3p Arm Loss and Survival in Head and Neck Cancer: An Analysis of TCGA Dataset
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Populations
2.2. Determining 3p Arm Status
2.3. Statistics
2.3.1. Clinical Features
2.3.2. Survival Analysis
2.3.3. Exome Sequence Mutations
2.3.4. Copy Number Alterations
2.3.5. mRNA Sequencing Counts
2.3.6. Integration of CNA and mRNA-Seq Results
2.3.7. Tumor Microevironment Estimation
2.3.8. Hypoxia Estimation
2.3.9. Reverse Phase Protein Array
2.3.10. miRNA Sequencing Analysis
3. Results
3.1. Definitions and Cohort Characteristics
3.1.1. Defining the Study and Validation Cohorts
3.1.2. Defining a Threshold for 3p Arm Deletion Status
3.1.3. Clinical Characteristics Including Stage at Presentation and Distant Metastasis Differ by 3p Deletion Status
3.2. Survival Outcomes by 3p Arm Deletion Status
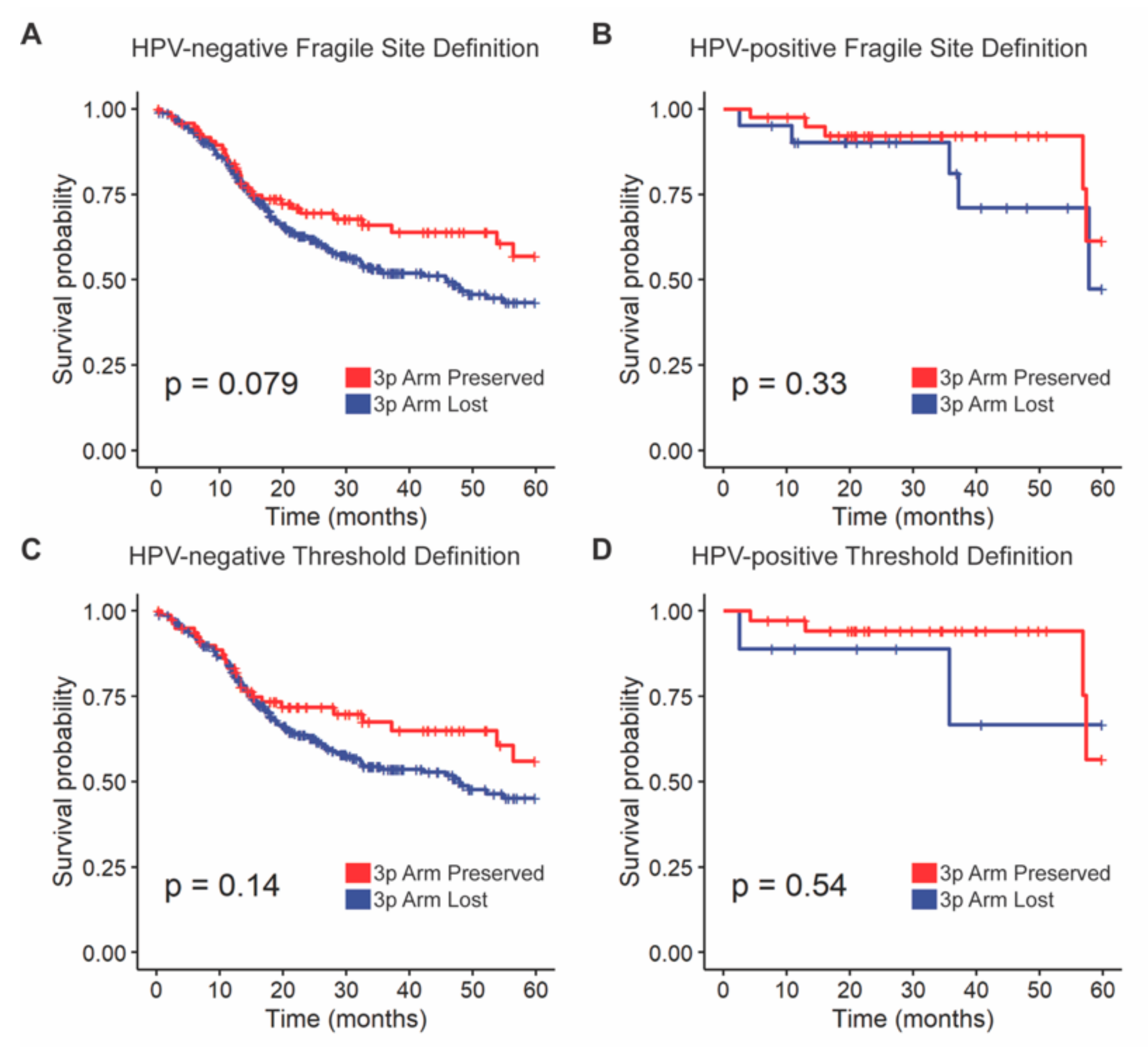
3.2.1. Overall Survival Outcomes Were Not Associated with 3p Arm Deletion Status
3.2.2. External Validation of Overall Survival Outcomes
3.3. Mutational Differences by 3p Arm Deletion Status
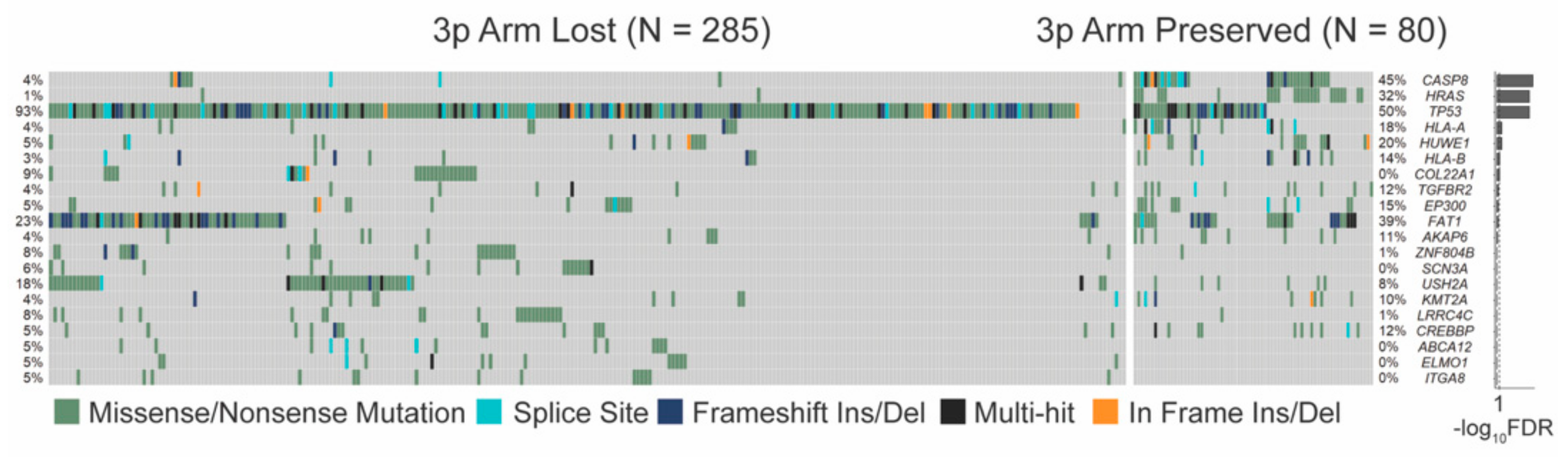
3.3.1. SNV Mutations Differ Significantly by 3p Arm Deletion Status
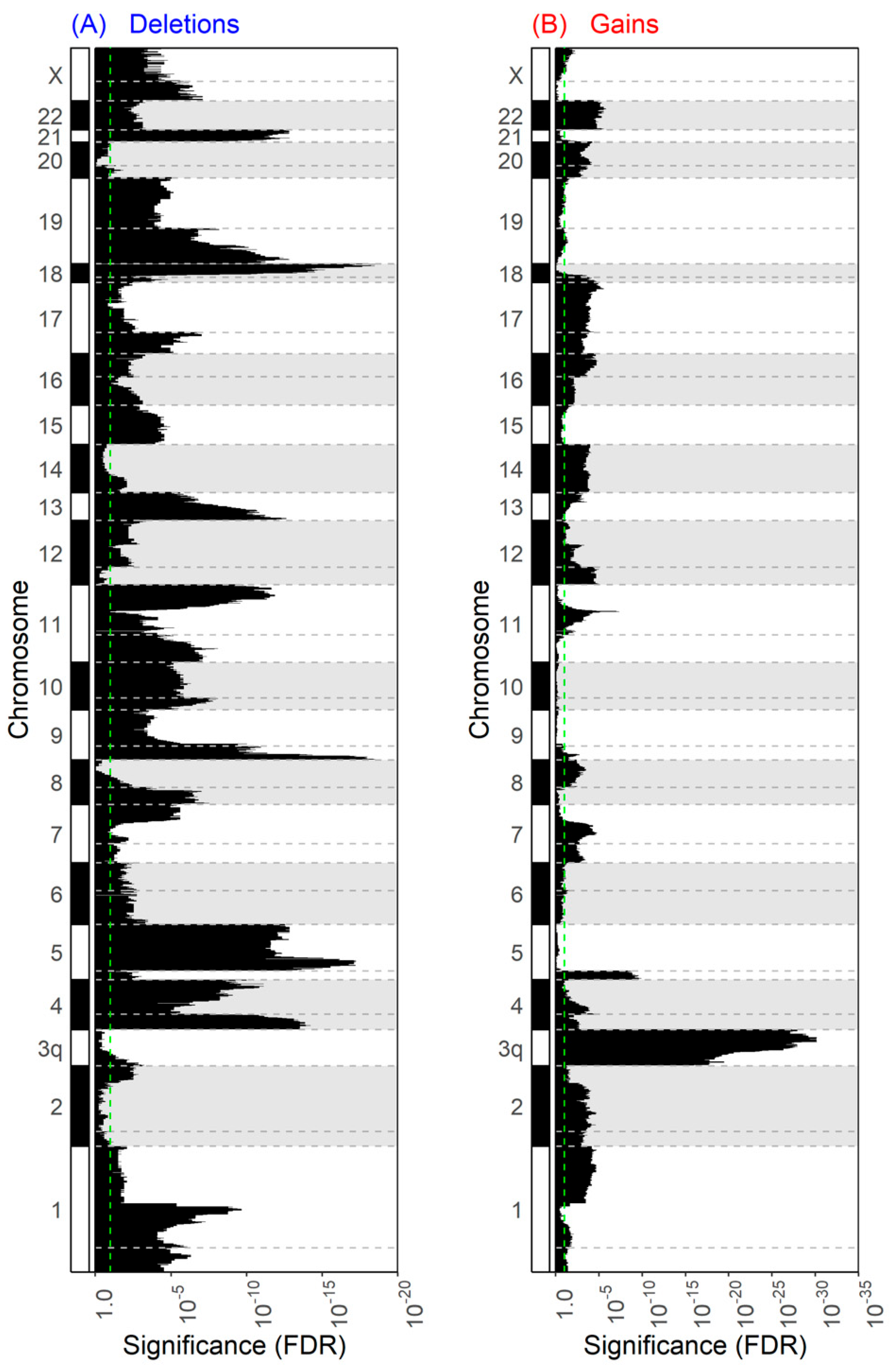
3.3.2. Copy Number Aberrations Are Widespread in 3p Deletion
3.3.3. Pathway Analysis of CNAs Integrated with mRNA Data
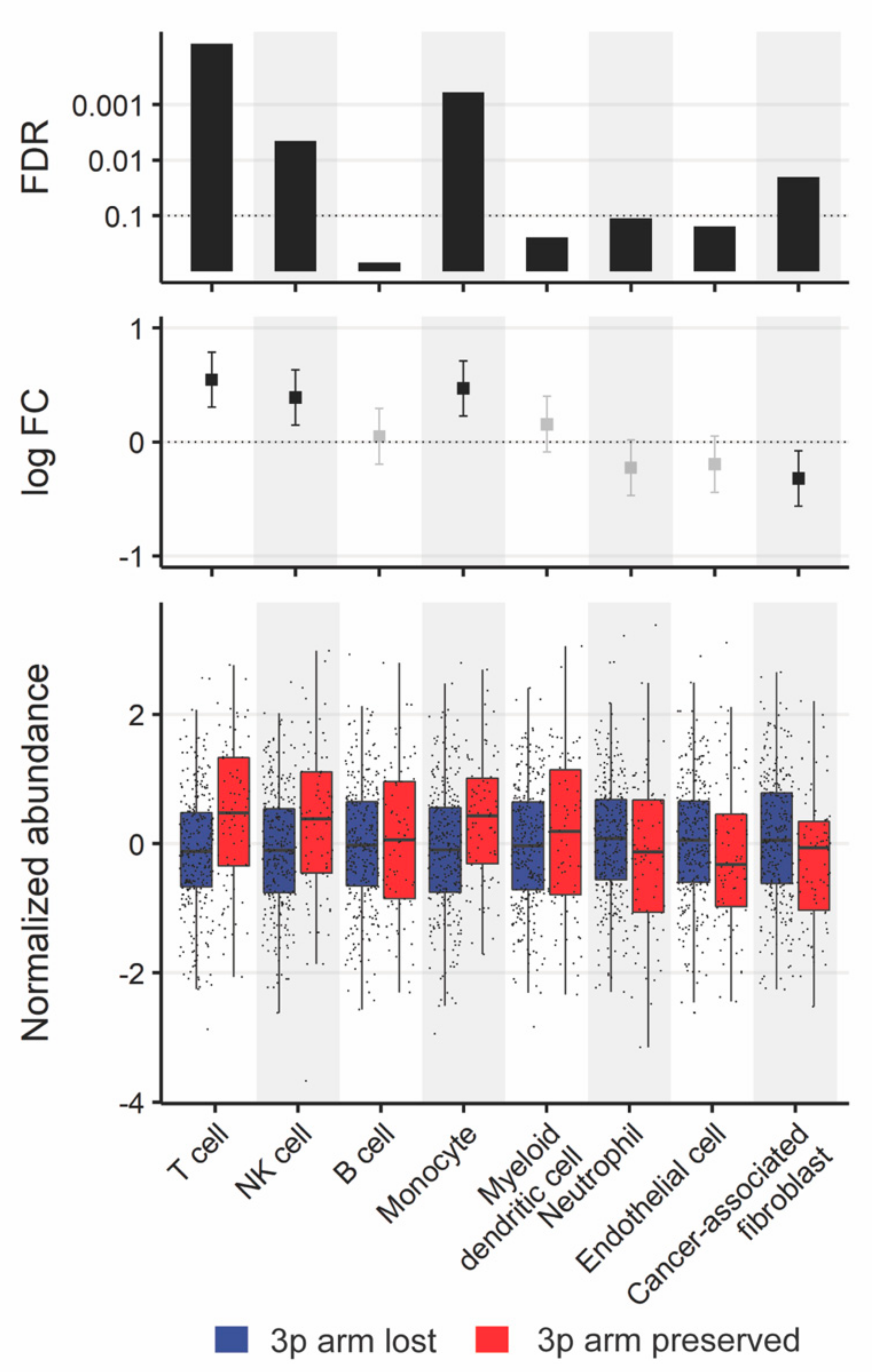
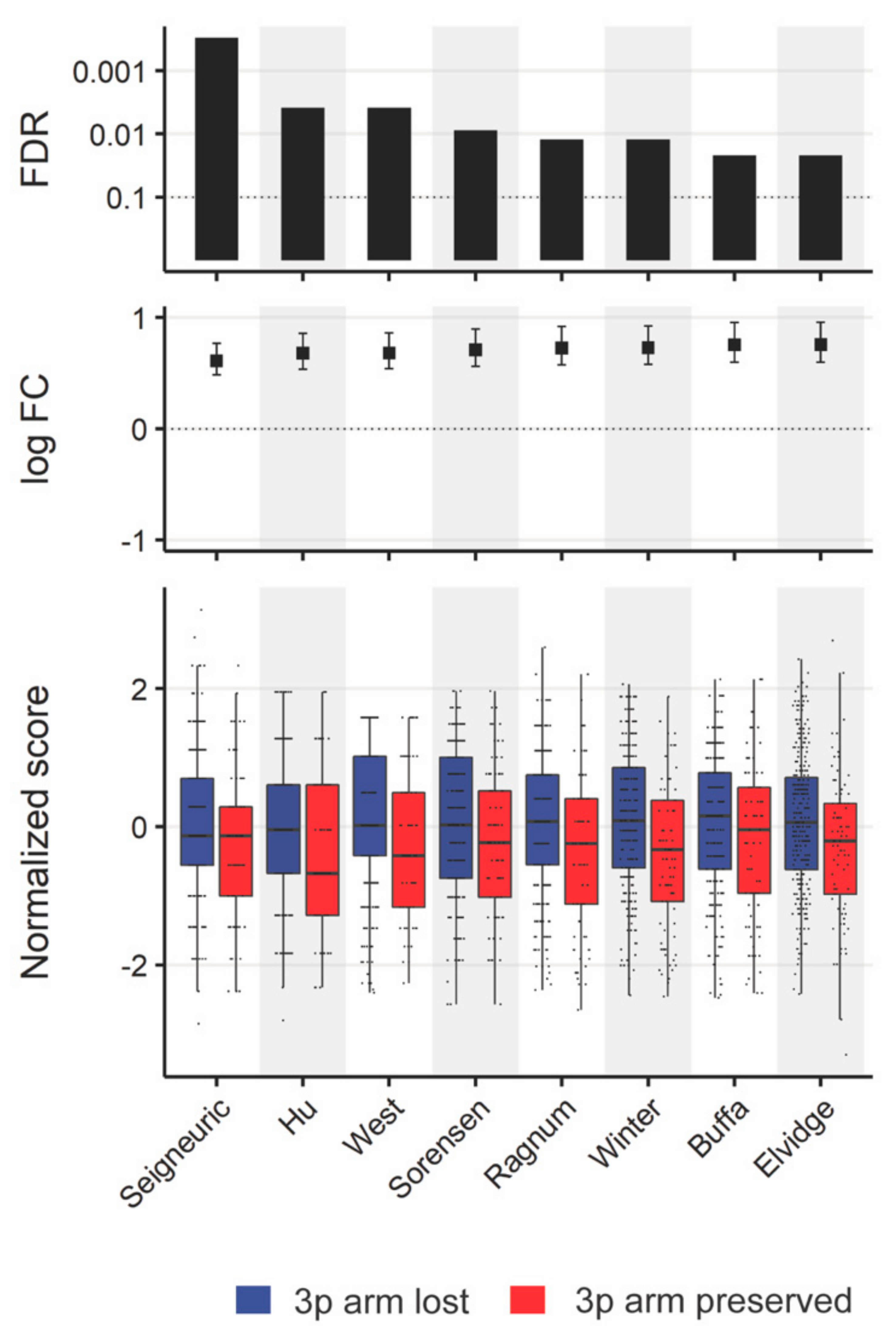
3.3.4. Comparison of Tumor Microenvironments
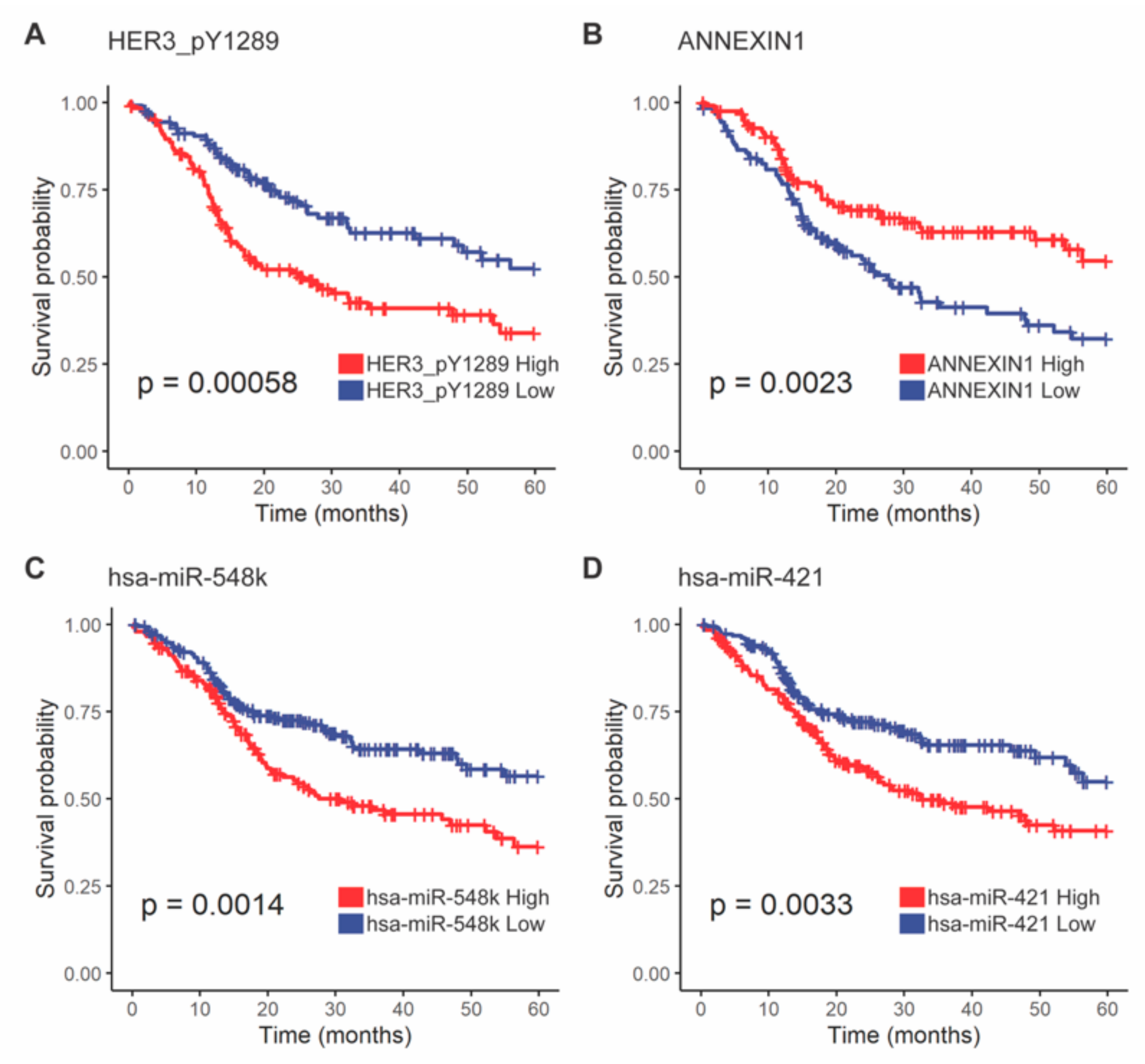
3.3.5. HER3-pY1289 and ANXA1 Proteins Were Less Abundant in 3p Lost Tumors and Were Associated with Survival Outcomes
3.3.6. hsa-miR-548k and hsa-miR-421 Were More Abundant in 3p Arm Loss Tumors and Were Associated with Poor Survival
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.M.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef]
- López-Verdín, S.; Lavalle-Carrasco, J.; Carreón-Burciaga, R.G.; Serafín-Higuera, N.; Molina-Frechero, N.; González-González, R.; Bologna-Molina, R. Molecular Markers of Anticancer Drug Resistance in Head and Neck Squamous Cell Carcinoma: A Literature Review. Cancers 2018, 10, 376. [Google Scholar] [CrossRef] [Green Version]
- Shaikh, M.H.; Barrett, J.W.; Khan, M.I.; Kim, H.A.J.; Zeng, P.Y.F.; Mymryk, J.S.; Nichols, A.C. Chromosome 3p loss in the progression and prognosis of head and neck cancer. Oral. Oncol. 2020, 109, 104944. [Google Scholar] [CrossRef]
- Campbell, P.J.; Getz, G.; Korbel, J.O.; Stuart, J.M.; Jennings, J.L.; Stein, L.D.; Perry, M.D.; Nahal-Bose, H.K.; Ouellette, B.F.F.; Li, C.H.; et al. Pan-cancer analysis of whole genomes. Nature 2020, 578, 82–93. [Google Scholar] [CrossRef] [Green Version]
- Gerstung, M.; Jolly, C.; Leshchiner, I.; Dentro, S.C.; Gonzalez, S.; Rosebrock, D.; Mitchell, T.J.; Rubanova, Y.; Anur, P.; Yu, K.; et al. The evolutionary history of 2,658 cancers. Nature 2020, 578, 122–128. [Google Scholar] [CrossRef] [Green Version]
- Weber, R.G.; Scheer, M.; Born, I.A.; Joos, S.; Cobbers, J.M.J.L.; Hofele, C.; Reifenberger, G.; Zöller, J.E.; Lichter, P. Recurrent Chromosomal Imbalances Detected in Biopsy Material from Oral Premalignant and Malignant Lesions by Combined Tissue Microdissection, Universal DNA Amplification, and Comparative Genomic Hybridization. Am. J. Pathol. 1998, 153, 295–303. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Cheung, K.J., Jr.; Lam, W.L.; Cheng, X.; Poh, C.; Priddy, R.; Epstein, J.; Le, N.D.; Rosin, M.P. Increased genetic damage in oral leukoplakia from high risk sites. Cancer 2001, 91, 2148–2155. [Google Scholar] [CrossRef]
- Gross, A.M.; Orosco, R.K.; Shen, J.P.; Egloff, A.M.; Carter, H.; Hofree, M.; Choueiri, M.; Coffey, C.S.; Lippman, S.M.; Hayes, D.N.; et al. Multi-tiered genomic analysis of head and neck cancer ties TP53 mutation to 3p loss. Nat. Genet. 2014, 46, 939–943. [Google Scholar] [CrossRef] [Green Version]
- Morris, L.G.T.; Chandramohan, R.; West, L.; Zehir, A.; Chakravarty, D.; Pfister, D.G.; Wong, R.J.; Lee, N.Y.; Sherman, E.J.; Baxi, S.S.; et al. The Molecular Landscape of Recurrent and Metastatic Head and Neck Cancers: Insights From a Precision Oncology Sequencing Platform. JAMA Oncol. 2017, 3, 244–255. [Google Scholar] [CrossRef]
- Shen, R.; Seshan, V.E. FACETS: Allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 2016, 44, e131-e131. [Google Scholar] [CrossRef] [Green Version]
- Broad Institute TCGA Genome Data Analysis Center. Analysis-Ready Standardized TCGA Data from Broad GDAC Firehose 2016_01_28 Run; Broad Institute of MIT and Harvard: Cambidge, MA, USA, 2016. [Google Scholar] [CrossRef]
- Liu, J.; Lichtenberg, T.; Hoadley, K.A.; Poisson, L.M.; Lazar, A.J.; Cherniack, A.D.; Kovatich, A.J.; Benz, C.C.; Levine, D.A.; Lee, A.V.; et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 2018, 173, 400–416. [Google Scholar] [CrossRef] [Green Version]
- Bratman, S.V.; Bruce, J.P.; O’Sullivan, B.; Pugh, T.J.; Xu, W.; Yip, K.W.; Liu, F.F. Human Papillomavirus Genotype Association With Survival in Head and Neck Squamous Cell Carcinoma. JAMA Oncol. 2016, 2, 823–826. [Google Scholar] [CrossRef] [Green Version]
- Therneau, T.; Grambsch, P. Modeling Survival Data: Extending the Cox Model, 2nd ed.; Springer: New York, NY, USA, 2000; ISBN 978-1-4757-3294-8. [Google Scholar]
- Mayakonda, A.; Lin, D.C.; Assenov, Y.; Plass, C.; Koeffler, H.P. Maftools: Efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018, 28, 1747–1756. [Google Scholar] [CrossRef] [Green Version]
- Tan, H.; Bao, J.; Zhou, X. Genome-wide mutational spectra analysis reveals significant cancer-specific heterogeneity. Sci. Rep. 2015, 5, 15266. [Google Scholar] [CrossRef] [Green Version]
- Ghasemi, F.; Prokopec, S.D.; MacNeil, D.; Mundi, N.; Gameiro, S.F.; Howlett, C.; Stecho, W.; Plantinga, P.; Pinto, N.; Ruicci, K.M.; et al. Mutational analysis of head and neck squamous cell carcinoma stratified by smoking status. JCI Insight 2019, 4, e123443. [Google Scholar] [CrossRef]
- Gameiro, S.F.; Ghasemi, F.; Barrett, J.W.; Nichols, A.C.; Mymryk, J.S. High Level Expression of MHC-II in HPV+ Head and Neck Cancers Suggests that Tumor Epithelial Cells Serve an Important Role as Accessory Antigen Presenting Cells. Cancers 2019, 11, 1129. [Google Scholar] [CrossRef] [Green Version]
- Gameiro, S.F.; Ghasemi, F.; Barrett, J.W.; Koropatnick, J.; Nichols, A.C.; Mymryk, J.S.; Maleki Vareki, S. Treatment-naive HPV+ head and neck cancers display a T-cell-inflamed phenotype distinct from their HPV- counterparts that has implications for immunotherapy. Oncoimmunology 2018, 7, e1498439. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Hammad, S. Highlight report: The relationship of DNA copy number alterations and mRNA levels in cancer. EXCLI J. 2017, 16, 1326–1327. [Google Scholar] [CrossRef]
- Song, X.; Ji, J.; Gleason, K.J.; Yang, F.; Martignetti, J.A.; Chen, L.S.; Wang, P. Insights into Impact of DNA Copy Number Alteration and Methylation on the Proteogenomic Landscape of Human Ovarian Cancer via a Multi-omics Integrative Analysis. Mol. Cell. Proteom. 2019, 18, S52–S65. [Google Scholar] [CrossRef] [Green Version]
- Creixell, P.; Reimand, J.; Haider, S.; Wu, G.; Shibata, T.; Vazquez, M.; Mustonen, V.; Gonzalez-Perez, A.; Pearson, J.; Sander, C.; et al. Pathway and network analysis of cancer genomes. Nat. Methods 2015, 12, 615–621. [Google Scholar] [CrossRef]
- Sturm, G.; Finotello, F.; Petitprez, F.; Zhang, J.D.; Baumbach, J.; Fridman, W.H.; List, M.; Aneichyk, T. Comprehensive evaluation of transcriptome-based cell-type quantification methods for immuno-oncology. Bioinformatics 2019, 35, i436–i445. [Google Scholar] [CrossRef]
- Becht, E.; Giraldo, N.A.; Lacroix, L.; Buttard, B.; Elarouci, N.; Petitprez, F.; Selves, J.; Laurent-Puig, P.; Sautès-Fridman, C.; Fridman, W.H.; et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016, 17, 218. [Google Scholar] [CrossRef]
- Davidson, M.A.; Shanks, E.J. 3q26-29 Amplification in head and neck squamous cell carcinoma: A review of established and prospective oncogenes. FEBS J. 2017, 284, 2705–2731. [Google Scholar] [CrossRef] [Green Version]
- Taylor, A.M.; Shih, J.; Ha, G.; Gao, G.F.; Zhang, X.; Berger, A.C.; Schumacher, S.E.; Wang, C.; Hu, H.; Liu, J.; et al. Genomic and Functional Approaches to Understanding Cancer Aneuploidy. Cancer Cell. 2018, 33, 676–689. [Google Scholar] [CrossRef] [Green Version]
- Bhandari, V.; Hoey, C.; Liu, L.Y.; Lalonde, E.; Ray, J.; Livingstone, J.; Lesurf, R.; Shiah, Y.-J.; Vujcic, T.; Huang, X.; et al. Molecular landmarks of tumor hypoxia across cancer types. Nat. Genet. 2019, 51, 308–318. [Google Scholar] [CrossRef]
- Li, J.; Lu, Y.; Akbani, R.; Ju, Z.; Roebuck, P.; Liu, W.; Yang, J.; Broom, B.; Verhaak, R.; Kane, D.; et al. TCPA: A Resource for Cancer Functional Proteomics Data. Nat. Methods 2013, 10, 1046–1047. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Akbani, R.; Zhao, W.; Lu, Y.; Weinstein, J.; Mills, G.; Liang, H. Explore, Visualize, and Analyze Functional Cancer Proteomic Data Using the Cancer Proteome Atlas. Cancer Res. 2017, 77, e51–e54. [Google Scholar] [CrossRef] [Green Version]
- Ru, Y.; Kechris, K.; Tabakoff, B.; Hoffman, P.; Radcliffe, R.; Bowler, R.; Mahaffey, S.; Rossi, S.; Calin, G.; Bemis, L.; et al. The multiMiR R Package and Database: Integration of microRNA-target Interactions Along With Their Disease and Drug Associations. Nucleic Acids Res. 2014, 42, e133. [Google Scholar] [CrossRef]
- Leemans, C.R.; Snijders, P.J.F.; Brakenhoff, R.H. The molecular landscape of head and neck cancer. Nat. Rev. Cancer 2018, 18, 269–282. [Google Scholar] [CrossRef]
- Cheng, J.; Demeulemeester, J.; Wedge, D.C.; Vollan, H.K.M.; Pitt, J.J.; Russnes, H.G.; Pandey, B.P.; Nilsen, G.; Nord, S.; Bignell, G.R.; et al. Pan-cancer analysis of homozygous deletions in primary tumours uncovers rare tumour suppressors. Nat. Commun. 2017, 8, 1221. [Google Scholar] [CrossRef] [PubMed]
- Pinto, N.; Prokopec, S.D.; Vizeacoumar, F.; Searle, K.; Lowerison, M.; Ruicci, K.M.; Yoo, J.; Fung, K.; MacNeil, D.; Lacefield, J.C.; et al. Lestaurtinib is a potent inhibitor of anaplastic thyroid cancer cell line models. PLoS ONE 2018, 13, e0207152. [Google Scholar] [CrossRef] [Green Version]
- Fridman, W.H.; Pagès, F.; Sautès-Fridman, C.; Galon, J. The immune contexture in human tumours: Impact on clinical outcome. Nat. Rev. Cancer 2012, 12, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Alsahafi, E.; Begg, K.; Amelio, I.; Raulf, N.; Lucarelli, P.; Sauter, T.; Tavassoli, M. Clinical update on head and neck cancer: Molecular biology and ongoing challenges. Cell. Death Dis. 2019, 10, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Beyer, A.; Aebersold, R. On the Dependency of Cellular Protein Levels on mRNA Abundance. Cell 2016, 165, 535–550. [Google Scholar] [CrossRef] [Green Version]
- Koenigs, M.B.; Lefranc-Torres, A.; Bonilla-Velez, J.; Patel, K.B.; Hayes, D.N.; Glomski, K.; Busse, P.M.; Chan, A.W.; Clark, J.R.; Deschler, D.G.; et al. Association of Estrogen Receptor Alpha Expression With Survival in Oropharyngeal Cancer Following Chemoradiation Therapy. J. Natl. Cancer Inst. 2019, 111, 933–942. [Google Scholar] [CrossRef] [PubMed]
- Liao, L.; Yan, W.-J.; Tian, C.-M.; Li, M.-Y.; Tian, Y.-Q.; Zeng, G.-Q. Knockdown of Annexin A1 Enhances Radioresistance and Inhibits Apoptosis in Nasopharyngeal Carcinoma. Technol. Cancer Res. Treat. 2018, 17, 17. [Google Scholar] [CrossRef] [Green Version]
- Campbell, M.R.; Amin, D.; Moasser, M.M. HER3 Comes of Age: New Insights into Its Functions and Role in Signaling, Tumor Biology, and Cancer Therapy. Clin. Cancer Res. 2010, 16, 1373. [Google Scholar] [CrossRef] [Green Version]
- Yonesaka, K.; Zejnullahu, K.; Okamoto, I.; Satoh, T.; Cappuzzo, F.; Souglakos, J.; Ercan, D.; Rogers, A.; Roncalli, M.; Takeda, M.; et al. Activation of ERBB2 signaling causes resistance to the EGFR-directed therapeutic antibody cetuximab. Sci. Transl. Med. 2011, 3, 99ra86. [Google Scholar] [CrossRef] [Green Version]
- Sergina, N.V.; Moasser, M.M. The HER family and cancer: Emerging molecular mechanisms and therapeutic targets. Trends Mol. Med. 2007, 13, 527–534. [Google Scholar] [CrossRef] [Green Version]
- Luhtala, S.; Staff, S.; Kallioniemi, A.; Tanner, M.; Isola, J. Clinicopathological and prognostic correlations of HER3 expression and its degradation regulators, NEDD4–1 and NRDP1, in primary breast cancer. BMC Cancer 2018, 18, 1045. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Wu, Y.; Feng, M.; Xue, X.; Fan, Y. A novel seven-miRNA prognostic model to predict overall survival in head and neck squamous cell carcinoma patients. Mol. Med. Rep. 2019, 20, 4340–4348. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Li, L.; Ou, Y.; Gao, Z.; Li, E.; Li, X.; Zhang, W.; Wang, J.; Xu, L.; Zhou, Y.; et al. Identification of genomic alterations in oesophageal squamous cell cancer. Nature 2014, 509, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Tang, Y.; Wang, J.; Yan, Z.; Xu, R. miR-421 induces cell proliferation and apoptosis resistance in human nasopharyngeal carcinoma via downregulation of FOXO4. Biochem. Biophys. Res. Commun. 2013, 435, 745–750. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Gao, Y.; Song, D.; Liu, T.; Feng, Y. Correlation between microRNA-421 expression level and prognosis of gastric cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 15128–15132. [Google Scholar] [PubMed]
- Zhou, S.; Wang, B.; Hu, J.; Zhou, Y.; Jiang, M.; Wu, M.; Qin, L.; Yang, X. miR-421 is a diagnostic and prognostic marker in patients with osteosarcoma. Tumour Biol. 2016, 37, 9001–9007. [Google Scholar] [CrossRef]
- Lin, A.B.; McNeely, S.C.; Beckmann, R.P. Achieving Precision Death with Cell-Cycle Inhibitors that Target DNA Replication and Repair. Clin. Cancer Res. 2017, 23, 3232. [Google Scholar] [CrossRef] [Green Version]
- van Harten, A.M.; Buijze, M.; van der Mast, R.; Rooimans, M.A.; Kemp, S.R.M.-d.; Bachas, C.; Brink, A.; Walsum, M.S.-v.; Wolthuis, R.M.F.; Brakenhoff, R.H. Targeting the cell cycle in head and neck cancer by Chk1 inhibition: A novel concept of bimodal cell death. Oncogene 2019, 8, 1–16. [Google Scholar] [CrossRef]
- Giacinti, C.; Giordano, A. RB and cell cycle progression. Oncogene 2006, 25, 5220–5227. [Google Scholar] [CrossRef] [Green Version]
- Redon, R.; Muller, D.; Caulee, K.; Wanherdrick, K.; Abecassis, J.; du Manoir, S. A Simple Specific Pattern of Chromosomal Aberrations at Early Stages of Head and Neck Squamous Cell Carcinomas. Cancer Res. 2001, 61, 4122. [Google Scholar]
- Cheng, H.; Yang, X.; Si, H.; Saleh, A.D.; Xiao, W.; Coupar, J.; Gollin, S.M.; Ferris, R.L.; Issaeva, N.; Yarbrough, W.G.; et al. Genomic and Transcriptomic Characterization Links Cell Lines with Aggressive Head and Neck Cancers. Cell Rep. 2018, 25, 1332–1345.e5. [Google Scholar] [CrossRef] [Green Version]
- Bockmühl, U.; Schlüns, K.; Küchler, I.; Petersen, S.; Petersen, I. Genetic imbalances with impact on survival in head and neck cancer patients. Am. J. Pathol. 2000, 157, 369–375. [Google Scholar] [CrossRef] [Green Version]
- Balermpas, P.; Michel, Y.; Wagenblast, J.; Seitz, O.; Weiss, C.; Rödel, F.; Rödel, C.; Fokas, E. Tumour-infiltrating lymphocytes predict response to definitive chemoradiotherapy in head and neck cancer. Br. J. Cancer 2014, 110, 501–509. [Google Scholar] [CrossRef]
- Fang, W.; Ma, Y.; Yin, J.C.; Hong, S.; Zhou, H.; Wang, A.; Wang, F.; Bao, H.; Wu, X.; Yang, Y.; et al. Comprehensive Genomic Profiling Identifies Novel Genetic Predictors of Response to Anti–PD-(L)1 Therapies in Non–Small Cell Lung Cancer. Clin. Cancer Res. 2019, 25, 5015. [Google Scholar] [CrossRef]
- Moser, B.; Loetscher, P. Lymphocyte traffic control by chemokines. Nat. Immunol. 2001, 2, 123–128. [Google Scholar] [CrossRef] [PubMed]
- William, W.N.; Zhao, X.; Bianchi, J.J.; Lin, H.Y.; Cheng, P.; Lee, J.J.; Carter, H.; Alexandrov, L.B.; Abraham, J.P.; Spetzler, D.B.; et al. Immune evasion in HPV—Head and neck precancer–cancer transition is driven by an aneuploid switch involving chromosome 9p loss. Proc. Natl. Acad. Sci. USA 2021, 118, e2022655118. [Google Scholar] [CrossRef]
- Ribas, A.; Hu-Lieskovan, S. What does PD-L1 positive or negative mean? J. Exp. Med. 2016, 213, 2835–2840. [Google Scholar] [CrossRef] [Green Version]
- Larkin, J.; Chiarion-Sileni, V.; Gonzalez, R.; Grob, J.; Cowey, C.; Lao, C.; Schadendorf, D.; Dummer, R.; Smylie, M.; Rutkowski, P.; et al. Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. N. Engl. J. Med. 2015, 373, 23–34. [Google Scholar] [CrossRef] [Green Version]
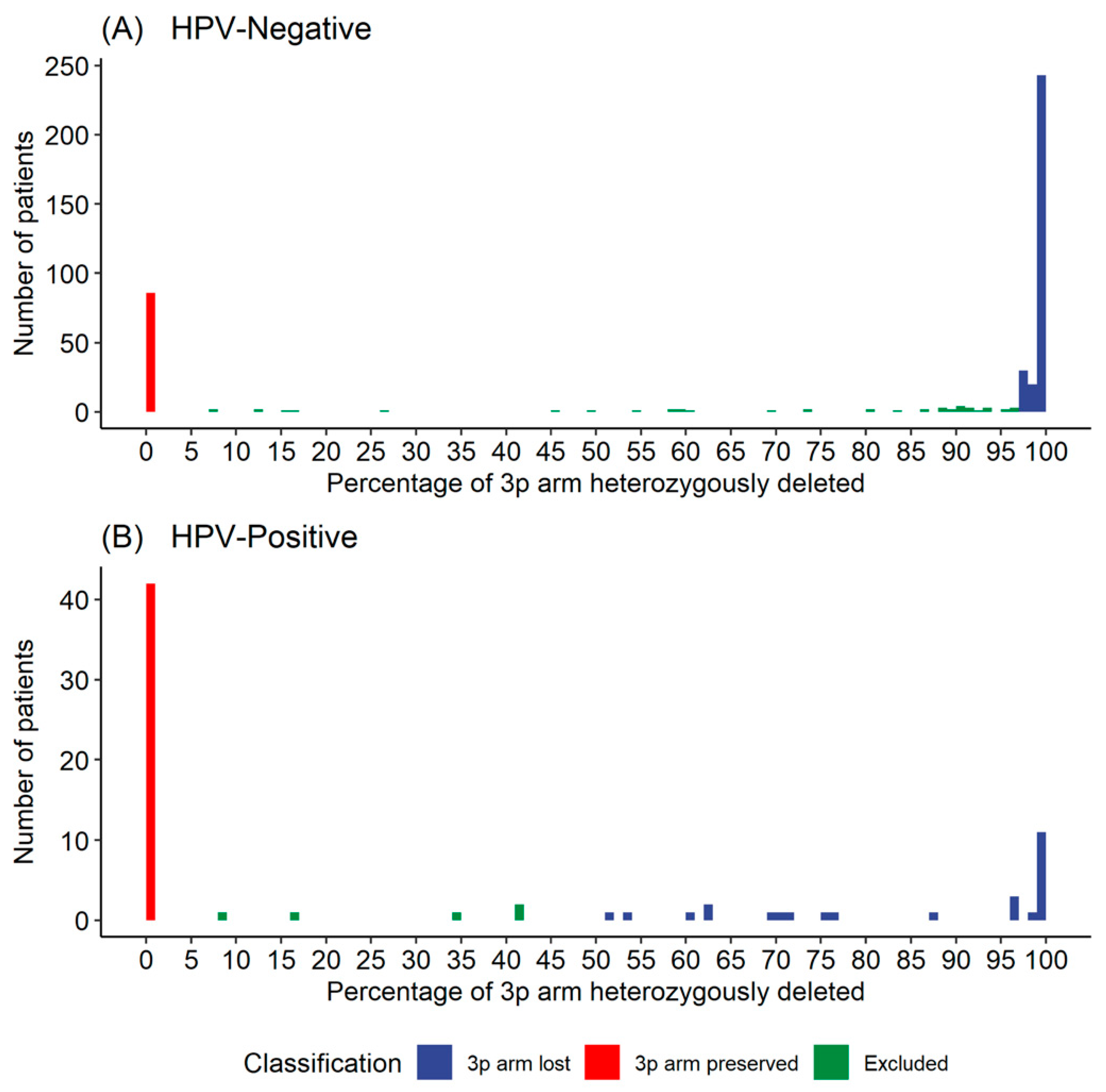
| HPV-Negative Samples, No. (%) (n = 375) | ||||
|---|---|---|---|---|
| Variables | Threshold Arm Lost (n = 293) | Threshold Arm Preserved (n = 82) | p-Value | |
| Age | Median (range) | 61 (19–84) | 64 (26–83) | 0.053 |
| Sex | Female | 69 (24) | 40 (49) | <10−4 |
| Male | 224 (76) | 42 (51) | ||
| Anatomical site | Oropharynx | 22 (8) | 1 (1) | <10−3 |
| Hypopharynx | 3 (1) | 1 (1) | ||
| Larynx | 82 (28) | 8 (10) | ||
| Oral cavity | 186 (63) | 72 (88) | ||
| Smoking history | Non-smoker | 51 (23) | 29 (41) | <10−3 |
| Light | 29 (13) | 14 (20) | ||
| Heavy | 144 (64) | 28 (39) | ||
| T category | T0-T2/TX | 112 (39) | 35 (45) | 0.30 |
| T3-T4 | 176 (61) | 42 (55) | ||
| N category | N0-N2a, NX | 176 (61) | 62 (81) | 0.0018 |
| N2b-N3 | 111 (39) | 15 (19) | ||
| Overall stage | I–III | 96 (37) | 38 (51) | 0.033 |
| IV | 165 (63) | 37 (49) | ||
| Adjuvant radiotherapy | No | 87 (34) | 32 (42) | 0.22 |
| Yes | 170 (66) | 44 (58) | ||
| Distant metastasis | No | 278 (95) | 82 (100) | 0.049 |
| Yes | 15 (5) | 0 (0) | ||
| HPV-Positive Samples, No. (%) (n = 66) | ||||
|---|---|---|---|---|
| Variables | Threshold Arm Lost (n = 26) | Threshold Arm Preserved (n = 40) | p-Value | |
| Age | Median (range) | 59 (40–82) | 56.5 (35–77) | 0.22 |
| Sex | Female | 2 (8) | 3 (8) | 1 |
| Male | 24 (92) | 37 (92) | ||
| Anatomical site | Oropharynx | 17 (65) | 31 (78) | 0.52 |
| Hypopharynx | 1 (4) | 1 (2) | ||
| Larynx | 1 (4) | 0 (0) | ||
| Oral cavity | 7 (27) | 8 (20) | ||
| Smoking history | Non-smoker | 6 (26) | 14 (41) | 0.015 |
| Light | 3 (13) | 12 (35) | ||
| Heavy | 14 (61) | 8 (24) | ||
| T category | T0-T2/TX | 16 (73) | 26 (76) | 0.76 |
| T3-T4 | 6 (27) | 8 (24) | ||
| N category | N0-N2a, NX | 17 (77) | 21 (64) | 0.38 |
| N2b-N3 | 5 (23) | 12 (36) | ||
| Overall stage | I–III | 6 (40) | 9 (38) | 1 |
| IV | 9 (60) | 15 (62) | ||
| Adjuvant radiotherapy | No | 8 (36) | 5 (14) | 0.055 |
| Yes | 14 (64) | 32 (86) | ||
| Distant metastasis | No | 25 (96) | 39 (98) | 1 |
| Yes | 1 (4) | 1 (2) | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.A.J.; Shaikh, M.H.; Lee, M.; Zeng, P.Y.F.; Sorgini, A.; Akintola, T.; Deng, X.; Jarycki, L.; Khan, H.; MacNeil, D.; et al. 3p Arm Loss and Survival in Head and Neck Cancer: An Analysis of TCGA Dataset. Cancers 2021, 13, 5313. https://doi.org/10.3390/cancers13215313
Kim HAJ, Shaikh MH, Lee M, Zeng PYF, Sorgini A, Akintola T, Deng X, Jarycki L, Khan H, MacNeil D, et al. 3p Arm Loss and Survival in Head and Neck Cancer: An Analysis of TCGA Dataset. Cancers. 2021; 13(21):5313. https://doi.org/10.3390/cancers13215313
Chicago/Turabian StyleKim, Hugh Andrew Jinwook, Mushfiq Hassan Shaikh, Mark Lee, Peter Y. F. Zeng, Alana Sorgini, Temitope Akintola, Xiaoxiao Deng, Laura Jarycki, Halema Khan, Danielle MacNeil, and et al. 2021. "3p Arm Loss and Survival in Head and Neck Cancer: An Analysis of TCGA Dataset" Cancers 13, no. 21: 5313. https://doi.org/10.3390/cancers13215313







