Current Technologies for RNA-Directed Liquid Diagnostics
Abstract
:Simple Summary
Abstract
1. Introduction
2. Sources of Circulating RNA Biomarkers
2.1. CTC and cf-DNA
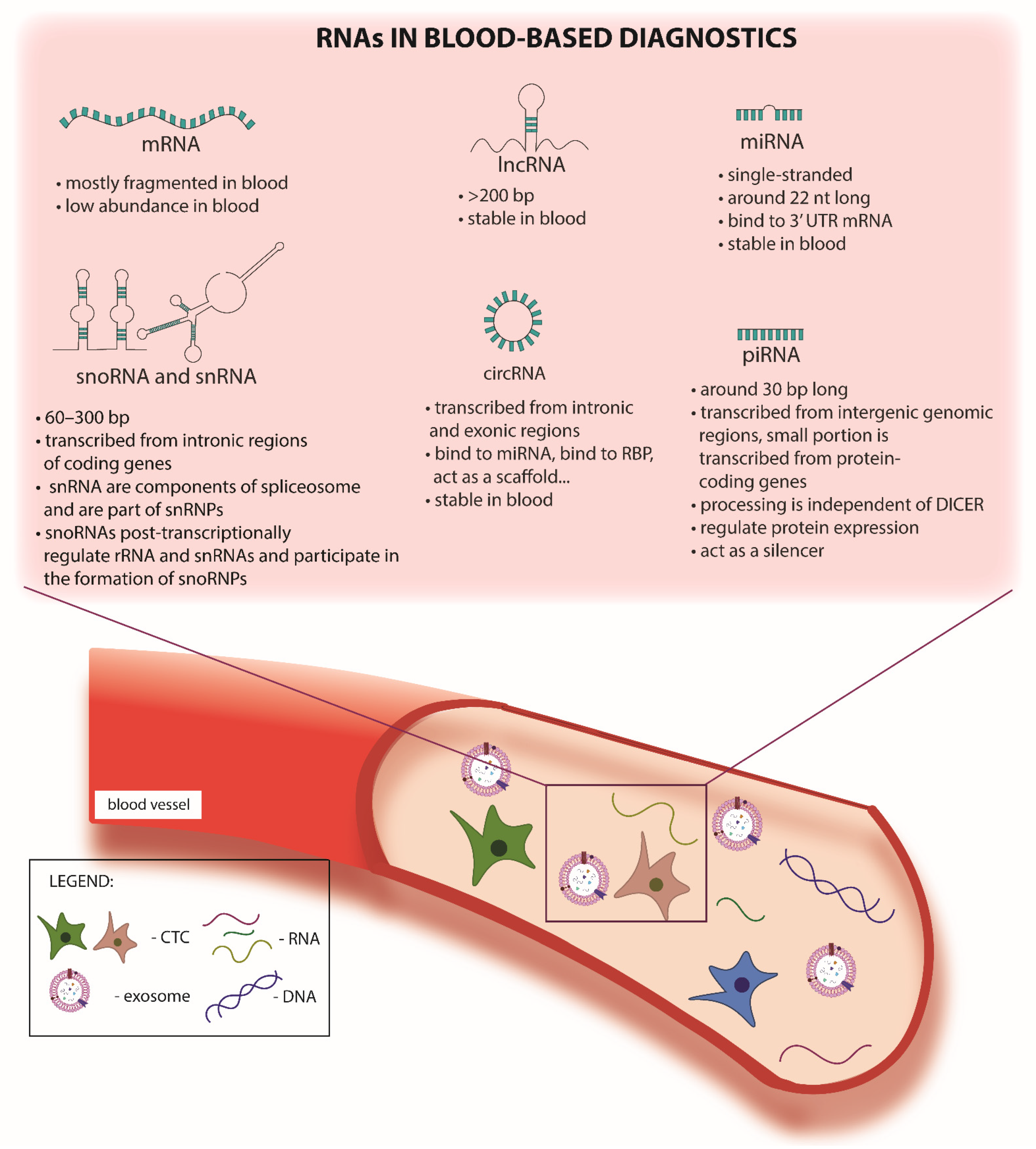
2.2. cf-RNA
2.3. Exosomes
3. Technologies in RNA Isolation and Detection
3.1. Blood Samples
3.1.1. cf-RNA
3.1.2. Exosomes
3.2. Other Body Fluids
3.3. Isolation of cf-RNA
3.3.1. TRIzol
3.3.2. Silica Membranes
3.3.3. Magnetic Beads
3.3.4. Isolation of cf-miRNA
3.4. Exosome Isolation Methods
3.4.1. Ultracentrifugation
3.4.2. Size-Exclusion Chromatography
3.4.3. Immunoaffinity-Based Methods
3.4.4. Precipitation Methods
3.4.5. Isolation of miRNA from Exosomes
3.5. RNA Detection and Quantification
3.5.1. Direct Detection and Quantification of RNA
3.5.2. RNA Sequencing
3.5.3. qPCR
3.5.4. ddPCR
3.5.5. Microarray
4. Clinical Relevance and Economic Aspect of Circulating Biomarkers
4.1. Clinical Relevance of Emerging Liquid Biopsy
4.2. Diagnostic Services for RNA Application
4.3. Market Value of miRNA Diagnosis and Research
4.4. Concerns Regarding the Clinical Aspect of Liquid Biopsy
5. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Reimers, N.; Pantel, K. Liquid biopsy: Novel technologies and clinical applications. Clin. Chem. Lab. Med. 2019, 57, 312–316. [Google Scholar] [CrossRef]
- Poulet, G.; Massias, J.; Taly, V. Liquid Biopsy: General Concepts. Acta Cytol. 2019, 63, 449–455. [Google Scholar] [CrossRef] [PubMed]
- Siravegna, G.; Mussolin, B.; Buscarino, M.; Corti, G.; Cassingena, A.; Crisafulli, G.; Ponzetti, A.; Cremolini, C.; Amatu, A.; Lauricella, C.; et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat. Med. 2015, 21, 795–801. [Google Scholar] [CrossRef] [Green Version]
- Scarlotta, M.; Simsek, C.; Kim, A.K. Liquid Biopsy in Solid Malignancy. Genet. Test Mol. Biomark. 2019, 23, 284–296. [Google Scholar] [CrossRef]
- Suehara, Y.; Sakata-Yanagimoto, M.; Hattori, K.; Kusakabe, M.; Nanmoku, T.; Sato, T.; Noguchi, M.; Chiba, S. Mutations found in cell-free DNAs of patients with malignant lymphoma at remission can derive from clonal hematopoiesis. Cancer Sci. 2019, 110, 3375–3381. [Google Scholar] [CrossRef] [Green Version]
- Bardelli, A.; Pantel, K. Liquid Biopsies, What We Do Not Know (Yet). Cancer Cell 2017, 31, 172–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, M.; Zhao, H. Next-generation sequencing in liquid biopsy: Cancer screening and early detection. Hum. Genom. 2019, 13, 34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Lin, X.; Huang, Y.; Wang, M.; Cen, C.; Tang, S.; Dique, M.R.; Cai, L.; Luis, M.A.; Smollar, J.; et al. Detection Methods and Clinical Applications of Circulating Tumor Cells in Breast Cancer. Front. Oncol. 2021, 11, 652253. [Google Scholar] [CrossRef]
- Russano, M.; Napolitano, A.; Ribelli, G.; Iuliani, M.; Simonetti, S.; Citarella, F.; Pantano, F.; Dell’Aquila, E.; Anesi, C.; Silvestris, N.; et al. Liquid biopsy and tumor heterogeneity in metastatic solid tumors: The potentiality of blood samples. J. Exp. Clin. Cancer Res. 2020, 39, 95. [Google Scholar] [CrossRef]
- Thiele, J.A.; Bethel, K.; Kralickova, M.; Kuhn, P. Circulating Tumor Cells: Fluid Surrogates of Solid Tumors. Annu. Rev. Pathol. 2017, 12, 419–447. [Google Scholar] [CrossRef]
- Meng, S.; Tripathy, D.; Frenkel, E.P.; Shete, S.; Naftalis, E.Z.; Huth, J.F.; Beitsch, P.D.; Leitch, M.; Hoover, S.; Euhus, D.; et al. Circulating tumor cells in patients with breast cancer dormancy. Clin. Cancer Res. 2004, 10, 8152–8162. [Google Scholar] [CrossRef] [Green Version]
- Alunni-Fabbroni, M.; Sandri, M.T. Circulating tumour cells in clinical practice: Methods of detection and possible characterization. Methods 2010, 50, 289–297. [Google Scholar] [CrossRef]
- Miller, M.C.; Doyle, G.V.; Terstappen, L.W. Significance of Circulating Tumor Cells Detected by the CellSearch System in Patients with Metastatic Breast Colorectal and Prostate Cancer. J. Oncol. 2010, 2010, 617421. [Google Scholar] [CrossRef]
- Diehl, F.; Schmidt, K.; Choti, M.A.; Romans, K.; Goodman, S.; Li, M.; Thornton, K.; Agrawal, N.; Sokoll, L.; Szabo, S.A.; et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 2008, 14, 985–990. [Google Scholar] [CrossRef]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014, 6, 224ra224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.; Xia, W.; Lv, Z.; Ni, C.; Xin, Y.; Yang, L. Liquid Biopsy for Cancer: Circulating Tumor Cells, Circulating Free DNA or Exosomes? Cell. Physiol. Biochem. 2017, 41, 755–768. [Google Scholar] [CrossRef]
- Skog, J.; Wurdinger, T.; van Rijn, S.; Meijer, D.H.; Gainche, L.; Sena-Esteves, M.; Curry, W.T., Jr.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell. Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef]
- Souza, M.F.; Kuasne, H.; Barros-Filho, M.C.; Ciliao, H.L.; Marchi, F.A.; Fuganti, P.E.; Paschoal, A.R.; Rogatto, S.R.; Colus, I.M.S. Circulating mRNAs and miRNAs as candidate markers for the diagnosis and prognosis of prostate cancer. PLoS ONE 2017, 12, e0184094. [Google Scholar] [CrossRef]
- Qu, F.; Xie, W.; Nakabayashi, M.; Zhang, H.; Jeong, S.H.; Wang, X.; Komura, K.; Sweeney, C.J.; Sartor, O.; Lee, G.M.; et al. Association of AR-V7 and Prostate-Specific Antigen RNA Levels in Blood with Efficacy of Abiraterone Acetate and Enzalutamide Treatment in Men with Prostate Cancer. Clin. Cancer Res. 2017, 23, 726–734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cuzick, J.; Swanson, G.P.; Fisher, G.; Brothman, A.R.; Berney, D.M.; Reid, J.E.; Mesher, D.; Speights, V.O.; Stankiewicz, E.; Foster, C.S.; et al. Prognostic value of an RNA expression signature derived from cell cycle proliferation genes in patients with prostate cancer: A retrospective study. Lancet Oncol. 2011, 12, 245–255. [Google Scholar] [CrossRef] [Green Version]
- Anfossi, S.; Babayan, A.; Pantel, K.; Calin, G.A. Clinical utility of circulating non-coding RNAs—An update. Nat. Rev. Clin. Oncol. 2018, 15, 541–563. [Google Scholar] [CrossRef]
- Papadaki, C.; Stoupis, G.; Tsalikis, L.; Monastirioti, A.; Papadaki, M.; Maliotis, N.; Stratigos, M.; Mastrostamatis, G.; Mavroudis, D.; Agelaki, S. Circulating miRNAs as a marker of metastatic disease and prognostic factor in metastatic breast cancer. Oncotarget 2019, 10, 966–981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giraldez, M.D.; Lozano, J.J.; Ramirez, G.; Hijona, E.; Bujanda, L.; Castells, A.; Gironella, M. Circulating microRNAs as biomarkers of colorectal cancer: Results from a genome-wide profiling and validation study. Clin. Gastroenterol. Hepatol. 2013, 11, 681–688 e683. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Komatsu, S. Proteomic approaches to uncover the flooding and drought stress response mechanisms in soybean. J. Proteom. 2018, 172, 201–215. [Google Scholar] [CrossRef] [PubMed]
- Tong, Y.S.; Wang, X.W.; Zhou, X.L.; Liu, Z.H.; Yang, T.X.; Shi, W.H.; Xie, H.W.; Lv, J.; Wu, Q.Q.; Cao, X.F. Identification of the long non-coding RNA POU3F3 in plasma as a novel biomarker for diagnosis of esophageal squamous cell carcinoma. Mol. Cancer 2015, 14, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nilsson, J.; Skog, J.; Nordstrand, A.; Baranov, V.; Mincheva-Nilsson, L.; Breakefield, X.O.; Widmark, A. Prostate cancer-derived urine exosomes: A novel approach to biomarkers for prostate cancer. Br. J. Cancer 2009, 100, 1603–1607. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Wu, Z.; Zhang, J.; Su, B. Salivary lncRNA as a potential marker for oral squamous cell carcinoma diagnosis. Mol. Med. Rep. 2013, 7, 761–766. [Google Scholar] [CrossRef] [Green Version]
- Ge, X.; Wang, Y.; Nie, J.; Li, Q.; Tang, L.; Deng, X.; Wang, F.; Xu, B.; Wu, X.; Zhang, X.; et al. The diagnostic/prognostic potential and molecular functions of long non-coding RNAs in the exosomes derived from the bile of human cholangiocarcinoma. Oncotarget 2017, 8, 69995–70005. [Google Scholar] [CrossRef] [Green Version]
- Shao, Y.; Ye, M.; Jiang, X.; Sun, W.; Ding, X.; Liu, Z.; Ye, G.; Zhang, X.; Xiao, B.; Guo, J. Gastric juice long noncoding RNA used as a tumor marker for screening gastric cancer. Cancer 2014, 120, 3320–3328. [Google Scholar] [CrossRef]
- Kamel, L.M.; Atef, D.M.; Mackawy, A.M.H.; Shalaby, S.M.; Abdelraheim, N. Circulating long non-coding RNA GAS5 and SOX2OT as potential biomarkers for diagnosis and prognosis of non-small cell lung cancer. Biotechnol. Appl. Biochem. 2019, 66, 634–642. [Google Scholar] [CrossRef]
- Wang, L.; Duan, W.; Yan, S.; Xie, Y.; Wang, C. Circulating long non-coding RNA colon cancer-associated transcript 2 protected by exosome as a potential biomarker for colorectal cancer. Biomed. Pharmacother. 2019, 113, 108758. [Google Scholar] [CrossRef] [PubMed]
- Lawrie, C.H.; Gal, S.; Dunlop, H.M.; Pushkaran, B.; Liggins, A.P.; Pulford, K.; Banham, A.H.; Pezzella, F.; Boultwood, J.; Wainscoat, J.S.; et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br. J. Haematol. 2008, 141, 672–675. [Google Scholar] [CrossRef] [PubMed]
- Anfossi, S.; Fu, X.; Nagvekar, R.; Calin, G.A. MicroRNAs, Regulatory Messengers Inside and Outside Cancer Cells. Adv. Exp. Med. Biol. 2018, 1056, 87–108. [Google Scholar] [CrossRef] [PubMed]
- Drokow, E.K.; Sun, K.; Ahmed, H.A.W.; Akpabla, G.S.; Song, J.; Shi, M. Circulating microRNA as diagnostic biomarkers for haematological cancers: A systematic review and meta-analysis. Cancer Manag. Res. 2019, 11, 4313–4326. [Google Scholar] [CrossRef] [Green Version]
- Yanez-Mo, M.; Siljander, P.R.; Andreu, Z.; Zavec, A.B.; Borras, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef] [Green Version]
- Thery, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [Green Version]
- Hu, T.; Wolfram, J.; Srivastava, S. Extracellular Vesicles in Cancer Detection: Hopes and Hypes. Trends Cancer 2021, 7, 122–133. [Google Scholar] [CrossRef]
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell. Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef]
- Li, P.; Kaslan, M.; Lee, S.H.; Yao, J.; Gao, Z. Progress in Exosome Isolation Techniques. Theranostics 2017, 7, 789–804. [Google Scholar] [CrossRef]
- Zhou, B.; Xu, K.; Zheng, X.; Chen, T.; Wang, J.; Song, Y.; Shao, Y.; Zheng, S. Application of exosomes as liquid biopsy in clinical diagnosis. Signal. Transduct. Target Ther. 2020, 5, 144. [Google Scholar] [CrossRef] [PubMed]
- Martellucci, S.; Orefice, N.S.; Angelucci, A.; Luce, A.; Caraglia, M.; Zappavigna, S. Extracellular Vesicles: New Endogenous Shuttles for miRNAs in Cancer Diagnosis and Therapy? Int. J. Mol. Sci. 2020, 21, 6486. [Google Scholar] [CrossRef] [PubMed]
- Thind, A.; Wilson, C. Exosomal miRNAs as cancer biomarkers and therapeutic targets. J. Extracell Vesicles 2016, 5, 31292. [Google Scholar] [CrossRef] [PubMed]
- Bryzgunova, O.; Konoshenko, M.; Zaporozhchenko, I.; Yakovlev, A.; Laktionov, P. Isolation of Cell-Free miRNA from Biological Fluids: Influencing Factors and Methods. Diagnostics 2021, 11, 865. [Google Scholar] [CrossRef] [PubMed]
- Witwer, K.W.; Buzas, E.I.; Bemis, L.T.; Bora, A.; Lasser, C.; Lotvall, J.; Nolte-’t Hoen, E.N.; Piper, M.G.; Sivaraman, S.; Skog, J.; et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J. Extracell Vesicles 2013, 2, 20360. [Google Scholar] [CrossRef]
- Ramirez, M.I.; Amorim, M.G.; Gadelha, C.; Milic, I.; Welsh, J.A.; Freitas, V.M.; Nawaz, M.; Akbar, N.; Couch, Y.; Makin, L.; et al. Technical challenges of working with extracellular vesicles. Nanoscale 2018, 10, 881–906. [Google Scholar] [CrossRef] [Green Version]
- Yokota, M.; Tatsumi, N.; Nathalang, O.; Yamada, T.; Tsuda, I. Effects of heparin on polymerase chain reaction for blood white cells. J. Clin. Lab. Anal. 1999, 13, 133–140. [Google Scholar] [CrossRef]
- Gyorgy, B.; Paloczi, K.; Kovacs, A.; Barabas, E.; Beko, G.; Varnai, K.; Pallinger, E.; Szabo-Taylor, K.; Szabo, T.G.; Kiss, A.A.; et al. Improved circulating microparticle analysis in acid-citrate dextrose (ACD) anticoagulant tube. Thromb. Res. 2014, 133, 285–292. [Google Scholar] [CrossRef] [Green Version]
- Arroyo, J.D.; Chevillet, J.R.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.S.; Bennett, C.F.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L.; et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003–5008. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Ba, Y.; Ma, L.; Cai, X.; Yin, Y.; Wang, K.; Guo, J.; Zhang, Y.; Chen, J.; Guo, X.; et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008, 18, 997–1006. [Google Scholar] [CrossRef] [Green Version]
- Munker, R.; Calin, G.A. MicroRNA profiling in cancer. Clin. Sci. 2011, 121, 141–158. [Google Scholar] [CrossRef] [PubMed]
- Gallo, A.; Tandon, M.; Alevizos, I.; Illei, G.G. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS ONE 2012, 7, e30679. [Google Scholar] [CrossRef] [Green Version]
- Pardini, B.; Cordero, F.; Naccarati, A.; Viberti, C.; Birolo, G.; Oderda, M.; Di Gaetano, C.; Arigoni, M.; Martina, F.; Calogero, R.A.; et al. microRNA profiles in urine by next-generation sequencing can stratify bladder cancer subtypes. Oncotarget 2018, 9, 20658–20669. [Google Scholar] [CrossRef]
- Pos, O.; Biro, O.; Szemes, T.; Nagy, B. Circulating cell-free nucleic acids: Characteristics and applications. Eur. J. Hum. Genet. 2018, 26, 937–945. [Google Scholar] [CrossRef] [Green Version]
- Boukouris, S.; Mathivanan, S. Exosomes in bodily fluids are a highly stable resource of disease biomarkers. Proteom. Clin. Appl. 2015, 9, 358–367. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Q.; Li, M.; Wang, X.; Li, Q.; Wang, T.; Zhu, Q.; Zhou, X.; Wang, X.; Gao, X.; Li, X. Immune-related microRNAs are abundant in breast milk exosomes. Int. J. Biol. Sci. 2012, 8, 118–123. [Google Scholar] [CrossRef]
- Cogswell, J.P.; Ward, J.; Taylor, I.A.; Waters, M.; Shi, Y.; Cannon, B.; Kelnar, K.; Kemppainen, J.; Brown, D.; Chen, C.; et al. Identification of miRNA changes in Alzheimer’s disease brain and CSF yields putative biomarkers and insights into disease pathways. J. Alzheimer’s Dis. 2008, 14, 27–41. [Google Scholar] [CrossRef] [PubMed]
- da Silveira, J.C.; Veeramachaneni, D.N.; Winger, Q.A.; Carnevale, E.M.; Bouma, G.J. Cell-secreted vesicles in equine ovarian follicular fluid contain miRNAs and proteins: A possible new form of cell communication within the ovarian follicle. Biol. Reprod. 2012, 86, 71. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, T.; Mori, K.; Sunayama, H.; Takano, E.; Kitayama, Y.; Shimizu, T.; Hirose, Y.; Inubushi, S.; Sasaki, R.; Tanino, H. Antibody-Conjugated Signaling Nanocavities Fabricated by Dynamic Molding for Detecting Cancers Using Small Extracellular Vesicle Markers from Tears. J. Am. Chem. Soc. 2020, 142, 6617–6624. [Google Scholar] [CrossRef] [PubMed]
- Vojtech, L.; Woo, S.; Hughes, S.; Levy, C.; Ballweber, L.; Sauteraud, R.P.; Strobl, J.; Westerberg, K.; Gottardo, R.; Tewari, M.; et al. Exosomes in human semen carry a distinctive repertoire of small non-coding RNAs with potential regulatory functions. Nucl. Acids Res. 2014, 42, 7290–7304. [Google Scholar] [CrossRef] [Green Version]
- Yagi, Y.; Ohkubo, T.; Kawaji, H.; Machida, A.; Miyata, H.; Goda, S.; Roy, S.; Hayashizaki, Y.; Suzuki, H.; Yokota, T. Next-generation sequencing-based small RNA profiling of cerebrospinal fluid exosomes. Neurosci. Lett. 2017, 636, 48–57. [Google Scholar] [CrossRef]
- Muraoka, S.; Jedrychowski, M.P.; Yanamandra, K.; Ikezu, S.; Gygi, S.P.; Ikezu, T. Proteomic Profiling of Extracellular Vesicles Derived from Cerebrospinal Fluid of Alzheimer’s Disease Patients: A Pilot Study. Cells 2020, 9, 1959. [Google Scholar] [CrossRef]
- Gui, Y.; Liu, H.; Zhang, L.; Lv, W.; Hu, X. Altered microRNA profiles in cerebrospinal fluid exosome in Parkinson disease and Alzheimer disease. Oncotarget 2015, 6, 37043–37053. [Google Scholar] [CrossRef] [Green Version]
- Pardini, B.; Sabo, A.A.; Birolo, G.; Calin, G.A. Noncoding RNAs in Extracellular Fluids as Cancer Biomarkers: The New Frontier of Liquid Biopsies. Cancers 2019, 11, 1170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, S.C.; Yiap, B.C. DNA, RNA, and protein extraction: The past and the present. J. Biomed. Biotechnol. 2009, 2009, 574398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diefenbach, R.J.; Lee, J.H.; Kefford, R.F.; Rizos, H. Evaluation of commercial kits for purification of circulating free DNA. Cancer Genet. 2018, 228, 21–27. [Google Scholar] [CrossRef]
- McAlexander, M.A.; Phillips, M.J.; Witwer, K.W. Comparison of Methods for miRNA Extraction from Plasma and Quantitative Recovery of RNA from Cerebrospinal Fluid. Front. Genet. 2013, 4, 83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Padhye, A.A.; Gutekunst, R.W.; Smith, D.J.; Punithalingam, E. Maxillary sinusitis caused by Pleurophomopsis lignicola. J. Clin. Microbiol. 1997, 35, 2136–2141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brunet-Vega, A.; Pericay, C.; Quilez, M.E.; Ramirez-Lazaro, M.J.; Calvet, X.; Lario, S. Variability in microRNA recovery from plasma: Comparison of five commercial kits. Anal. Biochem. 2015, 488, 28–35. [Google Scholar] [CrossRef]
- Williams, Z.; Ben-Dov, I.Z.; Elias, R.; Mihailovic, A.; Brown, M.; Rosenwaks, Z.; Tuschl, T. Comprehensive profiling of circulating microRNA via small RNA sequencing of cDNA libraries reveals biomarker potential and limitations. Proc. Natl. Acad. Sci. USA 2013, 110, 4255–4260. [Google Scholar] [CrossRef] [Green Version]
- Hantzsch, M.; Tolios, A.; Beutner, F.; Nagel, D.; Thiery, J.; Teupser, D.; Holdt, L.M. Comparison of whole blood RNA preservation tubes and novel generation RNA extraction kits for analysis of mRNA and MiRNA profiles. PLoS ONE 2014, 9, e113298. [Google Scholar] [CrossRef]
- Pezzi, H.M.; Niles, D.J.; Schehr, J.L.; Beebe, D.J.; Lang, J.M. Integration of Magnetic Bead-Based Cell Selection into Complex Isolations. ACS Omega 2018, 3, 3908–3917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.; Shiesh, S.C.; Lee, G.B.; Chen, C. Two-step magnetic bead-based (2MBB) techniques for immunocapture of extracellular vesicles and quantification of microRNAs for cardiovascular diseases: A pilot study. PLoS ONE 2020, 15, e0229610. [Google Scholar] [CrossRef] [Green Version]
- Smerkova, K.; Rypar, T.; Adam, V.; Vaculovicova, M. Direct Magnetic Bead-Based Extraction of MicroRNA from Urine with Capillary Electrophoretic Analysis Using Fluorescence Detection and Universal Label. J. Biomed. Nanotechnol. 2020, 16, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Niehof, M.; Reamon-Buettner, S.M.; Danov, O.; Hansen, T.; Sewald, K. A modified protocol for successful miRNA profiling in human precision-cut lung slices (PCLS). BMC Res. Notes 2021, 14, 255. [Google Scholar] [CrossRef] [PubMed]
- Wright, E.S. RNAconTest: Comparing tools for noncoding RNA multiple sequence alignment based on structural consistency. RNA 2020, 26, 531–540. [Google Scholar] [CrossRef] [PubMed]
- Sourvinou, I.S.; Markou, A.; Lianidou, E.S. Quantification of circulating miRNAs in plasma: Effect of preanalytical and analytical parameters on their isolation and stability. J. Mol. Diagn. 2013, 15, 827–834. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Kowdley, K.V. Method for microRNA isolation from clinical serum samples. Anal. Biochem. 2012, 431, 69–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gardiner, C.; Di Vizio, D.; Sahoo, S.; Thery, C.; Witwer, K.W.; Wauben, M.; Hill, A.F. Techniques used for the isolation and characterization of extracellular vesicles: Results of a worldwide survey. J. Extracell Vesicles 2016, 5, 32945. [Google Scholar] [CrossRef]
- Thery, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell. Biol. 2006. [Google Scholar] [CrossRef]
- Davies, R.T.; Kim, J.; Jang, S.C.; Choi, E.J.; Gho, Y.S.; Park, J. Microfluidic filtration system to isolate extracellular vesicles from blood. Lab Chip 2012, 12, 5202–5210. [Google Scholar] [CrossRef] [Green Version]
- Baranyai, T.; Herczeg, K.; Onodi, Z.; Voszka, I.; Modos, K.; Marton, N.; Nagy, G.; Mager, I.; Wood, M.J.; El Andaloussi, S.; et al. Isolation of Exosomes from Blood Plasma: Qualitative and Quantitative Comparison of Ultracentrifugation and Size Exclusion Chromatography Methods. PLoS ONE 2015, 10, e0145686. [Google Scholar] [CrossRef] [Green Version]
- Kalra, H.; Adda, C.G.; Liem, M.; Ang, C.S.; Mechler, A.; Simpson, R.J.; Hulett, M.D.; Mathivanan, S. Comparative proteomics evaluation of plasma exosome isolation techniques and assessment of the stability of exosomes in normal human blood plasma. Proteomics 2013, 13, 3354–3364. [Google Scholar] [CrossRef]
- Holcar, M.; Ferdin, J.; Sitar, S.; Tusek-Znidaric, M.; Dolzan, V.; Plemenitas, A.; Zagar, E.; Lenassi, M. Enrichment of plasma extracellular vesicles for reliable quantification of their size and concentration for biomarker discovery. Sci. Rep. 2020, 10, 21346. [Google Scholar] [CrossRef]
- Macias, M.; Rebmann, V.; Mateos, B.; Varo, N.; Perez-Gracia, J.L.; Alegre, E.; Gonzalez, A. Comparison of six commercial serum exosome isolation methods suitable for clinical laboratories. Effect in cytokine analysis. Clin. Chem. Lab. Med. 2019, 57, 1539–1545. [Google Scholar] [CrossRef]
- Boriachek, K.; Islam, M.N.; Moller, A.; Salomon, C.; Nguyen, N.T.; Hossain, M.S.A.; Yamauchi, Y.; Shiddiky, M.J.A. Biological Functions and Current Advances in Isolation and Detection Strategies for Exosome Nanovesicles. Small 2018, 14, 1702153. [Google Scholar] [CrossRef] [PubMed]
- Stranska, R.; Gysbrechts, L.; Wouters, J.; Vermeersch, P.; Bloch, K.; Dierickx, D.; Andrei, G.; Snoeck, R. Comparison of membrane affinity-based method with size-exclusion chromatography for isolation of exosome-like vesicles from human plasma. J. Transl. Med. 2018, 16, 1. [Google Scholar] [CrossRef] [PubMed]
- Lobb, R.J.; Becker, M.; Wen, S.W.; Wong, C.S.; Wiegmans, A.P.; Leimgruber, A.; Moller, A. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J. Extracell Vesicles 2015, 4, 27031. [Google Scholar] [CrossRef]
- Helwa, I.; Cai, J.; Drewry, M.D.; Zimmerman, A.; Dinkins, M.B.; Khaled, M.L.; Seremwe, M.; Dismuke, W.M.; Bieberich, E.; Stamer, W.D.; et al. A Comparative Study of Serum Exosome Isolation Using Differential Ultracentrifugation and Three Commercial Reagents. PLoS ONE 2017, 12, e0170628. [Google Scholar] [CrossRef]
- Pritchard, C.C.; Kroh, E.; Wood, B.; Arroyo, J.D.; Dougherty, K.J.; Miyaji, M.M.; Tait, J.F.; Tewari, M. Blood cell origin of circulating microRNAs: A cautionary note for cancer biomarker studies. Cancer Prev. Res. 2012, 5, 492–497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michell, D.L.; Allen, R.M.; Landstreet, S.R.; Zhao, S.; Toth, C.L.; Sheng, Q.; Vickers, K.C. Isolation of High-density Lipoproteins for Non-coding Small RNA Quantification. J. Vis. Exp. 2016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vickers, K.C.; Palmisano, B.T.; Shoucri, B.M.; Shamburek, R.D.; Remaley, A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell. Biol. 2011, 13, 423–433. [Google Scholar] [CrossRef] [Green Version]
- Mateescu, B.; Kowal, E.J.; van Balkom, B.W.; Bartel, S.; Bhattacharyya, S.N.; Buzas, E.I.; Buck, A.H.; de Candia, P.; Chow, F.W.; Das, S.; et al. Obstacles and opportunities in the functional analysis of extracellular vesicle RNA—An ISEV position paper. J. Extracell Vesicles 2017, 6, 1286095. [Google Scholar] [CrossRef] [Green Version]
- Chevillet, J.R.; Kang, Q.; Ruf, I.K.; Briggs, H.A.; Vojtech, L.N.; Hughes, S.M.; Cheng, H.H.; Arroyo, J.D.; Meredith, E.K.; Gallichotte, E.N.; et al. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc. Natl. Acad. Sci. USA 2014, 111, 14888–14893. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.K.; Yeo, J.; Kim, B.; Ha, M.; Kim, V.N. Short structured RNAs with low GC content are selectively lost during extraction from a small number of cells. Mol. Cell 2012, 46, 893–895. [Google Scholar] [CrossRef] [Green Version]
- Cheng, L.; Sharples, R.A.; Scicluna, B.J.; Hill, A.F. Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J. Extracell Vesicles 2014, 3, 23743. [Google Scholar] [CrossRef]
- Prendergast, E.N.; de Souza Fonseca, M.A.; Dezem, F.S.; Lester, J.; Karlan, B.Y.; Noushmehr, H.; Lin, X.; Lawrenson, K. Optimizing exosomal RNA isolation for RNA-Seq analyses of archival sera specimens. PLoS ONE 2018, 13, e0196913. [Google Scholar] [CrossRef] [Green Version]
- Tang, Y.T.; Huang, Y.Y.; Zheng, L.; Qin, S.H.; Xu, X.P.; An, T.X.; Xu, Y.; Wu, Y.S.; Hu, X.M.; Ping, B.H.; et al. Comparison of isolation methods of exosomes and exosomal RNA from cell culture medium and serum. Int. J. Mol. Med. 2017, 40, 834–844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buschmann, D.; Kirchner, B.; Hermann, S.; Marte, M.; Wurmser, C.; Brandes, F.; Kotschote, S.; Bonin, M.; Steinlein, O.K.; Pfaffl, M.W.; et al. Evaluation of serum extracellular vesicle isolation methods for profiling miRNAs by next-generation sequencing. J. Extracell Vesicles 2018, 7, 1481321. [Google Scholar] [CrossRef] [PubMed]
- Gobbini, E.; Swalduz, A.; Levra, M.G.; Ortiz-Cuaran, S.; Toffart, A.C.; Perol, M.; Moro-Sibilot, D.; Saintigny, P. Implementing ctDNA Analysis in the Clinic: Challenges and Opportunities in Non-Small Cell Lung Cancer. Cancers 2020, 12, 3112. [Google Scholar] [CrossRef]
- Mohanty, A.; Mohanty, S.K.; Rout, S.; Pani, C. Liquid Biopsy, the hype vs. hope in molecular and clinical oncology. Semin. Oncol. 2021. [Google Scholar] [CrossRef] [PubMed]
- CleanPlex. CleanPlex Amplicon Sequencing Technology for Targeted DNA and RNA-Seq. Available online: https://www.paragongenomics.com/targeted-sequencing/amplicon-sequencing/cleanplex-ngs-amplicon-sequencing/ (accessed on 20 July 2021).
- Stewart, C.M.; Tsui, D.W.Y. Circulating cell-free DNA for non-invasive cancer management. Cancer Genet. 2018, 228, 169–179. [Google Scholar] [CrossRef]
- Baker, M. MicroRNA profiling: Separating signal from noise. Nat. Methods 2010, 7, 687–692. [Google Scholar] [CrossRef]
- Saliminejad, K.; Khorram Khorshid, H.R.; Soleymani Fard, S.; Ghaffari, S.H. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J. Cell Physiol. 2019, 234, 5451–5465. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Xia, L.; Zhou, Q.; Wang, L.; Chen, D.; Gao, X.; Li, Y. Label-Free Detection of miRNA Using Surface-Enhanced Raman Spectroscopy. Anal. Chem. 2020, 92, 12769–12773. [Google Scholar] [CrossRef] [PubMed]
- Jaksik, R.; Iwanaszko, M.; Rzeszowska-Wolny, J.; Kimmel, M. Microarray experiments and factors which affect their reliability. Biol. Direct 2015, 10, 46. [Google Scholar] [CrossRef] [PubMed]
- Illumina. Key Differences between Next-Generation Sequencing and Sanger Sequencing. Available online: https://www.illumina.com/science/technology/next-generation-sequencing/ngs-vs-sanger-sequencing.html (accessed on 21 July 2021).
- Draghici, S.; Khatri, P.; Eklund, A.C.; Szallasi, Z. Reliability and reproducibility issues in DNA microarray measurements. Trends Genet. 2006, 22, 101–109. [Google Scholar] [CrossRef] [Green Version]
- Opitz, L.; Salinas-Riester, G.; Grade, M.; Jung, K.; Jo, P.; Emons, G.; Ghadimi, B.M.; Beissbarth, T.; Gaedcke, J. Impact of RNA degradation on gene expression profiling. BMC Med. Genom. 2010, 3, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madlener, S.; Gojo, J. Liquid Biomarkers for Pediatric Brain Tumors: Biological Features, Advantages and Perspectives. J. Pers. Med. 2020, 10, 254. [Google Scholar] [CrossRef]
- Androvic, P.; Valihrach, L.; Elling, J.; Sjoback, R.; Kubista, M. Two-tailed RT-qPCR: A novel method for highly accurate miRNA quantification. Nucl. Acids Res. 2017, 45, e144. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zou, Y.; Li, C.; Sicking, W.; Piantanida, I.; Yi, T.; Schmuck, C. A molecular peptide beacon for the ratiometric sensing of nucleic acids. J. Am. Chem. Soc. 2012, 134, 1958–1961. [Google Scholar] [CrossRef]
- Li, F.; Zhou, Y.Y.; Peng, T.; Xu, H.; Zhang, R.B.; Zhao, H.; Wang, Z.Y.; Lv, J.X.; Wu, Z.S.; Shen, Z.F. Highly sensitive detection of cancer-related genes based on complete fluorescence restoration of a molecular beacon with a functional overhang. Analyst 2016, 141, 4417–4423. [Google Scholar] [CrossRef]
- Duan, R.; Zuo, X.; Wang, S.; Quan, X.; Chen, D.; Chen, Z.; Jiang, L.; Fan, C.; Xia, F. Lab in a tube: Ultrasensitive detection of microRNAs at the single-cell level and in breast cancer patients using quadratic isothermal amplification. J. Am. Chem. Soc. 2013, 135, 4604–4607. [Google Scholar] [CrossRef]
- Vendrell, J.A.; Mau-Them, F.T.; Beganton, B.; Godreuil, S.; Coopman, P.; Solassol, J. Circulating Cell Free Tumor DNA Detection as a Routine Tool forLung Cancer Patient Management. Int. J. Mol. Sci. 2017, 18, 264. [Google Scholar] [CrossRef] [PubMed]
- Cirmena, G.; Dameri, M.; Ravera, F.; Fregatti, P.; Ballestrero, A.; Zoppoli, G. Assessment of Circulating Nucleic Acids in Cancer: From Current Status to Future Perspectives and Potential Clinical Applications. Cancers 2021, 13, 3460. [Google Scholar] [CrossRef] [PubMed]
- Bruch, R.; Baaske, J.; Chatelle, C.; Meirich, M.; Madlener, S.; Weber, W.; Dincer, C.; Urban, G.A. CRISPR/Cas13a-Powered Electrochemical Microfluidic Biosensor for Nucleic Acid Amplification-Free miRNA Diagnostics. Adv. Mater. 2019, 31, e1905311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Granados-Riveron, J.T.; Aquino-Jarquin, G. CRISPR/Cas13-Based Approaches for Ultrasensitive and Specific Detection of microRNAs. Cells 2021, 10, 1655. [Google Scholar] [CrossRef]
- Bruch, R.; Johnston, M.; Kling, A.; Mattmuller, T.; Baaske, J.; Partel, S.; Madlener, S.; Weber, W.; Urban, G.A.; Dincer, C. CRISPR-powered electrochemical microfluidic multiplexed biosensor for target amplification-free miRNA diagnostics. Biosens. Bioelectron. 2021, 177, 112887. [Google Scholar] [CrossRef]
- Becker, C.; Hammerle-Fickinger, A.; Riedmaier, I.; Pfaffl, M.W. mRNA and microRNA quality control for RT-qPCR analysis. Methods 2010, 50, 237–243. [Google Scholar] [CrossRef]
- Garcia-Elias, A.; Alloza, L.; Puigdecanet, E.; Nonell, L.; Tajes, M.; Curado, J.; Enjuanes, C.; Diaz, O.; Bruguera, J.; Marti-Almor, J.; et al. Defining quantification methods and optimizing protocols for microarray hybridization of circulating microRNAs. Sci. Rep. 2017, 7, 7725. [Google Scholar] [CrossRef]
- Hong, M.; Tao, S.; Zhang, L.; Diao, L.T.; Huang, X.; Huang, S.; Xie, S.J.; Xiao, Z.D.; Zhang, H. RNA sequencing: New technologies and applications in cancer research. J. Hematol. Oncol. 2020, 13, 166. [Google Scholar] [CrossRef]
- Chakraborty, C.; Bhattacharya, M.; Agoramoorthy, G. Single-cell sequencing of miRNAs: A modified technology. Cell Biol. Int. 2020, 44, 1773–1780. [Google Scholar] [CrossRef] [PubMed]
- Benesova, S.; Kubista, M.; Valihrach, L. Small RNA-Sequencing: Approaches and Considerations for miRNA Analysis. Diagnostics 2021, 11, 964. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, S.; Yeri, A.; Cheah, P.S.; Chung, A.; Danielson, K.; De Hoff, P.; Filant, J.; Laurent, C.D.; Laurent, L.D.; Magee, R.; et al. Small RNA Sequencing across Diverse Biofluids Identifies Optimal Methods for exRNA Isolation. Cell 2019, 177, 446–462 e416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, T.X.; Rothenberg, M.E. MicroRNA. J. Allergy Clin. Immunol. 2018, 141, 1202–1207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cirillo, P.D.R.; Margiotti, K.; Mesoraca, A.; Giorlandino, C. Quantification of circulating microRNAs by droplet digital PCR for cancer detection. BMC Res. Notes 2020, 13, 351. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Jiang, T.; Liu, Y.; Huai, G.; Lan, C.; Li, G.; Jia, G.; Wang, K.; Yang, M. Droplet digital PCR-based circulating microRNA detection serve as a promising diagnostic method for gastric cancer. BMC Cancer 2018, 18, 676. [Google Scholar] [CrossRef] [PubMed]
- Thomson, J.M.; Parker, J.S.; Hammond, S.M. Microarray analysis of miRNA gene expression. Methods Enzymol. 2007, 427, 107–122. [Google Scholar] [CrossRef]
- Callari, M.; Tiberio, P.; De Cecco, L.; Cavadini, E.; Dugo, M.; Ghimenti, C.; Daidone, M.G.; Canevari, S.; Appierto, V. Feasibility of circulating miRNA microarray analysis from archival plasma samples. Anal. Biochem. 2013, 437, 123–125. [Google Scholar] [CrossRef]
- Bratulic, S.; Gatto, F.; Nielsen, J. The Translational Status of Cancer Liquid Biopsies. Regen. Eng. Transl. Med. 2019, 1–41. [Google Scholar] [CrossRef] [Green Version]
- Kilgour, E.; Rothwell, D.G.; Brady, G.; Dive, C. Liquid Biopsy-Based Biomarkers of Treatment Response and Resistance. Cancer Cell 2020, 37, 485–495. [Google Scholar] [CrossRef]
- Pinzani, P.; D’Argenio, V.; Del Re, M.; Pellegrini, C.; Cucchiara, F.; Salvianti, F.; Galbiati, S. Updates on liquid biopsy: Current trends and future perspectives for clinical application in solid tumors. Clin. Chem. Lab. Med. 2021, 59, 1181–1200. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.C. Scanning for cancer genomic changes in plasma: Toward an era of personalized blood-based tumor markers. Clin. Chem. 2013, 59, 1553–1555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez, A.; Duyvejonck, H.; Van Belleghem, J.D.; Gryp, T.; Van Simaey, L.; Vermeulen, S.; Van Mechelen, E.; Vaneechoutte, M. Comparison of procedures for RNA-extraction from peripheral blood mononuclear cells. PLoS ONE 2020, 15, e0229423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, C.W.; Wu, H.C.; Terry, M.B.; Santella, R.M. microRNA Expression in Prospectively Collected Blood as a Potential Biomarker of Breast Cancer Risk in the BCFR. Anticancer Res. 2015, 35, 3969–3977. [Google Scholar] [PubMed]
- Mishra, S.; Srivastava, A.K.; Suman, S.; Kumar, V.; Shukla, Y. Circulating miRNAs revealed as surrogate molecular signatures for the early detection of breast cancer. Cancer Lett. 2015, 369, 67–75. [Google Scholar] [CrossRef]
- Aggarwal, P. Liquid-based cytology in lung adenocarcinoma: The way forward. Diagn. Cytopathol. 2019, 47, 1119. [Google Scholar] [CrossRef]
- Campos-Carrillo, A.; Weitzel, J.N.; Sahoo, P.; Rockne, R.; Mokhnatkin, J.V.; Murtaza, M.; Gray, S.W.; Goetz, L.; Goel, A.; Schork, N.; et al. Circulating tumor DNA as an early cancer detection tool. Pharmacol. Ther. 2020, 207, 107458. [Google Scholar] [CrossRef]
- Li, G.; Tang, W.; Yang, F. Cancer Liquid Biopsy Using Integrated Microfluidic Exosome Analysis Platforms. Biotechnol. J. 2020, 15, e1900225. [Google Scholar] [CrossRef]
- Cescon, D.W.; Bratman, S.V.; Chan, S.M.; Siu, L.L. Circulating tumor DNA and liquid biopsy in oncology. Nat. Cancer 2020, 1, 276–290. [Google Scholar] [CrossRef]
- Panagiotara, A.; Markou, A.; Lianidou, E.S.; Patrinos, G.P.; Katsila, T. Exosomes: A Cancer Theranostics Road Map. Public Health Genom. 2017, 20, 116–125. [Google Scholar] [CrossRef]
- Yu, W.; Hurley, J.; Roberts, D.; Chakrabortty, S.K.; Enderle, D.; Noerholm, M.; Breakefield, X.O.; Skog, J.K. Exosome-based liquid biopsies in cancer: Opportunities and challenges. Ann. Oncol. 2021, 32, 466–477. [Google Scholar] [CrossRef] [PubMed]
- Sherkar, K.; Choudhary, S.; Liquid Biopsy Market. Allied Market Research. Available online: https://www.alliedmarketresearch.com/liquid-biopsy-market (accessed on 23 August 2021).
- Bonneau, E.; Neveu, B.; Kostantin, E.; Tsongalis, G.J.; De Guire, V. How close are miRNAs from clinical practice? A perspective on the diagnostic and therapeutic market. EJIFCC 2019, 30, 114–127. [Google Scholar] [PubMed]
- US Food and Drug Administration. Nucleic Acid Based Tests. Available online: https://www.fda.gov/medical-devices/in-vitro-diagnostics/nucleic-acid-based-tests (accessed on 10 July 2021).
- Oosta, G.; Razvi, E. Analysis of miRNA market trends reveals hotspots of research activity. Epigenomics 2012, 4, 237–240. [Google Scholar] [CrossRef]
- Swellam, M.; Ramadan, A.; El-Hussieny, E.A.; Bakr, N.M.; Hassan, N.M.; Sobeih, M.E.; EzzElArab, L.R. Clinical significance of blood-based miRNAs as diagnostic and prognostic nucleic acid markers in breast cancer: Comparative to conventional tumor markers. J. Cell. Biochem. 2019, 120, 12321–12330. [Google Scholar] [CrossRef]
- Shah, S.; Jadhav, K.; Shah, V.; Gupta, N.; Dagrus, K. miRNA 21: Diagnostic Prognostic and Therapeutic Marker for Oral Cancer. Microrna 2016, 5, 175–179. [Google Scholar] [CrossRef]
- Schwarzenbach, H.; Nishida, N.; Calin, G.A.; Pantel, K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat. Rev. Clin. Oncol. 2014, 11, 145–156. [Google Scholar] [CrossRef]
- Schwarzenbach, H. Clinical Relevance of Circulating, Cell-Free and Exosomal microRNAs in Plasma and Serum of Breast Cancer Patients. Oncol. Res. Treat. 2017, 40, 423–429. [Google Scholar] [CrossRef]
- Volinia, S.; Calin, G.A.; Liu, C.G.; Ambs, S.; Cimmino, A.; Petrocca, F.; Visone, R.; Iorio, M.; Roldo, C.; Ferracin, M.; et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 2006, 103, 2257–2261. [Google Scholar] [CrossRef] [Green Version]
- Quirico, L.; Orso, F. The power of microRNAs as diagnostic and prognostic biomarkers in liquid biopsies. Cancer Drug Resist. 2020, 3, 117–139. [Google Scholar] [CrossRef] [Green Version]
- Terrinoni, A.; Calabrese, C.; Basso, D.; Aita, A.; Caporali, S.; Plebani, M.; Bernardini, S. The circulating miRNAs as diagnostic and prognostic markers. Clin. Chem. Lab. Med. 2019, 57, 932–953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jain, K.K. Commercial potential of RNAi. Mol. Biosyst. 2006, 2, 523–526. [Google Scholar] [CrossRef]
- FortuneBusinessInsights. MicroRNA Market Worth Size USD 613.2 Million at 13.4% CAGR by 2027 Backed by Increasing Number of Clinical Trials in North America: Fortune Business Insights™. Available online: https://www.globenewswire.com/news-release/2021/01/25/2163343/0/en/MicroRNA-Market-Worth-Size-USD-613-2-Million-at-13-4-CAGR-by-2027-Backed-by-Increasing-Number-of-Clinical-Trials-in-North-America-Fortune-Business-Insights.html (accessed on 6 September 2021).
- ReportLinker. The Global microRNA (Abbreviated miRNA) Market Size Is Expected to Reach USD 626.27 Million by 2025. Available online: https://www.prnewswire.com/news-releases/the-global-microrna-abbreviated-mirna-market-size-is-expected-to-reach-usd-626-27-million-by-2025--300672448.html (accessed on 23 April 2021).
- GrandViewResearch. microRNA Market Size, Share & Trends Analysis Report by Products, by Services, by Application (Cancer, Neurological Disease, Infectious Diseases), By End-use, By Region, And Segment Forecasts, 2021–2028. Available online: https://www.grandviewresearch.com/industry-analysis/microrna-market (accessed on 14 May 2021).
- Chakraborty, C.; Sharma, A.R.; Sharma, G.; Lee, S.S. Therapeutic advances of miRNAs: A preclinical and clinical update. J. Adv. Res. 2021, 28, 127–138. [Google Scholar] [CrossRef] [PubMed]
- GlobeNewswire. MicroRNA (miRNA) Market to Reach USD 415.1 Million By 2026|Reports And Data. Available online: https://www.globenewswire.com/news-release/2019/05/20/1829098/0/en/MicroRNA-miRNA-Market-To-Reach-USD-415-1-Million-By-2026-Reports-And-Data.html (accessed on 23 April 2021).
- Sheridan, C. Exosome cancer diagnostic reaches market. Nat. Biotechnol. 2016, 34, 359–360. [Google Scholar] [CrossRef]
- Shigeyasu, K.; Toden, S.; Zumwalt, T.J.; Okugawa, Y.; Goel, A. Emerging Role of MicroRNAs as Liquid Biopsy Biomarkers in Gastrointestinal Cancers. Clin. Cancer Res. 2017, 23, 2391–2399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neumann, M.H.D.; Bender, S.; Krahn, T.; Schlange, T. ctDNA and CTCs in Liquid Biopsy—Current Status and Where We Need to Progress. Comput. Struct. Biotechnol. J. 2018, 16, 190–195. [Google Scholar] [CrossRef]
- Arneth, B. Update on the types and usage of liquid biopsies in the clinical setting: A systematic review. BMC Cancer 2018, 18, 527. [Google Scholar] [CrossRef] [Green Version]
- Marrugo-Ramirez, J.; Mir, M.; Samitier, J. Blood-Based Cancer Biomarkers in Liquid Biopsy: A Promising Non-Invasive Alternative to Tissue Biopsy. Int. J. Mol. Sci. 2018, 19, 2877. [Google Scholar] [CrossRef] [Green Version]
- Komatsu, S.; Kiuchi, J.; Imamura, T.; Ichikawa, D.; Otsuji, E. Circulating microRNAs as a liquid biopsy: A next-generation clinical biomarker for diagnosis of gastric cancer. J. Cancer Metast. Treat. 2018, 4, 36. [Google Scholar] [CrossRef] [Green Version]
| Terminology | Full Name and Brief Description |
|---|---|
| cf-DNA: distinguishing between both terms (cell-free DNA and circulating free DNA) is often not possible in literature. | Most of the time it means circulating free DNA. Degraded DNA fragments (50–200 base pairs) are released to the bloodstream. |
| Cell-free DNA refers to the DNA which is not contained inside the cell nuclei. It is obtained by blood separation without cell lysis. | |
| ct-DNA | Circulating tumour DNA: originates from tumour cells and is released to the bloodstream. |
| CTC | Circulating tumour cells: tumour cells released to the bloodstream. |
| cf-RNA: distinguishing between both terms (cell-free RNA and circulating free RNA) is even more complicated than with cf-DNA. The full names are often mixed and used indistinctly in literature. | Most of the times it means circulating free RNA. Degraded RNA fragments released to the bloodstream. |
| Cell-free RNA: refers to the RNA not contained in the cell. Released to the bloodstream. | |
| ccfNA | Circulating cell-free nucleic acids are a mixture of single- or double-stranded nucleic acids released into the blood plasma/serum by different tissues via apoptosis, necrosis and secretions. |
| EV | Extracellular vesicle: membrane particles derived from various cell types. Usually classified into exosomes, microvesicles, oncosomes and apoptotic bodies. |
| Exosome | The smallest extracellular vesicles (approximately 30–150 nm) released from cells. They are formed as intraluminal vesicles inside multivesicular bodies (MVB). |
| Small extracellular vesicle | According to MISEV guidelines 2018, other terms are suggested instead of exosomes. The terms can be based on different characteristics such as size, e.g., small and medium/large EVs together with the size range (e.g., <100 nM or <200 nM for small EVs) |
| Methods | Price | Duration | Throughput | Advantages | Disadvantages |
|---|---|---|---|---|---|
| Silica membranes | Low | 1–2 h | High |
|
|
| Magnetic beads and precipitation | Low | 1–2 h | High |
|
|
| TRIzol reagent | Low | 1–2 h | High |
|
|
| Method | Price | Duration | Throughput | Advantage | Disadvantage |
|---|---|---|---|---|---|
| Standard ultracentrifugation 1 | High | 3–4 h | Low |
|
|
| Sucrose cushion ultracentrifugation 1 | High | 3–4 h | Low |
|
|
| Sucrose gradient ultracentrifugation 1 | High | ≥16 h | Low |
|
|
| Size-exclusion chromatography | Low | 1 h | Intermediate |
|
|
| Immunoaffinity | Medium | 3 h | Intermediate |
|
|
| Precipitation | Low | 1.5 ≥ 14 h 2 | High |
|
|
| Method | Price | Duration | Performance | Advantages | Disadvantages |
|---|---|---|---|---|---|
| NGS-RNA Seq | Hundreds of dollars [99] | 3–5 days [99] |
|
| |
| miRNA sequencing | Hundreds of dollars [103] | 1–2 weeks [103] |
|
|
|
| Microarray | Hundreds of dollars [103] | 2–4 days [103] |
|
|
|
| Sanger Sequencing | High costs for a single experiment [100] | 2–3 days preparation |
|
| |
| RT-qPCR | Less than a hundred dollars | 4–7 h |
|
|
|
| ddPCR | Less than a hundred dollars [99] | 4–7 h [99] |
|
|
|
| BEAMing (beads, emulsion, amplification and magnet) | Hundreds of dollars [99] | 2 days [99] |
|
|
|
| CRISPR-Cas diagnostics (electrochemical biosensors for microRNA detection) | Low costs [117] | <4 h [117] |
|
|
|
| Trade Name | Manufacturer | Sample | Indication for Use | Method |
|---|---|---|---|---|
| Quantidex qPCR BCR-ABL IS Kit | ASURAGEN, Inc. | Whole blood | Monitoring patients with CML during treatment with tyrosine kinase inhibitors | RT-PCR detecting BCR-ABL1 and ABL1 |
| Xpert BCR-ABL Ultra, GeneXpert Dx System, GeneXpert Infinity-48s and GeneXpert Infinity-80 Systems | Cepheid | Whole blood | Monitoring patients with CML during treatment with tyrosine kinase inhibitors | RT-PCR detecting BCR-ABL1 and ABL1 |
| QXDx BCR-ABL %IS Kit for use on the QXDx Auto DG ddPCR System | Bio-Rad Laboratories, Inc. | Whole blood | Monitoring patients with CML during treatment with tyrosine kinase inhibitors | RT-PCR detecting BCR-ABL1 and ABL1 |
| MRDx BCR-ABL Test, MRDx BCR-ABL Test Software | MolecularMD Corporation | Whole blood | Monitoring patients with CML during treatment with tyrosine kinase inhibitors | RT-PCR detecting BCR-ABL1 and ABL1 |
| PROGENSA PCA3 assay | Gen-Probe, Inc. | Urine | To aid in the decision for repeated prostatic biopsy | Detecting PCA3 and PSA using Transcription Mediated Amplification and Hybridization Protection Assay (HPA) method |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martinez-Dominguez, M.V.; Zottel, A.; Šamec, N.; Jovčevska, I.; Dincer, C.; Kahlert, U.D.; Nickel, A.-C. Current Technologies for RNA-Directed Liquid Diagnostics. Cancers 2021, 13, 5060. https://doi.org/10.3390/cancers13205060
Martinez-Dominguez MV, Zottel A, Šamec N, Jovčevska I, Dincer C, Kahlert UD, Nickel A-C. Current Technologies for RNA-Directed Liquid Diagnostics. Cancers. 2021; 13(20):5060. https://doi.org/10.3390/cancers13205060
Chicago/Turabian StyleMartinez-Dominguez, Maria Victoria, Alja Zottel, Neja Šamec, Ivana Jovčevska, Can Dincer, Ulf Dietrich Kahlert, and Ann-Christin Nickel. 2021. "Current Technologies for RNA-Directed Liquid Diagnostics" Cancers 13, no. 20: 5060. https://doi.org/10.3390/cancers13205060
APA StyleMartinez-Dominguez, M. V., Zottel, A., Šamec, N., Jovčevska, I., Dincer, C., Kahlert, U. D., & Nickel, A.-C. (2021). Current Technologies for RNA-Directed Liquid Diagnostics. Cancers, 13(20), 5060. https://doi.org/10.3390/cancers13205060









