo, S.; Fofana, I.; Matter, M.; Blumer, T.; Calabrese, D.; Boldanova, T.; Piscuoglio, S.; Wieland, S.; Ringnalda, F.; Schwank, G.; et al. Organoid Models of Human Liver Cancers Derived from Tumor Needle Biopsies. Cell Rep. 2018, 24, 1363–1376. [Google Scholar] [CrossRef] [Green Version]
- Saito, Y.; Muramatsu, T.; Kanai, Y.; Ojima, H.; Sukeda, A.; Hiraoka, N.; Arai, E.; Sugiyama, Y.; Matsuzaki, J.; Uchida, R.; et al. Establishment of Patient-Derived Organoids and Drug Screening for Biliary Tract Carcinoma. Cell Rep. 2019, 27, 1265–1276.e4. [Google Scholar] [CrossRef] [Green Version]
- Driehuis, E.; van Hoeck, A.; Moore, K.; Kolders, S.; Francies, H.E.; Gulersonmez, M.C.; Stigter, E.C.A.; Burgering, B.; Geurts, V.; Gracanin, A.; et al. Pancreatic cancer organoids recapitulate disease and allow personalized drug screening. Proc. Natl. Acad. Sci. USA 2019, 116, 26580–26590. [Google Scholar] [CrossRef] [PubMed]
- Tiriac, H.; Belleau, P.; Engle, D.D.; Plenker, D.; Deschênes, A.; Somerville, T.D.D.; Froeling, F.E.M.; Burkhart, R.A.; Denroche, R.E.; Jang, G.-H.; et al. Organoid Profiling Identifies Common Responders to Chemotherapy in Pancreatic Cancer. Cancer Discov. 2018, 8, 1112–1129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, B.; Trujillo, M.A.; Fujikura, K.; Qiu, M.; Chen, F.; Felsenstein, M.; Zhou, C.; Skaro, M.; Gauthier, C.; Macgregor-Das, A.; et al. Molecular characterization of organoids derived from pancreatic intraductal papillary mucinous neoplasms. J. Pathol. 2020, 252, 252–262. [Google Scholar] [CrossRef] [PubMed]
- Beato, F.; Reverón, D.; Dezsi, K.B.; Ortiz, A.; Johnson, J.O.; Chen, D.-T.; Ali, K.; Yoder, S.J.; Jeong, D.; Malafa, M.; et al. Establishing a living biobank of patient-derived organoids of intraductal papillary mucinous neoplasms of the pancreas. Lab. Investig. 2021, 101, 204–217. [Google Scholar] [CrossRef]
- Kawasaki, K.; Toshimitsu, K.; Matano, M.; Fujita, M.; Fujii, M.; Togasaki, K.; Ebisudani, T.; Shimokawa, M.; Takano, A.; Takahashi, S.; et al. An Organoid Biobank of Neuroendocrine Neoplasms Enables Genotype-Phenotype Mapping. Cell 2020, 183, 1420–1435.e21. [Google Scholar] [CrossRef]
- Li, Y.F.; Gao, Y.; Liang, B.W.; Cao, X.Q.; Sun, Z.J.; Yu, J.H.; Liu, Z.D.; Han, Y. Patient-derived organoids of non-small cells lung cancer and their application for drug screening. Neoplasma 2020, 67, 430–437. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.; Mun, H.; Sung, C.O.; Cho, E.J.; Jeon, H.-J.; Chun, S.-M.; Jung, D.J.; Shin, T.H.; Jeong, G.S.; Kim, D.K.; et al. Patient-derived lung cancer organoids as in vitro cancer models for therapeutic screening. Nat. Commun. 2019, 10, 1–15. [Google Scholar] [CrossRef]
- Li, Z.; Qian, Y.; Li, W.; Liu, L.; Yu, L.; Liu, X.; Wu, G.; Wang, Y.; Luo, W.; Fang, F.; et al. Human Lung Adenocarcinoma-Derived Organoid Models for Drug Screening. iScience 2020, 23, 101411. [Google Scholar] [CrossRef]
- Hu, Y.; Sui, X.; Song, F.; Li, Y.; Li, K.; Chen, Z.; Yang, F.; Chen, X.; Zhang, Y.; Wang, X.; et al. Lung cancer organoids analyzed on microwell arrays predict drug responses of patients within a week. Nat. Commun. 2021, 12, 1–14. [Google Scholar] [CrossRef]
- Jacob, F.; Salinas, R.D.; Zhang, D.Y.; Nguyen, P.T.; Schnoll, J.G.; Wong, S.Z.H.; Thokala, R.; Sheikh, S.; Saxena, D.; Prokop, S.; et al. A Patient-Derived Glioblastoma Organoid Model and Biobank Recapitulates Inter- and Intra-tumoral Heterogeneity. Cell 2019, 180, 188–204.e22. [Google Scholar] [CrossRef]
- Lee, S.H.; Hu, W.; Matulay, J.; Silva, M.V.; Owczarek, T.B.; Kim, K.; Chua, C.W.; Barlow, L.J.; Kandoth, C.; Williams, A.B.; et al. Tumor Evolution and Drug Response in Patient-Derived Organoid Models of Bladder Cancer. Cell 2018, 173, 515–528.e17. [Google Scholar] [CrossRef] [Green Version]
- Calandrini, C.; Schutgens, F.; Oka, R.; Margaritis, T.; Candelli, T.; Mathijsen, L.; Ammerlaan, C.; Van Ineveld, R.L.; Derakhshan, S.; De Haan, S.; et al. An organoid biobank for childhood kidney cancers that captures disease and tissue heterogeneity. Nat. Commun. 2020, 11, 1–14. [Google Scholar] [CrossRef]
- Mazzucchelli, S.; Piccotti, F.; Allevi, R.; Truffi, M.; Sorrentino, L.; Russo, L.; Agozzino, M.; Signati, L.; Bonizzi, A.; Villani, L.; et al. Establishment and Morphological Characterization of Patient-Derived Organoids from Breast Cancer. Biol. Proced. Online 2019, 21, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Yu, D.; Kwon, Y.; Lee, K.S.; Sim, S.H.; Kong, S.-Y.; Lee, E.S.; Park, I.H.; Park, C. Genomic Characteristics of Triple-Negative Breast Cancer Nominate Molecular Subtypes That Predict Chemotherapy Response. Mol. Cancer Res. 2019, 18, 253–263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campaner, E.; Zannini, A.; Santorsola, M.; Bonazza, D.; Bottin, C.; Cancila, V.; Tripodo, C.; Bortul, M.; Zanconati, F.; Schoeftner, S.; et al. Breast Cancer Organoids Model Patient-Specific Response to Drug Treatment. Cancers 2020, 12, 3869. [Google Scholar] [CrossRef]
- Botti, G.; di Bonito, M.; Cantile, M. Organoid Biobanks as a New Tool for Pre-Clinical Validation of Candidate Drug Effi-cacy and Safety. Int. J. Physiol. Pathophysiol. Pharmacol. 2021, 13, 17. [Google Scholar]
- Vlachogiannis, G.; Hedayat, S.; Vatsiou, A.; Jamin, Y.; Fernández-Mateos, J.; Khan, K.; Lampis, A.; Eason, K.; Huntingford, I.; Burke, R.; et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 2018, 359, 920–926. [Google Scholar] [CrossRef] [Green Version]
- Ooft, S.N.; Weeber, F.; Dijkstra, K.K.; McLean, C.M.; Kaing, S.; van Werkhoven, E.; Schipper, L.; Hoes, L.; Vis, D.J.; van de Haar, J.; et al. Patient-derived organoids can predict response to chemotherapy in metastatic colorectal cancer patients. Sci. Transl. Med. 2019, 11, eaay2574. [Google Scholar] [CrossRef]
- Yao, Y.; Xu, X.; Yang, L.; Zhu, J.; Wan, J.; Shen, L.; Xia, F.; Fu, G.; Deng, Y.; Pan, M.; et al. Patient-Derived Organoids (PDO) As the Potential Model to Predict Treatment Outcome of Rectal Cancer Patients Underwent Neo-Adjuvant Chemoradiotherapy. Int. J. Radiat. Oncol. 2019, 105, S106. [Google Scholar] [CrossRef]
- Tiriac, H.; Plenker, D.; Baker, L.; Tuveson, D.A. Organoid models for translational pancreatic cancer research. Curr. Opin. Genet. Dev. 2019, 54, 7–11. [Google Scholar] [CrossRef]
- Romero-Calvo, I.; Weber, C.R.; Ray, M.; Brown, M.; Kirby, K.; Nandi, R.K.; Long, T.M.; Sparrow, S.M.; Ugolkov, A.; Qiang, W.; et al. Human Organoids Share Structural and Genetic Features with Primary Pancreatic Adenocarcinoma Tumors. Mol. Cancer Res. 2019, 17, 70–83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Driehuis, E.; Kolders, S.; Spelier, S.; Lõhmussaar, K.; Willems, S.M.; Devriese, L.A.; De Bree, R.; De Ruiter, E.J.; Korving, J.; Begthel, H.; et al. Oral Mucosal Organoids as a Potential Platform for Personalized Cancer Therapy. Cancer Discov. 2019, 9, 852–871. [Google Scholar] [CrossRef] [PubMed]
- Beutel, A.; Schütte, L.; Scheible, J.; Roger, E.; Müller, M.; Perkhofer, L.; Kestler, A.; Kraus, J.; Kestler, H.; Barth, T.; et al. A Prospective Feasibility Trial to Challenge Patient–Derived Pancreatic Cancer Organoids in Predicting Treatment Response. Cancers 2021, 13, 2539. [Google Scholar] [CrossRef] [PubMed]
- Narasimhan, V.; Wright, J.A.; Churchill, M.; Wang, T.; Rosati, R.; Lannagan, T.R.; Vrbanac, L.; Richardson, A.B.; Kobayashi, H.; Price, T.; et al. Medium-throughput Drug Screening of Patient-derived Organoids from Colorectal Peritoneal Metastases to Direct Personalized Therapy. Clin. Cancer Res. 2020, 26, 3662–3670. [Google Scholar] [CrossRef]
- Ooft, S.; Weeber, F.; Schipper, L.; Dijkstra, K.; McLean, C.; Kaing, S.; van de Haar, J.; Prevoo, W.; van Werkhoven, E.; Snaebjornsson, P.; et al. Prospective experimental treatment of colorectal cancer patients based on organoid drug responses. ESMO Open 2021, 6, 100103. [Google Scholar] [CrossRef]
- Verduin, M.; Hoeben, A.; De Ruysscher, D.; Vooijs, M. Patient-Derived Cancer Organoids as Predictors of Treatment Response. Front. Oncol. 2021, 11, 820. [Google Scholar] [CrossRef]
- Derouet, M.F.; Allen, J.; Wilson, G.W.; Ng, C.; Radulovich, N.; Kalimuthu, S.; Tsao, M.-S.; Darling, G.E.; Yeung, J.C. Towards personalized induction therapy for esophageal adenocarcinoma: Organoids derived from endoscopic biopsy recapitulate the pre-treatment tumor. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef]
- Steele, N.G.; Chakrabarti, J.; Wang, J.; Biesiada, J.; Holokai, L.; Chang, J.; Nowacki, L.M.; Hawkins, J.; Mahe, M.; Sundaram, N.; et al. An Organoid-Based Preclinical Model of Human Gastric Cancer. Cell. Mol. Gastroenterol. Hepatol. 2019, 7, 161–184. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Xu, H.; Zhang, L.; Song, L.; Feng, D.; Peng, X.; Wu, M.; Zou, Y.; Wang, B.; Hua, G.; et al. Malignant ascites-derived organoid (MADO) cultures for gastric cancer in vitro modelling and drug screening. J. Cancer Res. Clin. Oncol. 2019, 145, 2637–2647. [Google Scholar] [CrossRef]
- Yao, Y.; Xu, X.; Yang, L.; Zhu, J.; Wan, J.; Shen, L.; Xia, F.; Fu, G.; Deng, Y.; Pan, M.; et al. Patient-Derived Organoids Predict Chemoradiation Responses of Locally Advanced Rectal Cancer. Cell Stem Cell 2020, 26, 17–26.e6. [Google Scholar] [CrossRef]
- Frappart, P.; Walter, K.; Gout, J.; Beutel, A.K.; Morawe, M.; Arnold, F.; Breunig, M.; Barth, T.F.; Marienfeld, R.; Schulte, L.; et al. Pancreatic cancer-derived organoids—A disease modeling tool to predict drug response. United Eur. Gastroenterol. J. 2020, 8, 594–606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guillen, K.; Fujita, M.; Butterfield, A.; Scherer, S. A Breast Cancer Patient-Derived Xenograft and Organoid Platform for Drug Discovery and Precision Oncology. bioRxiv 2021. [Google Scholar] [CrossRef]
- De Witte, C.J.; Valle-Inclan, J.E.; Hami, N.; Lõhmussaar, K.; Kopper, O.; Vreuls, C.P.H.; Jonges, G.N.; van Diest, P.; Nguyen, L.; Clevers, H.; et al. Patient-Derived Ovarian Cancer Organoids Mimic Clinical Response and Exhibit Heterogeneous Inter- and Intrapatient Drug Responses. Cell Rep. 2020, 31, 107762. [Google Scholar] [CrossRef]
- Votanopoulos, K.I.; Forsythe, S.; Msc, H.S.; Mazzocchi, A.; Aleman, J.; Miller, L.; Levine, E.; Triozzi, P.; Skardal, A. Model of Patient-Specific Immune-Enhanced Organoids for Immunotherapy Screening: Feasibility Study. Ann. Surg. Oncol. 2020, 27, 1956–1967. [Google Scholar] [CrossRef]
- Kopper, O.; de Witte, C.J.; Lõhmussaar, K.; Valle-Inclan, J.E.; Hami, N.; Kester, L.; Balgobind, A.V.; Korving, J.; Proost, N.; Begthel, H.; et al. An organoid platform for ovarian cancer captures intra- and interpatient heterogeneity. Nat. Med. 2019, 25, 838–849. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Demirci, U.; Chen, P. Emerging organoid models: Leaping forward in cancer research. J. Hematol. Oncol. 2019, 12, 1–10. [Google Scholar] [CrossRef] [Green Version]
- ClinicalTrials.gov. U.S. National Library of Medicine. Available online: https://Clinicaltrials.Gov (accessed on 1 April 2021).
- Verissimo, C.S.; Overmeer, R.M.; Ponsioen, B.; Drost, J.; Mertens, S.; Verlaan-Klink, I.; Van Gerwen, B.; Van Der Ven, M.; Van De Wetering, M.; Egan, D.A.; et al. Targeting mutant RAS in patient-derived colorectal cancer organoids by combinatorial drug screening. eLife 2016, 5, e18489. [Google Scholar] [CrossRef] [Green Version]
- Ponz-Sarvise, M.; Corbo, V.; Tiriac, H.; Engle, D.; Frese, K.K.; Oni, T.E.; Hwang, C.-I.; Öhlund, D.; Chio, I.I.C.; Baker, L.; et al. Identification of Resistance Pathways Specific to Malignancy Using Organoid Models of Pancreatic Cancer. Clin. Cancer Res. 2019, 25, 6742–6755. [Google Scholar] [CrossRef] [Green Version]
- Pauli, C.; Hopkins, B.D.; Prandi, D.; Shaw, R.; Fedrizzi, T.; Sboner, A.; Sailer, V.; Augello, M.; Puca, L.; Rosati, R.; et al. Personalized In Vitro and In Vivo Cancer Models to Guide Precision Medicine. Cancer Discov. 2017, 7, 462–477. [Google Scholar] [CrossRef] [Green Version]
- Fiore, D.; Ramesh, P.; Proto, M.C.; Piscopo, C.; Franceschelli, S.; Anzelmo, S.; Medema, J.P.; Bifulco, M.; Gazzerro, P. Rimonabant Kills Colon Cancer Stem Cells without Inducing Toxicity in Normal Colon Organoids. Front. Pharmacol. 2018, 8, 949. [Google Scholar] [CrossRef] [Green Version]
- Lu, W.; Rettenmeier, E.; Paszek, M.; Yueh, M.-F.; Tukey, R.H.; Trottier, J.; Barbier, O.; Chen, S. Crypt Organoid Culture as an in Vitro Model in Drug Metabolism and Cytotoxicity Studies. Drug Metab. Dispos. 2017, 45, 748–754. [Google Scholar] [CrossRef] [PubMed]
- Pamies, D.; Block, K.; Lau, P.; Gribaldo, L.; Pardo, C.A.; Barreras, P.; Smirnova, L.; Wiersma, D.; Zhao, L.; Harris, G.; et al. Rotenone exerts developmental neurotoxicity in a human brain spheroid model. Toxicol. Appl. Pharmacol. 2018, 354, 101–114. [Google Scholar] [CrossRef] [PubMed]
- Chhibber, T.; Bagchi, S.; Lahooti, B.; Verma, A.; Al-Ahmad, A.; Paul, M.K.; Pendyala, G.; Jayant, R.D. CNS organoids: An innovative tool for neurological disease modeling and drug neurotoxicity screening. Drug Discov. Today 2019, 25, 456–465. [Google Scholar] [CrossRef]
- Schielke, C.; Hartel, C.; Durante, M.; Ritter, S.; Schroeder, I.S. Solving the Issue of Ionizing Radiation Induced Neurotoxicity by Using Novel Cell Models and State of the Art Accelerator Facilities. Front. Phys. 2020, 8, 417. [Google Scholar] [CrossRef]
- Liu, F.; Huang, J.; Liu, Z. Vincristine Impairs Microtubules and Causes Neurotoxicity in Cerebral Organoids. Neuroscience 2019, 404, 530–540. [Google Scholar] [CrossRef] [PubMed]
- Leite, S.B.; Roosens, T.; El Taghdouini, A.; Mannaerts, I.; Smout, A.J.; Najimi, M.; Sokal, E.; Noor, F.; Chesne, C.; van Grunsven, L.A. Novel human hepatic organoid model enables testing of drug-induced liver fibrosis in vitro. Biomaterials 2016, 78, 1–10. [Google Scholar] [CrossRef]
- Mun, S.J.; Ryu, J.-S.; Lee, M.-O.; Son, Y.S.; Oh, S.J.; Cho, H.-S.; Son, M.-Y.; Kim, D.-S.; Kim, S.J.; Yoo, H.J.; et al. Generation of expandable human pluripotent stem cell-derived hepatocyte-like liver organoids. J. Hepatol. 2019, 71, 970–985. [Google Scholar] [CrossRef] [PubMed]
- Driehuis, E.; Oosterom, N.; Heil, S.G.; Muller, I.B.; Lin, M.; Kolders, S.; Jansen, G.; De Jonge, R.; Pieters, R.; Clevers, H.; et al. Patient-derived oral mucosa organoids as an in vitro model for methotrexate induced toxicity in pediatric acute lymphoblastic leukemia. PLoS ONE 2020, 15, e0231588. [Google Scholar] [CrossRef]
- Clevers, H.; Tuveson, D.A. Organoid Models for Cancer Research. Annu. Rev. Cancer Biol. 2019, 3, 223–234. [Google Scholar] [CrossRef]
- Yuki, K.; Cheng, N.; Nakano, M.; Kuo, C.J. Organoid Models of Tumor Immunology. Trends Immunol. 2020, 41, 652–664. [Google Scholar] [CrossRef]
- Cattaneo, C.M.; Dijkstra, K.K.; Fanchi, L.F.; Kelderman, S.; Kaing, S.; van Rooij, N.; Brink, S.V.D.; Schumacher, T.; Voest, E.E. Tumor organoid–T-cell coculture systems. Nat. Protoc. 2020, 15, 15–39. [Google Scholar] [CrossRef]
- Dijkstra, K.; Cattaneo, C.M.; Weeber, F.; Chalabi, M.; Van De Haar, J.; Fanchi, L.F.; Slagter, M.; Van Der Velden, D.L.; Kaing, S.; Kelderman, S.; et al. Generation of Tumor-Reactive T Cells by Co-culture of Peripheral Blood Lymphocytes and Tumor Organoids. Cell 2018, 174, 1586–1598.e12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, S.; McOlash, L.; Palen, K.; Johnson, B.; Duris, C.; Yang, Q.; Dwinell, M.B.; Hunt, B.; Evans, D.B.; Gershan, J.; et al. Development of primary human pancreatic cancer organoids, matched stromal and immune cells and 3D tumor microenvironment models. BMC Cancer 2018, 18, 335. [Google Scholar] [CrossRef]
- Chakrabarti, J.; Holokai, L.; Syu, L.; Steele, N.G.; Chang, J.; Wang, J.; Ahmed, S.; Dlugosz, A.; Zavros, Y. Hedgehog signaling induces PD-L1 expression and tumor cell proliferation in gastric cancer. Oncotarget 2018, 9, 37439–37457. [Google Scholar] [CrossRef] [Green Version]
- Finnberg, N.K.; Gokare, P.; Lev, A.; Grivennikov, S.I.; MacFarlane, A.W., IV; Campbell, K.S.; Winters, R.M.; Kaputa, K.; Far-ma, J.M.; Abbas, A.E.; et al. Application of 3D tumoroid systems to define immune and cytotoxic therapeutic responses based on tumoroid and tissue slice culture molecular signatures. Oncotarget 2017, 8, 66747–66757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grebenyuk, S.; Ranga, A. Engineering Organoid Vascularization. Front. Bioeng. Biotechnol. 2019, 7, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Wan, Z.; Kamm, R.D. Vascularized organoids on a chip: Strategies for engineering organoids with functional vasculature. Lab Chip 2021, 21, 473–488. [Google Scholar] [CrossRef] [PubMed]
- Hofer, M.; Lutolf, M.P. Engineering organoids. Nat. Rev. Mater. 2021, 6, 402–420. [Google Scholar] [CrossRef]
| Tumor type | Sample Size | Type of Specimen | Additional Information | Reference |
|---|---|---|---|---|
| Colorectal Cancer | 22 tumor organoid lines from 20 different colon cancer patients and matched organoids from adjacent untransformed tissue of 19 patients | surgially resected tissue only primary lesions | first organoid biobank described only untreated patients | [64] |
| 49 organoids from primary lesions (15 premalignant lesions (tubular/tubulovillous/serrated), 32 adenocarcinoma, 2 neuroendocrine carcinoma) and 6 organoid lines from metastatic lesions of adenocarcinomam; additionally 41 counterpart organoids from normal colorectal mucosa | endoscopic biopsy specimen or surgically resected sample primary as well as metastatic lesions | including rare histological subtypes such as poorly differentiated or mucinous adenocarcinoma or neuroendocrine neoplasms | [55] | |
| Gastric Cancer | 46 organoid culture lines from tumor or dysplastic lesions of 34 patients and 17 organoid lines from adjacent untransformed mucosa | surgically rescted tissue primary lesion and lymph node metastases | predominantly untreated patients (three patients with neoadjuvant chemotherapy) | [70] |
| Liver Cancer | 10 HCC-derived organoid lines of 8 patients and corresponding normal liver organoids from all of the patients | needle biopsy specimen primary lesions (for 5 patients 2 different nodules were biopsied) | HCC tumors of different etologies (viral hepatitis, NAFLD, ALD) successful establishment in only 26% of patient samples | [72] |
| tumor organoids of 3 patients with IHCC and 1 patient with GBC (additionally 1 organoid line from PDAC and 1 organoid line from neuroendocrine tumor of ampulla vateri) | surgically resected tissue | sucessful longterm culture of organoids in 6 out of 18 cases | [73] | |
| Pancreatic cancer | 52 (31 analyzed) organoid lines from different subtypes of pancreatic cancer or distal bile duct carcinoma (63% PDAC, 10% CC, 6.67% ACC, 3.33% adenosquamous PDAC, 10% IPMN derived PDAC, 6.67% papilla vateri AC); matched normal coontrol organoids of tumor-adjacent normal tissue from 5 patients | predominantly surgically resected tissue, only 2 biopsy samples | [74] | |
| 49 PDAC organoid lines and normal pancreatic organoids of adjacent untransformed tissue whenever possible to establish | fine-needle aspiration, ascites specimen or surgically resected tissue only primary lesions (or ascites) | tumor stage of all patinets was III or IV except for 2 patients (IIA and IIB) only 3 patients were pre-treted before sampeling | [49] | |
| 114 organoid cultures from 101 PDAC patients; additionally 11 human normal pancreatic ductal organoids from healthy normal pancreata obtained from islet transplant centers | surgically resected tissue, FNB samples or specimen from rapid autopsies primary as well as metastatic lesions | [75] | ||
| 10 (8 analyzed) organoid cultures from patients diagnosed with IPMN and 7 additional normal pancreatic duct organoids | surgically resected specimen | [76] | ||
| 15 organoid lines from patients with IPMN and normal pancreatic organoids of matched adjacent normal mucosa | surgically resected specimen | comprising 3 low-grade IPMNs, 2 moderate-grade IPMNs, 7 high-grade IPMNs, and 3 IPMNs associated with invasive carcinoma | [77] | |
| Neuroendocrine Tumors | 25 organoid lines from pastients with gastroenteropancreatic neuroendocrine neoplasms (NEN) | surgically resected tissue and biopsy samples | comprising NEN from esophagus, stomach, duodenum, colon, liver, pancreas and biliary tract | [78] |
| Lung Cancer | organoid lines from 10 patients with NSCLC | mainly surgically resected tissue | [79] | |
| 80 organoid lines from patients with different subtypes of lung cancer (66.25% adenocarcinoma, 6.25% small cell lung cancer, 3.75% large cell carcinoma, 3.75% adenosquamous carcinoma, 20% squamous cell carcinoma) and 5 organoid lines from normal bronchial tissue | surgically resected tissue | small number of banked organoid lines from pulmonal metastatic lesions of colonic adenocarcinoma | [80] | |
| 12 orgnoid lines from 15 patients with lung adenocarcinomas of different subtypes | surgically resected tissue | [81] | ||
| organoid lines from 103 surgically resected specimens (71 ACS, 23 SCCs, 4 SCLCs and 5 other lung cancer types); among these 103 specimens 42 pairs of tumor and corresponding normal tissues processed in paralle | surgically resected tissue 3 samples gained by endobronchial ultrasound-guided transbronchial needle aspiration | samples from all tumor stages | [82] | |
| Glioblastoma | 70 organoid lines from patients with glioblastoma | surgically resected tissue | [83] | |
| Bladder Cancer | tumor organoid lines from 53 patients with bladder cancer (basal and luminal bladder cancer subtypes); whenever possible corresponding normal organoid lines from untransformed mucosa | surgically resected tissue (cystectomy or transurethral resection (TUR)) | [71] | |
| 22 PDO lines from 16 patients ranging from low-grade non-muscle invasive disease to muscle-invasive cancer | surgically resected tissue (TUR) | 1 patient was systemically treted before sampeling, 6 organoid lines established from patients with a prior intravesical treatment before sampeling | [84] | |
| Kidney cancer | 54 organoid lines from different subtypes of pediatric kidney cancer (40 Wilms tumors, 7 MRTKs, 3 RCCs, 2 CMNs, 1 metanephric adenoma and 1 nephrogenic rest); whenever possible organoid line from corresponding healthy tissue | surgically resected tissue (nephrectomy) or biopsy sample mainly primary lesion, but als metastatic lesions | first pediatric organoid biobank chemo-naïve as well as chemo-treated specimens | [85] |
| Breast Cancer | >100 breast cancer organoid lines representing distribution of ductal, lobular, adeno- and carcinoma in situ as well as all types of receptor combination (ER, PR, HER2) | surgically resected tissue | chemo-naïve as well as chemo-treated specimens | [63] |
| 33 organoids from 33 patients with breast cancer; 84.84% invasive ductal carcinoma and 15.15% invasive lobular carcinoma | surgically resected tissue and core biopsy specimens | [86] | ||
| 64 organoid lines from patients with triple negative breast cancer | [87] | |||
| 17 tumor organoid lines from 32 patients with invasive ductal or lobular carcinoma as well as organoid lines from tumor-adjacent normal tissue | surgically resected tissue | only of treatment-naïve patients | [88] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kastner, C.; Hendricks, A.; Deinlein, H.; Hankir, M.; Germer, C.-T.; Schmidt, S.; Wiegering, A. Organoid Models for Cancer Research—From Bed to Bench Side and Back. Cancers 2021, 13, 4812. https://doi.org/10.3390/cancers13194812
Kastner C, Hendricks A, Deinlein H, Hankir M, Germer C-T, Schmidt S, Wiegering A. Organoid Models for Cancer Research—From Bed to Bench Side and Back. Cancers. 2021; 13(19):4812. https://doi.org/10.3390/cancers13194812
Chicago/Turabian StyleKastner, Carolin, Anne Hendricks, Hanna Deinlein, Mohammed Hankir, Christoph-Thomas Germer, Stefanie Schmidt, and Armin Wiegering. 2021. "Organoid Models for Cancer Research—From Bed to Bench Side and Back" Cancers 13, no. 19: 4812. https://doi.org/10.3390/cancers13194812
APA StyleKastner, C., Hendricks, A., Deinlein, H., Hankir, M., Germer, C.-T., Schmidt, S., & Wiegering, A. (2021). Organoid Models for Cancer Research—From Bed to Bench Side and Back. Cancers, 13(19), 4812. https://doi.org/10.3390/cancers13194812







