UBE2M Drives Hepatocellular Cancer Progression as a p53 Negative Regulator by Binding to MDM2 and Ribosomal Protein L11
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Tissue Microarray and Immunohistochemistry
2.3. RNA Interference and Plasmid Transfection
2.4. Next Generation Sequence (NGS) Analysis
2.5. Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
2.6. Western Blotting
2.7. Fractionation of Nuclear and Cytoplasmic Extract
2.8. Cycloheximide Assay
2.9. Immunoprecipitation
2.10. Ubiquitylation Assay
2.11. Immunofluorescence
2.12. Establishment of UBE2M shRNA HCC Cell Lines
2.13. In Vivo Xenograft Model
2.14. Statistical Analysis
3. Results
3.1. UBE2M Is Overexpressed in HCCs with Poor Prognosis: Its Depletion Exerts Antiproliferative and Apoptotic Effect in HCCs
3.2. p53/MDM2 and RPL Related Genes Were More Associated in UBE2M-Depleted HCCs
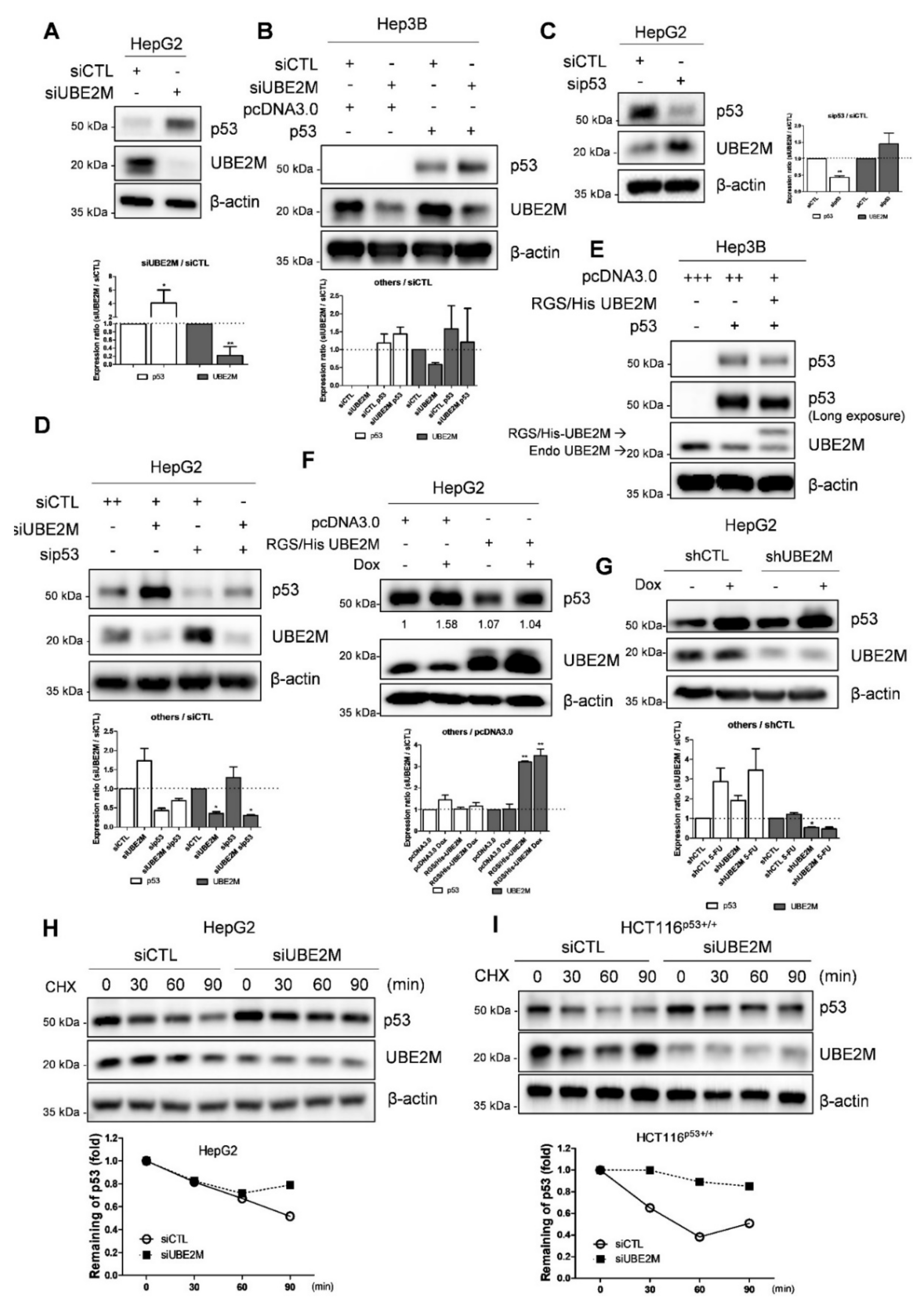
3.3. UBE2M Depletion Activates p53 and Maintains Its Stability in HCCs
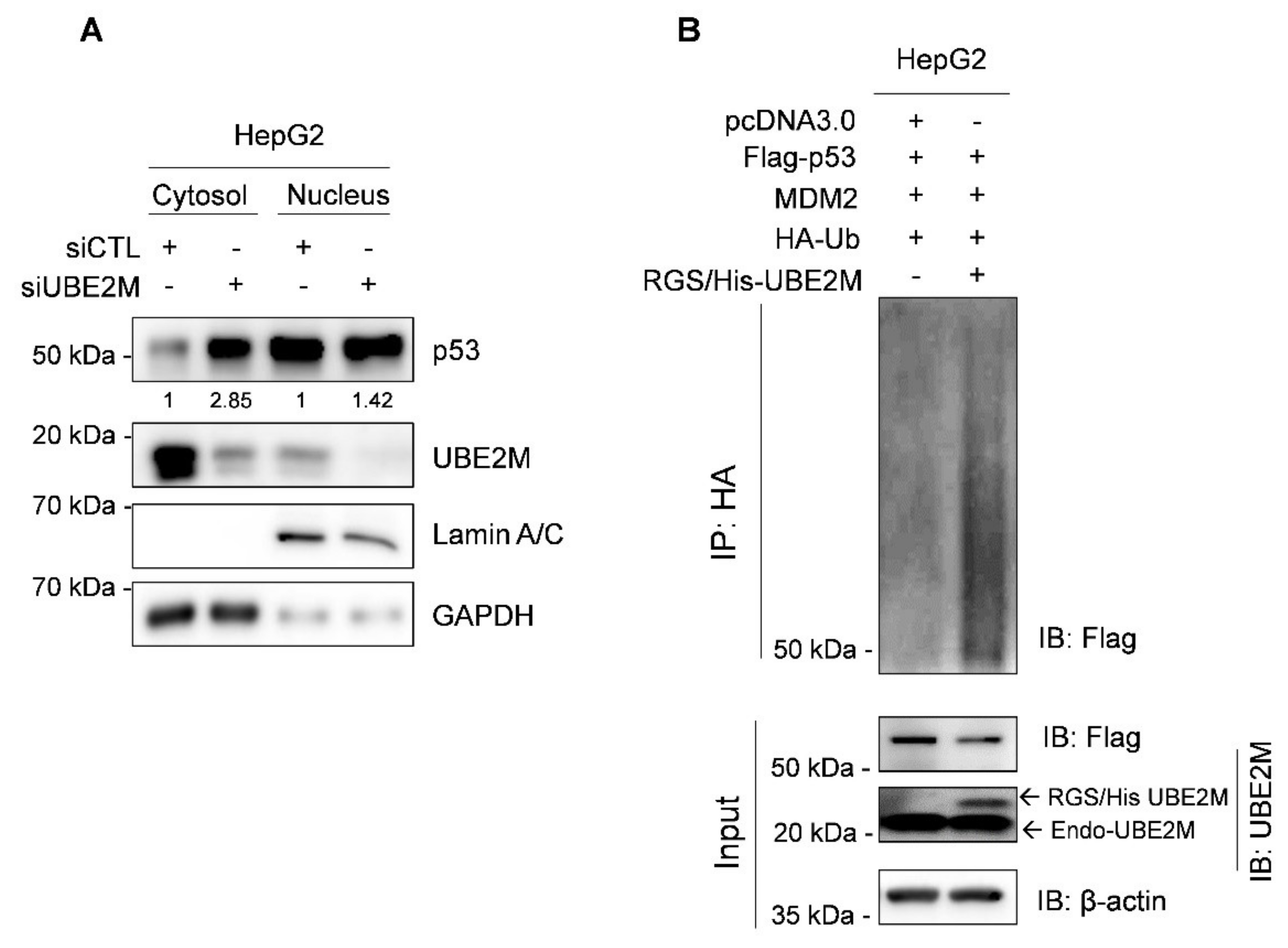
3.4. Ectopic Expression of UBE2M Enhances Degradation of Exogenous P53 Mediated by MDM2 in HepG2 Cells
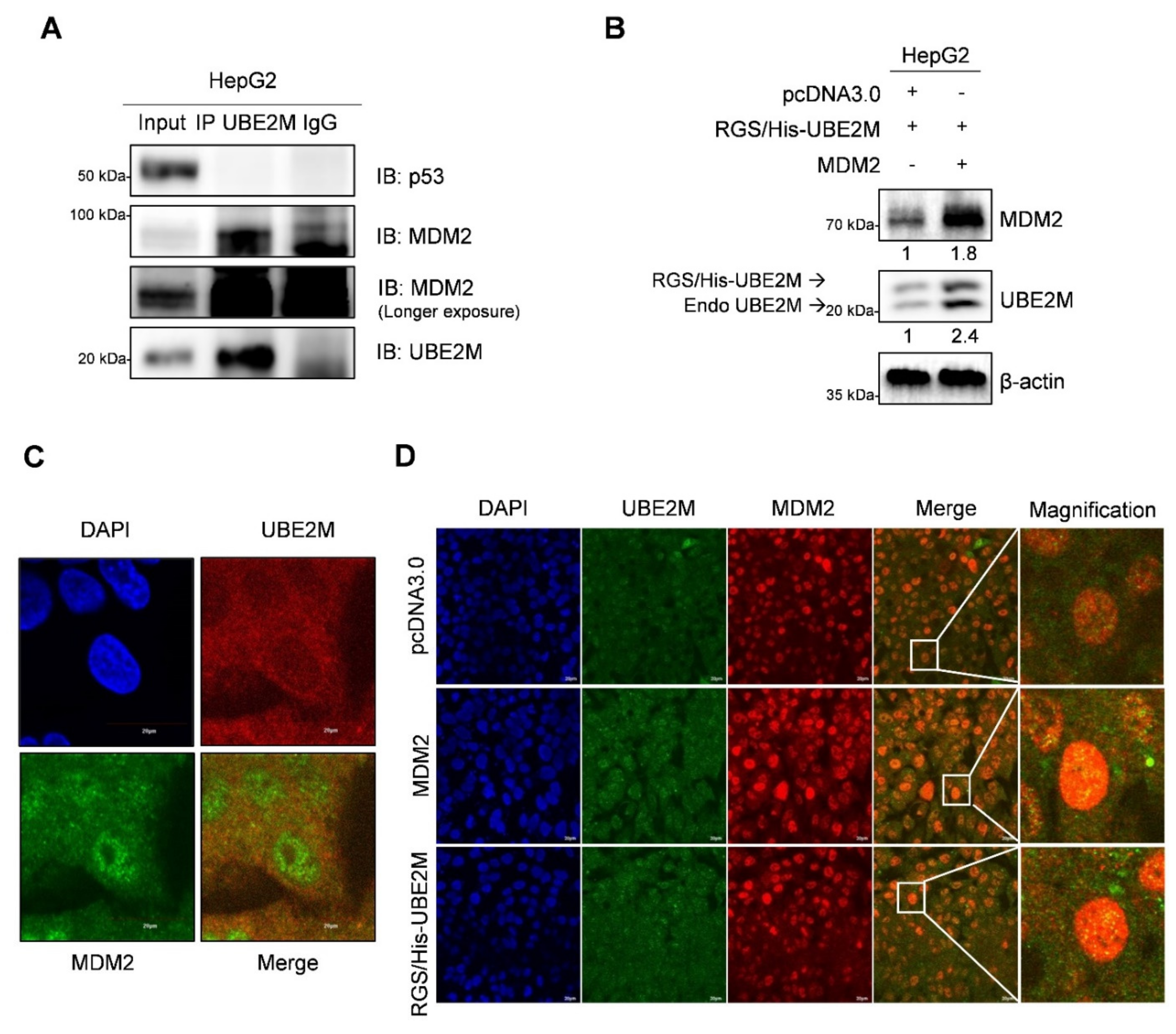
3.5. UBE2M Binds to MDM2, but Regulates p53 via Their Crosstalk in HepG2 Cells
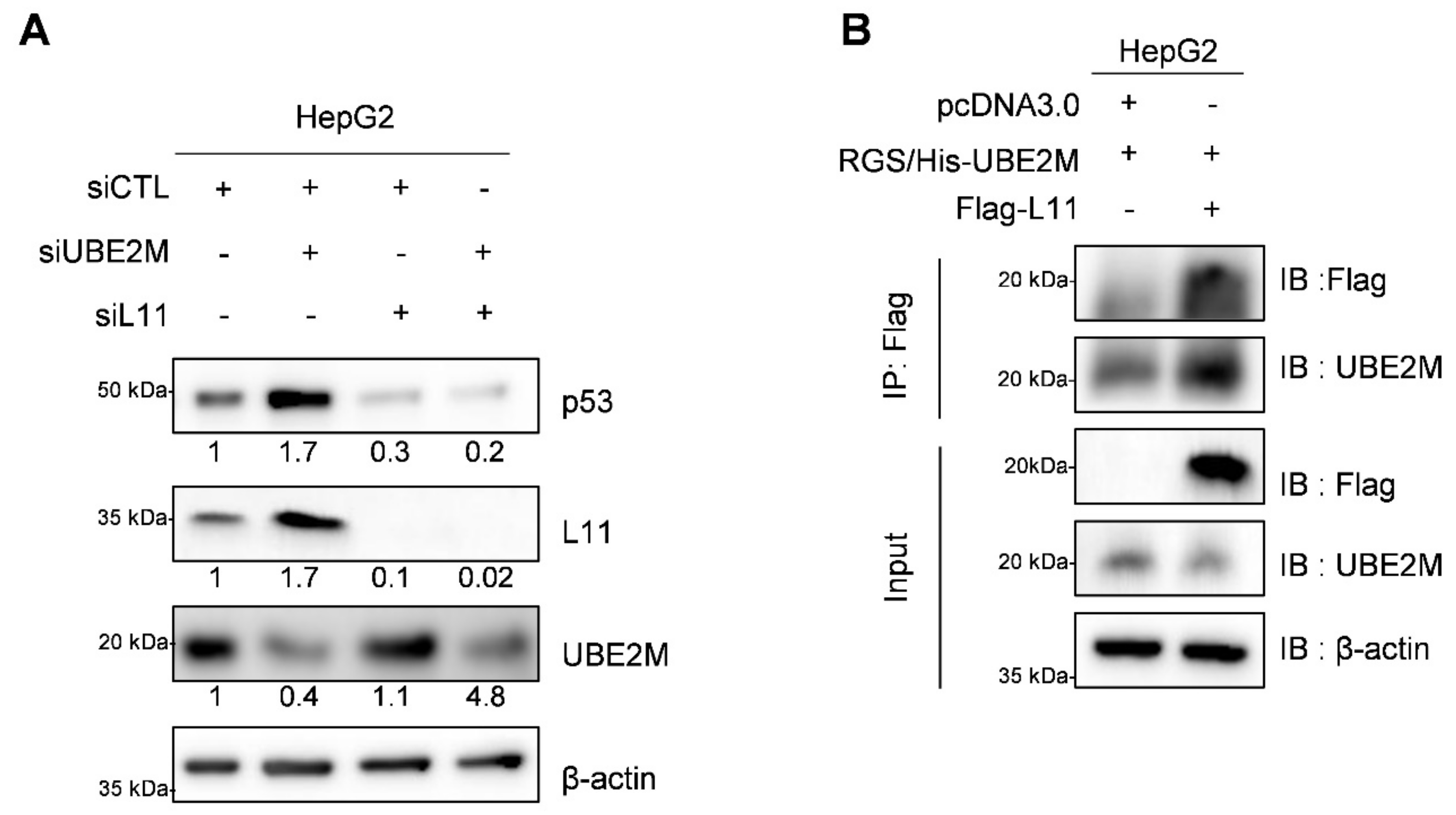
3.6. UBE2M Depletion Activates p53 and Ribosomal Protein L11 in HepG2 Cells
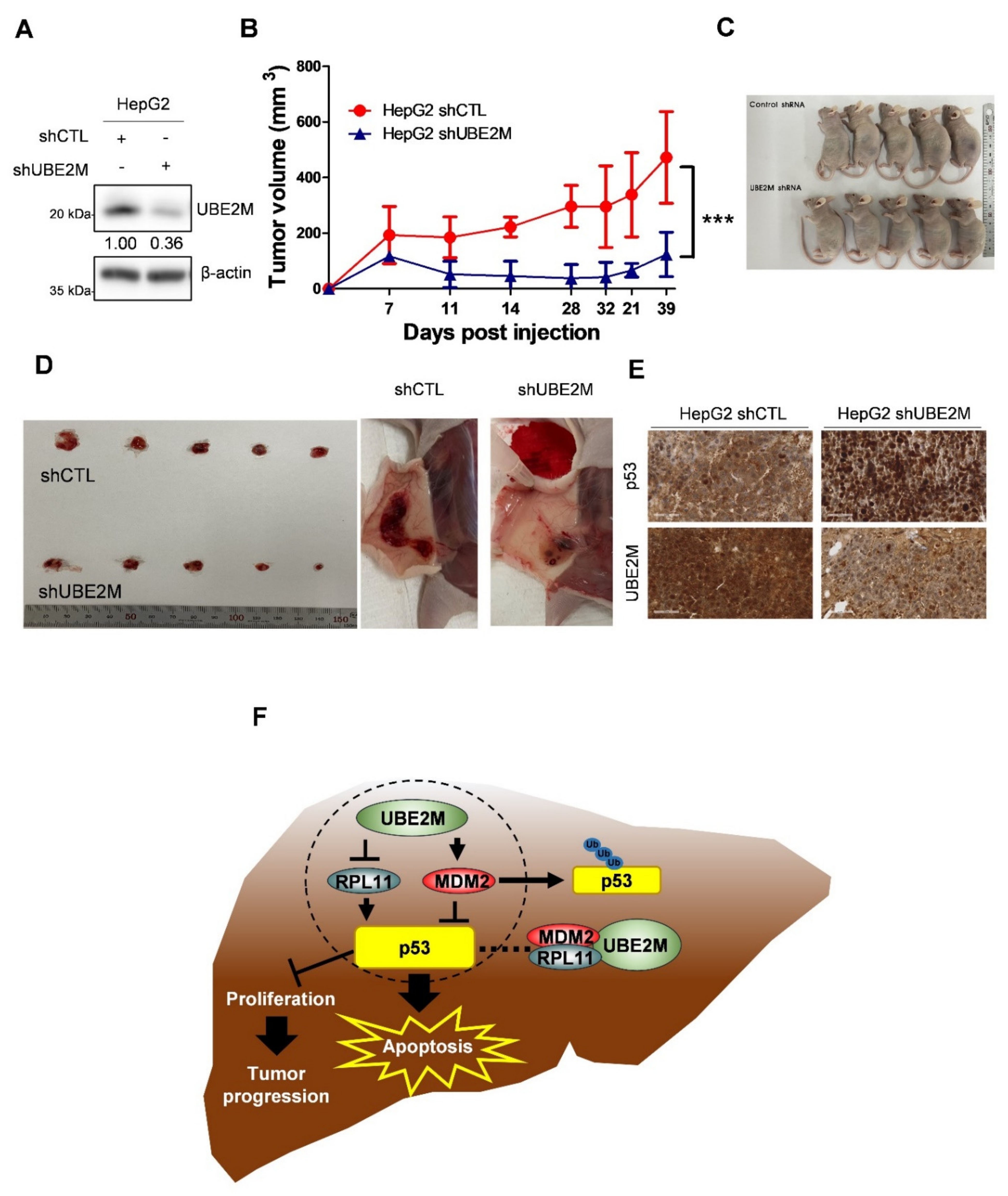
3.7. UBE2M Depletion Retards the Growth of HepG2 Cells Implanted in Balb/c Male Athymic Nude Mouse along with Elevated p53 and Decreased UBE2M Expression by IHC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mattiuzzi, C.; Lippi, G. Cancer statistics: A comparison between World Health Organization (WHO) and Global Burden of Disease (GBD). Eur. J. Public Health 2020, 30, 1026–1027. [Google Scholar] [CrossRef] [PubMed]
- Delgado, T.C.; Barbier-Torres, L.; Zubiete-Franco, I.; Lopitz-Otsoa, F.; Varela-Rey, M.; Fernandez-Ramos, D.; Martinez-Chantar, M.L. Neddylation, a novel paradigm in liver cancer. Transl. Gastroenterol. Hepatol. 2018, 3, 37. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Hu, T.; Liang, Y.; Jiang, Y.; Pan, Y.; Li, C.; Zhang, P.; Wei, D.; Li, P.; Jeong, L.S.; et al. Synergistic inhibition of autophagy and neddylation pathways as a novel therapeutic approach for targeting liver cancer. Oncotarget 2015, 6, 9002–9017. [Google Scholar] [CrossRef] [Green Version]
- Fernandez-Ramos, D.; Martinez-Chantar, M.L. NEDDylation in liver cancer: The regulation of the RNA binding protein Hu antigen R. Pancreatology 2015, 15, S49–S54. [Google Scholar] [CrossRef]
- Zhou, L.; Jiang, Y.; Luo, Q.; Li, L.; Jia, L. Neddylation: A novel modulator of the tumor microenvironment. Mol. Cancer. 2019, 18, 77. [Google Scholar] [CrossRef] [Green Version]
- Ying, J.; Zhang, M.; Qiu, X.; Lu, Y. Targeting the neddylation pathway in cells as a potential therapeutic approach for diseases. Cancer Chemother. Pharmacol. 2018, 81, 797–808. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Zhang, W.; Sun, Y.; Jia, L. Protein neddylation and its alterations in human cancers for targeted therapy. Cell Signal. 2018, 44, 92–102. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Sun, Y. Cullin-RING Ligases as attractive anti-cancer targets. Curr. Pharm. Des. 2013, 19, 3215–3225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Hamid, O.; Carvajal, R.D. The Need for Neddylation: A Key to Achieving NED in Uveal Melanoma. Clin. Cancer Res. 2018, 24, 3477–3479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, L.; Jiang, Y.; Liu, X.; Li, L.; Yang, X.; Dong, C.; Lin, Y.; Li, Y.; Yu, J.; He, R.; et al. Promotion of tumor-associated macrophages infiltration by elevated neddylation pathway via NF-kappaB-CCL2 signaling in lung cancer. Oncogene 2019, 38, 5792–5804. [Google Scholar] [CrossRef]
- Chen, Y.J.; Wu, H.; Shen, X.Z. The ubiquitin-proteasome system and its potential application in hepatocellular carcinoma therapy. Cancer Lett. 2016, 379, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Ohh, M.; Kim, W.Y.; Moslehi, J.J.; Chen, Y.; Chau, V.; Read, M.A.; Kaelin, W.G., Jr. An intact NEDD8 pathway is required for Cullin-dependent ubiquitylation in mammalian cells. EMBO Rep. 2002, 3, 177–182. [Google Scholar] [CrossRef] [Green Version]
- Rabut, G.; Peter, M. Function and regulation of protein neddylation. “Protein modifications: Beyond the usual suspects” review series. EMBO Rep. 2008, 9, 969–976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Morgan, M.A.; Sun, Y. Targeting Neddylation pathways to inactivate cullin-RING ligases for anticancer therapy. Antioxid. Redox Signal. 2014, 21, 2383–2400. [Google Scholar] [CrossRef] [Green Version]
- Liakopoulos, D.; Doenges, G.; Matuschewski, K.; Jentsch, S. A novel protein modification pathway related to the ubiquitin system. EMBO J. 1998, 17, 2208–2214. [Google Scholar] [CrossRef] [Green Version]
- Gong, L.; Yeh, E.T. Identification of the activating and conjugating enzymes of the NEDD8 conjugation pathway. J. Biol. Chem. 1999, 274, 12036–12042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, W.; Xu, J.; Tan, M.; Li, H.; Wei, W.; Sun, Y. UBE2M is a Stress-Inducible Dual E2 for Neddylation and Ubiquitylation that Promotes Targeted Degradation of UBE2F. Mol. Cell 2018, 70, 1008–1024.e6. [Google Scholar] [CrossRef] [Green Version]
- Huang, A.M.; Kao, Y.T.; Toh, S.; Lin, P.Y.; Chou, C.H.; Hu, H.T.; Lu, C.Y.; Liou, J.Y.; Chao, S.Y.; Hour, T.C.; et al. UBE2M-mediated p27(Kip1) degradation in gemcitabine cytotoxicity. Biochem. Pharmacol. 2011, 82, 35–42. [Google Scholar] [CrossRef]
- Sun, X.X.; Dai, M.S.; Lu, H. 5-fluorouracil activation of p53 involves an MDM2-ribosomal protein interaction. J. Biol. Chem. 2007, 282, 8052–8059. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.C.; Yu, X.N.; Sun, J.L.; Xiong, J.; Yang, Y.J.; Jiang, X.M.; Zhu, J.M. UBE2M promotes cell proliferation via the β-catenin/cyclin D1 signaling in hepatocellular carcinoma. Aging 2020, 12, 2373–2392. [Google Scholar] [CrossRef]
- Yu, J.; Huang, W.L.; Xu, Q.G.; Zhang, L.; Sun, S.H.; Zhou, W.P.; Yang, F. Overactivated neddylation pathway in human hepatocellular carcinoma. Cancer Med. 2018, 7, 3363–3372. [Google Scholar] [CrossRef]
- Li, L.; Kang, J.; Zhang, W.; Cai, L.; Wang, S.; Liang, Y.; Jiang, Y.; Liu, X.; Zhang, Y.; Ruan, H.; et al. Validation of NEDD8-conjugating enzyme UBC12 as a new therapeutic target in lung cancer. EBioMedicine 2019, 45, 81–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Shi, C.C.; Zhang, H.P.; Li, G.Q.; Li, S.S. MLN4924 suppresses neddylation and induces cell cycle arrest, senescence, and apoptosis in human osteosarcoma. Oncotarget 2016, 7, 45263–45274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyashita, T.; Krajewski, S.; Krajewska, M.; Wang, H.G.; Lin, H.K.; Liebermann, D.A.; Hoffman, B.; Reed, J.C. Tumor suppressor p53 is a regulator of bcl-2 and bax gene expression in vitro and in vivo. Oncogene 1994, 9, 1799–1805. [Google Scholar]
- Duffy, M.J.; Synnott, N.C.; McGowan, P.M.; Crown, J.; O’Connor, D.; Gallagher, W.M. p53 as a target for the treatment of cancer. Cancer Treat. Rev. 2014, 40, 1153–1160. [Google Scholar] [CrossRef]
- Jung, J.H.; Lee, H.; Zeng, S.X.; Lu, H. RBM10, a New Regulator of p53. Cells 2020, 9, 2107. [Google Scholar] [CrossRef]
- Harris, C.C. Structure and function of the p53 tumor suppressor gene: Clues for rational cancer therapeutic strategies. J. Natl. Cancer Inst. 1996, 88, 1442–1455. [Google Scholar] [CrossRef] [Green Version]
- Lacroix, M.; Riscal, R.; Arena, G.; Linares, L.K.; Le Cam, L. Metabolic functions of the tumor suppressor p53: Implications in normal physiology, metabolic disorders, and cancer. Mol. Metab. 2019, 33, 2–22. [Google Scholar] [CrossRef]
- Kussie, P.H.; Gorina, S.; Marechal, V.; Elenbaas, B.; Moreau, J.; Levine, A.J.; Pavletich, N.P. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science 1996, 274, 948–953. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Franklin, D.A.; Dong, J.; Zhang, Y. MDM2-p53 pathway in hepatocellular carcinoma. Cancer Res. 2014, 74, 7161–7167. [Google Scholar] [CrossRef] [Green Version]
- Barak, Y.; Gottlieb, E.; Juven-Gershon, T.; Oren, M. Regulation of mdm2 expression by p53: Alternative promoters produce transcripts with nonidentical translation potential. Genes Dev. 1994, 8, 1739–1749. [Google Scholar] [CrossRef] [Green Version]
- Deisenroth, C.; Franklin, D.A.; Zhang, Y. The Evolution of the Ribosomal Protein-MDM2-p53 Pathway. Cold Spring Harb. Perspect. Med. 2016, 6, a026138. [Google Scholar] [CrossRef] [Green Version]
- Jung, J.H.; Lee, H.; Kim, J.H.; Sim, D.Y.; Ahn, H.; Kim, B.; Chang, S.; Kim, S.H. p53-Dependent Apoptotic Effect of Puromycin via Binding of Ribosomal Protein L5 and L11 to MDM2 and its Combination Effect with RITA or Doxorubicin. Cancers 2019, 11, 582. [Google Scholar] [CrossRef] [Green Version]
- Macias, E.; Jin, A.; Deisenroth, C.; Bhat, K.; Mao, H.; Lindstrom, M.S.; Zhang, Y. An ARF-independent c-MYC-activated tumor suppression pathway mediated by ribosomal protein-Mdm2 Interaction. Cancer Cell 2010, 18, 231–243. [Google Scholar] [CrossRef] [Green Version]
- Jung, J.H.; Lee, H.; Cao, B.; Liao, P.; Zeng, S.X.; Lu, H. RNA-binding motif protein 10 induces apoptosis and suppresses proliferation by activating p53. Oncogene 2020, 39, 1031–1040. [Google Scholar] [CrossRef]
- Aubrey, B.J.; Kelly, G.L.; Janic, A.; Herold, M.J.; Strasser, A. How does p53 induce apoptosis and how does this relate to p53-mediated tumour suppression? Cell Death Differ. 2018, 25, 104–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J. The Cell-Cycle Arrest and Apoptotic Functions of p53 in Tumor Initiation and Progression. Cold Spring Harb. Perspect. Med. 2016, 6, a026104. [Google Scholar] [CrossRef]
- Cukras, S.; Morffy, N.; Ohn, T.; Kee, Y. Inactivating UBE2M impacts the DNA damage response and genome integrity involving multiple cullin ligases. PLoS ONE 2014, 9, e101844. [Google Scholar] [CrossRef]
- Nag, S.; Zhang, X.; Srivenugopal, K.S.; Wang, M.H.; Wang, W.; Zhang, R. Targeting MDM2-p53 interaction for cancer therapy: Are we there yet? Curr. Med. Chem. 2014, 21, 553–574. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; He, Y.; Jin, A.; Tikunov, A.P.; Zhou, L.; Tollini, L.A.; Leslie, P.; Kim, T.H.; Li, L.O.; Coleman, R.A.; et al. Ribosomal protein-Mdm2-p53 pathway coordinates nutrient stress with lipid metabolism by regulating MCD and promoting fatty acid oxidation. Proc. Natl. Acad. Sci. USA 2014, 111, E2414–E2422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, X.; Liao, J.M.; Liao, W.J.; Lu, H. Scission of the p53-MDM2 Loop by Ribosomal Proteins. Genes Cancer 2012, 3, 298–310. [Google Scholar] [CrossRef] [PubMed]
- Deisenroth, C.; Zhang, Y. The Ribosomal Protein-Mdm2-p53 Pathway and Energy Metabolism: Bridging the Gap between Feast and Famine. Genes Cancer 2011, 2, 392–403. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.-H.; Jung, J.H.; Lee, H.-J.; Sim, D.-Y.; Im, E.; Park, J.; Park, W.-Y.; Ahn, C.-H.; Shim, B.-S.; Kim, B.; et al. UBE2M Drives Hepatocellular Cancer Progression as a p53 Negative Regulator by Binding to MDM2 and Ribosomal Protein L11. Cancers 2021, 13, 4901. https://doi.org/10.3390/cancers13194901
Kim J-H, Jung JH, Lee H-J, Sim D-Y, Im E, Park J, Park W-Y, Ahn C-H, Shim B-S, Kim B, et al. UBE2M Drives Hepatocellular Cancer Progression as a p53 Negative Regulator by Binding to MDM2 and Ribosomal Protein L11. Cancers. 2021; 13(19):4901. https://doi.org/10.3390/cancers13194901
Chicago/Turabian StyleKim, Ju-Ha, Ji Hoon Jung, Hyo-Jung Lee, Deok-Yong Sim, Eunji Im, Jieon Park, Woon-Yi Park, Chi-Hoon Ahn, Bum-Sang Shim, Bonglee Kim, and et al. 2021. "UBE2M Drives Hepatocellular Cancer Progression as a p53 Negative Regulator by Binding to MDM2 and Ribosomal Protein L11" Cancers 13, no. 19: 4901. https://doi.org/10.3390/cancers13194901
APA StyleKim, J.-H., Jung, J. H., Lee, H.-J., Sim, D.-Y., Im, E., Park, J., Park, W.-Y., Ahn, C.-H., Shim, B.-S., Kim, B., & Kim, S.-H. (2021). UBE2M Drives Hepatocellular Cancer Progression as a p53 Negative Regulator by Binding to MDM2 and Ribosomal Protein L11. Cancers, 13(19), 4901. https://doi.org/10.3390/cancers13194901









