Single-Cell Proteomic Analysis Dissects the Complexity of Tumor Microenvironment in Muscle Invasive Bladder Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Results
2.1. TME Landscape in MIBC
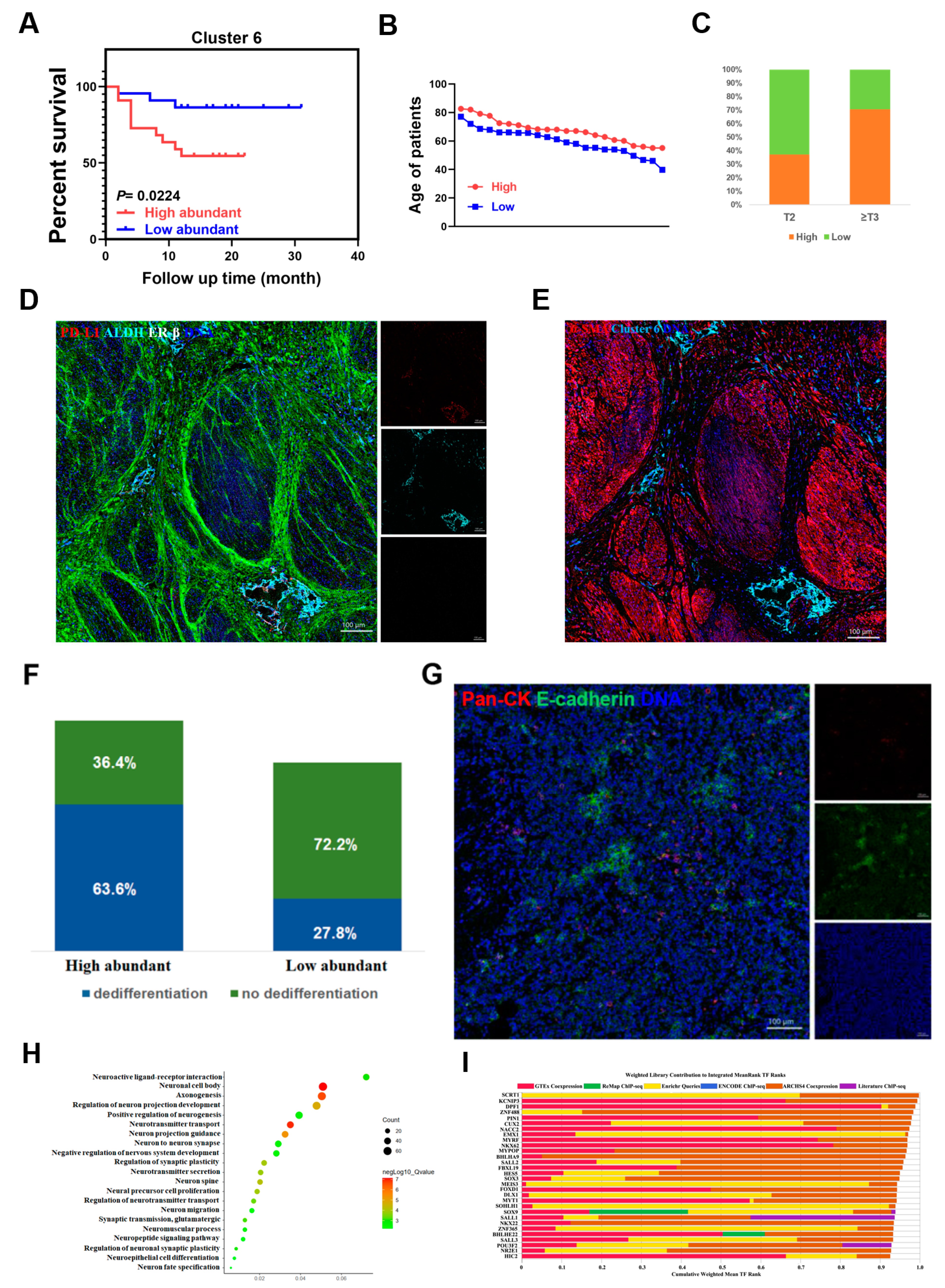
2.2. A Specific Cancer Stem-Like Cell Cluster Associates with Poor Prognosis
2.3. The Heterogeneous Phenotypes of Immune Cells in the MIBC TME
2.4. The Spatial Resolution-Based Phenotypes of the MIBC TME
3. Discussion
4. Materials and Methods
4.1. Patients and Samples
4.2. Cell Isolation
4.3. RNA-Sequencing
4.4. Antibodies and Antibody Labeling
4.5. Mass Cytometry
4.6. Imaging Mass Cytometry
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MIBC | muscle invasive bladder cancer |
| TME | tumor microenvironment |
| NAC | neoadjuvant chemotherapy |
| ConMC | consensus molecular classification |
| CyTOF | mass cytometry |
| IMC | imaging mass cytometry |
| CA | cancer |
| CP | para-carcinoma |
| EMT | epithelial–mesenchymal transition |
| IHC | immunohistochemical |
| H&E | hematoxylin-eosin |
| CSC | cancer stem cell |
| Tregs | regulatory T cells |
| scRNA-seq | single-cell RNA sequencing |
| AR | androgen receptor |
| BLCA | bladder urothelial carcinoma |
| RBC | red blood cell |
| PCA | principal component analysis |
| DEGs | differentially expressed genes |
| GO | gene ontology |
| t-SNE | t-distributed stochastic neighbor embedding |
| ROI | region of interest |
| TCGA | the cancer genome atlas |
| ECM | extra cellular matrix |
References
- Grayson, M. Bladder cancer. Nature 2017, 551, S33. [Google Scholar] [CrossRef] [Green Version]
- Antoni, S.; Ferlay, J.; Soerjomataram, I.; Znaor, A.; Jemal, A.; Bray, F. Bladder Cancer Incidence and Mortality: A Global Overview and Recent Trends. Eur. Urol. 2017, 71, 96–108. [Google Scholar] [CrossRef] [PubMed]
- Meeks, J.J.; Al-Ahmadie, H.; Faltas, B.M.; Taylor, J.A., 3rd; Flaig, T.W.; DeGraff, D.J.; Christensen, E.; Woolbright, B.L.; McConkey, D.J.; Dyrskjot, L. Genomic heterogeneity in bladder cancer: Challenges and possible solutions to improve outcomes. Nat. Rev. Urol. 2020, 17, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Prasad, S.M.; Decastro, G.J.; Steinberg, G.D.; Medscape. Urothelial carcinoma of the bladder: Definition, treatment and future efforts. Nat. Rev. Urol. 2011, 8, 631–642. [Google Scholar] [CrossRef] [PubMed]
- Gakis, G.; Efstathiou, J.; Lerner, S.P.; Cookson, M.S.; Keegan, K.A.; Guru, K.A.; Shipley, W.U.; Heidenreich, A.; Schoenberg, M.P.; Sagaloswky, A.I.; et al. ICUD-EAU International Consultation on Bladder Cancer 2012: Radical cystectomy and bladder preservation for muscle-invasive urothelial carcinoma of the bladder. Eur. Urol. 2013, 63, 45–57. [Google Scholar] [CrossRef] [PubMed]
- Zargar, H.; Espiritu, P.N.; Fairey, A.S.; Mertens, L.S.; Dinney, C.P.; Mir, M.C.; Krabbe, L.M.; Cookson, M.S.; Jacobsen, N.E.; Gandhi, N.M.; et al. Multicenter assessment of neoadjuvant chemotherapy for muscle-invasive bladder cancer. Eur. Urol. 2015, 67, 241–249. [Google Scholar] [CrossRef] [Green Version]
- Yuan, Y. Spatial Heterogeneity in the Tumor Microenvironment. Cold Spring Harb. Perspect. Med. 2016, 6, a026583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfannstiel, C.; Strissel, P.L.; Chiappinelli, K.B.; Sikic, D.; Wach, S.; Wirtz, R.M.; Wullweber, A.; Taubert, H.; Breyer, J.; Otto, W.; et al. The Tumor Immune Microenvironment Drives a Prognostic Relevance That Correlates with Bladder Cancer Subtypes. Cancer Immunol. Res. 2019, 7, 923–938. [Google Scholar] [CrossRef]
- Yuan, S.; Norgard, R.J.; Stanger, B.Z. Cellular Plasticity in Cancer. Cancer Discov. 2019, 9, 837–851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Da Silva-Diz, V.; Lorenzo-Sanz, L.; Bernat-Peguera, A.; Lopez-Cerda, M.; Munoz, P. Cancer cell plasticity: Impact on tumor progression and therapy response. Semin. Cancer Biol. 2018, 53, 48–58. [Google Scholar] [CrossRef]
- Gupta, P.B.; Pastushenko, I.; Skibinski, A.; Blanpain, C.; Kuperwasser, C. Phenotypic Plasticity: Driver of Cancer Initiation, Progression, and Therapy Resistance. Cell Stem Cell 2019, 24, 65–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lei, X.; Lei, Y.; Li, J.K.; Du, W.X.; Li, R.G.; Yang, J.; Li, J.; Li, F.; Tan, H.B. Immune cells within the tumor microenvironment: Biological functions and roles in cancer immunotherapy. Cancer Lett. 2020, 470, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Wang, Z.; Zeng, H.; Zhang, H.; Liu, Z.; Huang, Q.; Wang, J.; Chang, Y.; Bai, Q.; Liu, L.; et al. Identification and validation of poor prognosis immunoevasive subtype of muscle-invasive bladder cancer with tumor-infiltrating podoplanin(+) cell abundance. Oncoimmunology 2020, 9, 1747333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zarour, H.M. Reversing T-cell Dysfunction and Exhaustion in Cancer. Clin. Cancer Res. 2016, 22, 1856–1864. [Google Scholar] [CrossRef] [Green Version]
- Sakaguchi, S.; Mikami, N.; Wing, J.B.; Tanaka, A.; Ichiyama, K.; Ohkura, N. Regulatory T Cells and Human Disease. Annu. Rev. Immunol. 2020, 38, 541–566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Jiang, P.; Wei, S.; Xu, X.; Wang, J. Regulatory T cells in tumor microenvironment: New mechanisms, potential therapeutic strategies and future prospects. Mol. Cancer 2020, 19, 116. [Google Scholar] [CrossRef]
- Lee, H.W.; Chung, W.; Lee, H.O.; Jeong, D.E.; Jo, A.; Lim, J.E.; Hong, J.H.; Nam, D.H.; Jeong, B.C.; Park, S.H.; et al. Single-cell RNA sequencing reveals the tumor microenvironment and facilitates strategic choices to circumvent treatment failure in a chemorefractory bladder cancer patient. Genome Med. 2020, 12, 47. [Google Scholar] [CrossRef]
- Sfakianos, J.P.; Daza, J.; Hu, Y.; Anastos, H.; Bryant, G.; Bareja, R.; Badani, K.K.; Galsky, M.D.; Elemento, O.; Faltas, B.M.; et al. Epithelial plasticity can generate multi-lineage phenotypes in human and murine bladder cancers. Nat. Commun. 2020, 11, 2540. [Google Scholar] [CrossRef]
- Chen, Z.; Zhou, L.; Liu, L.; Hou, Y.; Xiong, M.; Yang, Y.; Hu, J.; Chen, K. Single-cell RNA sequencing highlights the role of inflammatory cancer-associated fibroblasts in bladder urothelial carcinoma. Nat. Commun. 2020, 11, 5077. [Google Scholar] [CrossRef]
- Wagner, J.; Rapsomaniki, M.A.; Chevrier, S.; Anzeneder, T.; Langwieder, C.; Dykgers, A.; Rees, M.; Ramaswamy, A.; Muenst, S.; Soysal, S.D.; et al. A Single-Cell Atlas of the Tumor and Immune Ecosystem of Human Breast Cancer. Cell 2019, 177, 1330–1345. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.J.; Traum, D.; Schug, J.; Gao, L.; Liu, C.; Consortium, H.; Atkinson, M.A.; Powers, A.C.; Feldman, M.D.; Naji, A.; et al. Multiplexed In Situ Imaging Mass Cytometry Analysis of the Human Endocrine Pancreas and Immune System in Type 1 Diabetes. Cell Metab. 2019, 29, 769–783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asakura, N.; Nakamura, N.; Muroi, A.; Nojima, Y.; Yamashita, T.; Kaneko, S.; Ikeda, K.; Koshikawa, N.; Suzuki, T. Expression of Cancer Stem Cell Markers EpCAM and CD90 Is Correlated with Anti- and Pro-Oncogenic EphA2 Signaling in Hepatocellular Carcinoma. Int. J. Mol. Sci. 2021, 22, 8652. [Google Scholar] [CrossRef]
- Elbadawy, M.; Hayashi, K.; Ayame, H.; Ishihara, Y.; Abugomaa, A.; Shibutani, M.; Hayashi, S.M.; Hazama, S.; Takenouchi, H.; Nakajima, M.; et al. Anti-cancer activity of amorphous curcumin preparation in patient-derived colorectal cancer organoids. Biomed. Pharmacother. 2021, 142, 112043. [Google Scholar] [CrossRef]
- Vassalli, G. Aldehyde Dehydrogenases: Not Just Markers, but Functional Regulators of Stem Cells. Stem Cells Int. 2019, 2019, 3904645. [Google Scholar] [CrossRef] [Green Version]
- Su, Y.; Qiu, Q.; Zhang, X.; Jiang, Z.; Leng, Q.; Liu, Z.; Stass, S.A.; Jiang, F. Aldehyde dehydrogenase 1 A1-positive cell population is enriched in tumor-initiating cells and associated with progression of bladder cancer. Cancer Epidemiol. Biomark. Prev. 2010, 19, 327–337. [Google Scholar] [CrossRef] [Green Version]
- Lugli, A.; Zlobec, I.; Berger, M.D.; Kirsch, R.; Nagtegaal, I.D. Tumour budding in solid cancers. Nat. Rev. Clin. Oncol. 2021, 18, 101–115. [Google Scholar] [CrossRef] [PubMed]
- Lauber, C.; Klink, B.; Seifert, M. Comparative analysis of histologically classified oligodendrogliomas reveals characteristic molecular differences between subgroups. BMC Cancer 2018, 18, 399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biswas, S.; Chung, S.H.; Jiang, P.; Dehghan, S.; Deng, W. Development of glial restricted human neural stem cells for oligodendrocyte differentiation in vitro and in vivo. Sci. Rep. 2019, 9, 9013. [Google Scholar] [CrossRef]
- Chatron, N.; Moller, R.S.; Champaigne, N.L.; Schneider, A.L.; Kuechler, A.; Labalme, A.; Simonet, T.; Baggett, L.; Bardel, C.; Kamsteeg, E.J.; et al. The epilepsy phenotypic spectrum associated with a recurrent CUX2 variant. Ann. Neurol. 2018, 83, 926–934. [Google Scholar] [CrossRef]
- Kobeissy, F.H.; Hansen, K.; Neumann, M.; Fu, S.; Jin, K.; Liu, J. Deciphering the Role of Emx1 in Neurogenesis: A Neuroproteomics Approach. Front. Mol. Neurosci. 2016, 9, 98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.; Choi, J.O.; Fan, C.; Shearer, R.S.; Sharif, M.; Busch, P.; Park, Y. Homo-trimerization is essential for the transcription factor function of Myrf for oligodendrocyte differentiation. Nucleic Acids Res. 2017, 45, 5112–5125. [Google Scholar] [CrossRef] [Green Version]
- Villanueva-Chimal, E.; Salinas, L.S.; Fernandez-Cardenas, L.P.; Huelgas-Morales, G.; Cabrera-Wrooman, A.; Navarro, R.E. DPFF-1 transcription factor deficiency causes the aberrant activation of MPK-1 and meiotic defects in the Caenorhabditis elegans germline. Genesis 2017, 55, e23072. [Google Scholar] [CrossRef]
- Fagiani, F.; Govoni, S.; Racchi, M.; Lanni, C. The Peptidyl-prolyl Isomerase Pin1 in Neuronal Signaling: From Neurodevelopment to Neurodegeneration. Mol. Neurobiol. 2021, 58, 1062–1073. [Google Scholar] [CrossRef]
- Liang, Y.J.; Ding, Y.; Levery, S.B.; Lobaton, M.; Handa, K.; Hakomori, S.I. Differential expression profiles of glycosphingolipids in human breast cancer stem cells vs. cancer non-stem cells. Proc. Natl. Acad. Sci. USA 2013, 110, 4968–4973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wingerter, A.; El Malki, K.; Sandhoff, R.; Seidmann, L.; Wagner, D.C.; Lehmann, N.; Vewinger, N.; Frauenknecht, K.B.M.; Sommer, C.J.; Traub, F.; et al. Exploiting Gangliosides for the Therapy of Ewing’s Sarcoma and H3K27M-Mutant Diffuse Midline Glioma. Cancers 2021, 13, 520. [Google Scholar] [CrossRef] [PubMed]
- Skotland, T.; Hessvik, N.P.; Sandvig, K.; Llorente, A. Exosomal lipid composition and the role of ether lipids and phosphoinositides in exosome biology. J. Lipid Res. 2019, 60, 9–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lambrechts, D.; Wauters, E.; Boeckx, B.; Aibar, S.; Nittner, D.; Burton, O.; Bassez, A.; Decaluwe, H.; Pircher, A.; Van den Eynde, K.; et al. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat. Med. 2018, 24, 1277–1289. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, aau6977. [Google Scholar] [CrossRef] [PubMed]
- Ono, K.; Eguchi, T.; Sogawa, C.; Calderwood, S.K.; Futagawa, J.; Kasai, T.; Seno, M.; Okamoto, K.; Sasaki, A.; Kozaki, K.I. HSP-enriched properties of extracellular vesicles involve survival of metastatic oral cancer cells. J. Cell. Biochem. 2018, 119, 7350–7362. [Google Scholar] [CrossRef] [Green Version]
- Sheng, J.; Zhang, J.; Wang, L.; Tano, V.; Tang, J.; Wang, X.; Wu, J.; Song, J.; Zhao, Y.; Rong, J.; et al. Topological analysis of hepatocellular carcinoma tumour microenvironment based on imaging mass cytometry reveals cellular neighbourhood regulated reversely by macrophages with different ontogeny. Gut 2021. [Google Scholar] [CrossRef]
- Tanoue, T.; Morita, S.; Plichta, D.R.; Skelly, A.N.; Suda, W.; Sugiura, Y.; Narushima, S.; Vlamakis, H.; Motoo, I.; Sugita, K.; et al. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature 2019, 565, 600–605. [Google Scholar] [CrossRef]
- Ahrends, T.; Spanjaard, A.; Pilzecker, B.; Babala, N.; Bovens, A.; Xiao, Y.; Jacobs, H.; Borst, J. CD4(+) T Cell Help Confers a Cytotoxic T Cell Effector Program Including Coinhibitory Receptor Downregulation and Increased Tissue Invasiveness. Immunity 2017, 47, 848–861.e5. [Google Scholar] [CrossRef] [Green Version]
- Sade-Feldman, M.; Yizhak, K.; Bjorgaard, S.L.; Ray, J.P.; de Boer, C.G.; Jenkins, R.W.; Lieb, D.J.; Chen, J.H.; Frederick, D.T.; Barzily-Rokni, M.; et al. Defining T Cell States Associated with Response to Checkpoint Immunotherapy in Melanoma. Cell 2018, 175, 998–1013.e20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Provenzano, E.; Ulaner, G.A.; Chin, S.F. Molecular Classification of Breast Cancer. PET Clin. 2018, 13, 325–338. [Google Scholar] [CrossRef]
- Kamoun, A.; de Reynies, A.; Allory, Y.; Sjodahl, G.; Robertson, A.G.; Seiler, R.; Hoadley, K.A.; Groeneveld, C.S.; Al-Ahmadie, H.; Choi, W.; et al. A Consensus Molecular Classification of Muscle-invasive Bladder Cancer. Eur. Urol. 2020, 77, 420–433. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Teng, H.; Liu, M.; Liu, B.; Zhang, D.; Xu, Z.; Wang, Y.; Zhou, H. Prognostic Value of Immune-Related Genes in the Tumor Microenvironment of Bladder Cancer. Front. Oncol. 2020, 10, 1302. [Google Scholar] [CrossRef] [PubMed]
- Zlobec, I.; Lugli, A. Tumour budding in colorectal cancer: Molecular rationale for clinical translation. Nat. Rev. Cancer 2018, 18, 203–204. [Google Scholar] [CrossRef] [PubMed]
- Karamitopoulou, E.; Wartenberg, M.; Zlobec, I.; Cibin, S.; Worni, M.; Gloor, B.; Lugli, A. Tumour budding in pancreatic cancer revisited: Validation of the ITBCC scoring system. Histopathology 2018, 73, 137–146. [Google Scholar] [CrossRef]
- Kemi, N.; Eskuri, M.; Ikalainen, J.; Karttunen, T.J.; Kauppila, J.H. Tumor Budding and Prognosis in Gastric Adenocarcinoma. Am. J. Surg. Pathol. 2019, 43, 229–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukumoto, K.; Kikuchi, E.; Mikami, S.; Ogihara, K.; Matsumoto, K.; Miyajima, A.; Oya, M. Tumor budding, a novel prognostic indicator for predicting stage progression in T1 bladder cancers. Cancer Sci. 2016, 107, 1338–1344. [Google Scholar] [CrossRef]
- Faulkner, S.; Jobling, P.; March, B.; Jiang, C.C.; Hondermarck, H. Tumor Neurobiology and the War of Nerves in Cancer. Cancer Discov. 2019, 9, 702–710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hutchings, C.; Phillips, J.A.; Djamgoz, M.B.A. Nerve input to tumours: Pathophysiological consequences of a dynamic relationship. Biochim. Biophys. Acta Rev. Cancer 2020, 1874, 188411. [Google Scholar] [CrossRef]
- Jung, E.; Alfonso, J.; Monyer, H.; Wick, W.; Winkler, F. Neuronal signatures in cancer. Int. J. Cancer 2020, 147, 3281–3291. [Google Scholar] [CrossRef] [PubMed]
- Fischer-Valuck, B.W.; Rao, Y.J.; Henke, L.E.; Rudra, S.; Hui, C.; Baumann, B.C.; Gay, H.A.; Michalski, J.M. Treatment Patterns and Survival Outcomes for Patients with Small Cell Carcinoma of the Bladder. Eur. Urol. Focus 2018, 4, 900–906. [Google Scholar] [CrossRef]
- Moschini, M.; D’Andrea, D.; Korn, S.; Irmak, Y.; Soria, F.; Comperat, E.; Shariat, S.F. Characteristics and clinical significance of histological variants of bladder cancer. Nat. Rev. Urol. 2017, 14, 651–668. [Google Scholar] [CrossRef] [PubMed]
- Long, X.; Xiong, W.; Zeng, X.; Qi, L.; Cai, Y.; Mo, M.; Jiang, H.; Zhu, B.; Chen, Z.; Li, Y. Cancer-associated fibroblasts promote cisplatin resistance in bladder cancer cells by increasing IGF-1/ERbeta/Bcl-2 signalling. Cell Death Dis. 2019, 10, 375. [Google Scholar] [CrossRef]
- Goulet, C.R.; Champagne, A.; Bernard, G.; Vandal, D.; Chabaud, S.; Pouliot, F.; Bolduc, S. Cancer-associated fibroblasts induce epithelial-mesenchymal transition of bladder cancer cells through paracrine IL-6 signalling. BMC Cancer 2019, 19, 137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lauwers, E.; Wang, Y.C.; Gallardo, R.; Van der Kant, R.; Michiels, E.; Swerts, J.; Baatsen, P.; Zaiter, S.S.; McAlpine, S.R.; Gounko, N.V.; et al. Hsp90 Mediates Membrane Deformation and Exosome Release. Mol. Cell 2018, 71, 689–702.e9. [Google Scholar] [CrossRef] [Green Version]
- Madeo, M.; Colbert, P.L.; Vermeer, D.W.; Lucido, C.T.; Cain, J.T.; Vichaya, E.G.; Grossberg, A.J.; Muirhead, D.; Rickel, A.P.; Hong, Z.; et al. Cancer exosomes induce tumor innervation. Nat. Commun. 2018, 9, 4284. [Google Scholar] [CrossRef] [Green Version]
- Azizi, E.; Carr, A.J.; Plitas, G.; Cornish, A.E.; Konopacki, C.; Prabhakaran, S.; Nainys, J.; Wu, K.; Kiseliovas, V.; Setty, M.; et al. Single-Cell Map of Diverse Immune Phenotypes in the Breast Tumor Microenvironment. Cell 2018, 174, 1293–1308.e36. [Google Scholar] [CrossRef] [Green Version]
- Lamballe, F.; Ahmad, F.; Vinik, Y.; Castellanet, O.; Daian, F.; Muller, A.K.; Kohler, U.A.; Bailly, A.L.; Josselin, E.; Castellano, R.; et al. Modeling Heterogeneity of Triple-Negative Breast Cancer Uncovers a Novel Combinatorial Treatment Overcoming Primary Drug Resistance. Adv. Sci. 2021, 8, 2003049. [Google Scholar] [CrossRef]
- Huang, K.K.; Huang, J.; Wu, J.K.L.; Lee, M.; Tay, S.T.; Kumar, V.; Ramnarayanan, K.; Padmanabhan, N.; Xu, C.; Tan, A.L.K.; et al. Long-read transcriptome sequencing reveals abundant promoter diversity in distinct molecular subtypes of gastric cancer. Genome Biol. 2021, 22, 44. [Google Scholar] [CrossRef]
- Arneth, B. Tumor Microenvironment. Medicina 2019, 56, 15. [Google Scholar] [CrossRef] [Green Version]
- Desbois, M.; Udyavar, A.R.; Ryner, L.; Kozlowski, C.; Guan, Y.; Durrbaum, M.; Lu, S.; Fortin, J.P.; Koeppen, H.; Ziai, J.; et al. Integrated digital pathology and transcriptome analysis identifies molecular mediators of T-cell exclusion in ovarian cancer. Nat. Commun. 2020, 11, 5583. [Google Scholar] [CrossRef]
- Li, J.; Stanger, B.Z. How Tumor Cell Dedifferentiation Drives Immune Evasion and Resistance to Immunotherapy. Cancer Res. 2020, 80, 4037–4041. [Google Scholar] [CrossRef] [PubMed]
- Provenzano, P.P.; Eliceiri, K.W.; Campbell, J.M.; Inman, D.R.; White, J.G.; Keely, P.J. Collagen reorganization at the tumor-stromal interface facilitates local invasion. BMC Med. 2006, 4, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xi, G.; Guo, W.; Kang, D.; Ma, J.; Fu, F.; Qiu, L.; Zheng, L.; He, J.; Fang, N.; Chen, J.; et al. Large-scale tumor-associated collagen signatures identify high-risk breast cancer patients. Theranostics 2021, 11, 3229–3243. [Google Scholar] [CrossRef]
- Negri, S.; Wang, Y.; Sono, T.; Lee, S.; Hsu, G.C.; Xu, J.; Meyers, C.A.; Qin, Q.; Broderick, K.; Witwer, K.W.; et al. Human perivascular stem cells prevent bone graft resorption in osteoporotic contexts by inhibiting osteoclast formation. Stem Cells Transl. Med. 2020, 9, 1617–1630. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [Green Version]
- Keenan, A.B.; Torre, D.; Lachmann, A.; Leong, A.K.; Wojciechowicz, M.L.; Utti, V.; Jagodnik, K.M.; Kropiwnicki, E.; Wang, Z.; Ma’ayan, A. ChEA3: Transcription factor enrichment analysis by orthogonal omics integration. Nucleic Acids Res. 2019, 47, W212–W224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perdiguero, P.; Morel, E.; Tafalla, C. Diversity of Rainbow Trout Blood B Cells Revealed by Single Cell RNA Sequencing. Biology 2021, 10, 511. [Google Scholar] [CrossRef]
- Lai, L.; Ong, R.; Li, J.; Albani, S. A CD45-based barcoding approach to multiplex mass-cytometry (CyTOF). Cytometry A 2015, 87, 369–374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chew, V.; Lai, L.; Pan, L.; Lim, C.J.; Li, J.; Ong, R.; Chua, C.; Leong, J.Y.; Lim, K.H.; Toh, H.C.; et al. Delineation of an immunosuppressive gradient in hepatocellular carcinoma using high-dimensional proteomic and transcriptomic analyses. Proc. Natl. Acad. Sci. USA 2017, 114, E5900–E5909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Lau, M.C.; Wong, M.T.; Newell, E.W.; Poidinger, M.; Chen, J. Cytofkit: A Bioconductor Package for an Integrated Mass Cytometry Data Analysis Pipeline. PLoS Comput. Biol. 2016, 12, e1005112. [Google Scholar] [CrossRef]
- De Vries, N.L.; van Unen, V.; Ijsselsteijn, M.E.; Abdelaal, T.; van der Breggen, R.; Sarasqueta, A.F.; Mahfouz, A.; Peeters, K.; Hollt, T.; Lelieveldt, B.P.F.; et al. High-dimensional cytometric analysis of colorectal cancer reveals novel mediators of antitumour immunity. Gut 2020, 69, 691–703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, T.R.; Kang, I.H.; Wheeler, D.B.; Lindquist, R.A.; Papallo, A.; Sabatini, D.M.; Golland, P.; Carpenter, A.E. CellProfiler Analyst: Data exploration and analysis software for complex image-based screens. BMC Bioinform. 2008, 9, 482. [Google Scholar] [CrossRef] [Green Version]
- Schapiro, D.; Jackson, H.W.; Raghuraman, S.; Fischer, J.R.; Zanotelli, V.R.T.; Schulz, D.; Giesen, C.; Catena, R.; Varga, Z.; Bodenmiller, B. histoCAT: Analysis of cell phenotypes and interactions in multiplex image cytometry data. Nat. Methods 2017, 14, 873–876. [Google Scholar] [CrossRef] [Green Version]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, C.; Wang, X.; Tao, Y.; Xie, Y.; Lai, Z.; Li, Z.; Hu, J.; Tang, S.; Pan, L.; He, L.; et al. Single-Cell Proteomic Analysis Dissects the Complexity of Tumor Microenvironment in Muscle Invasive Bladder Cancer. Cancers 2021, 13, 5440. https://doi.org/10.3390/cancers13215440
Feng C, Wang X, Tao Y, Xie Y, Lai Z, Li Z, Hu J, Tang S, Pan L, He L, et al. Single-Cell Proteomic Analysis Dissects the Complexity of Tumor Microenvironment in Muscle Invasive Bladder Cancer. Cancers. 2021; 13(21):5440. https://doi.org/10.3390/cancers13215440
Chicago/Turabian StyleFeng, Chao, Xi Wang, Yuting Tao, Yuanliang Xie, Zhiyong Lai, Zhijian Li, Jiaxin Hu, Shaomei Tang, Lixin Pan, Liangyu He, and et al. 2021. "Single-Cell Proteomic Analysis Dissects the Complexity of Tumor Microenvironment in Muscle Invasive Bladder Cancer" Cancers 13, no. 21: 5440. https://doi.org/10.3390/cancers13215440





