CD95L Inhibition Impacts Gemcitabine-Mediated Effects and Non-Apoptotic Signaling of TNF-α and TRAIL in Pancreatic Tumor Cells
Abstract
Simple Summary
Abstract
- (1)
- The CD95/CD95L system plays an important role in non-apoptotic signaling;
- (2)
- The CD95/CD95L system influences TNF-α/TRAIL-induced pro-inflammatory signaling;
- (3)
- The blockade of CD95L may enhance the chemotherapeutic effects of gemcitabine by lowering the drug-induced pro-inflammatory signaling;
- (4)
- The respective inhibitor sCD95Fc reduces in vivo pancreatic tumor growth in an adjuvant setting.
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Isolation of Total Cellular RNA
2.3. Synthesis of Complementary DNA (cDNA)
2.4. PerfectProbe Real Time PCR Assay
2.5. Cell Surface Protein Analysis by Flow Cytometry
2.6. WST-1 Proliferation Assay
2.7. Analysis of Pancreatic Tumor Cell Conditioned Media by IL8 ELISA
2.8. Laboratory Animals
2.9. Orthotopic Xenotransplantation of Human PDAC Cells and Treatment in the Palliative Setting
2.10. Adjuvant Setting: Relaparotomy, Tumor Resection and Treatment
2.11. Tissue Staining by Immunohistochemistry
2.11.1. Cryo-Tissues
2.11.2. FFPE Material
2.12. Statistical Analysis
3. Results
3.1. Expression of CD95 and CD95L in Pancreatic Tumor Cell Lines
3.2. Impact of CD95L Inhibition on the Gemcitabine Induced Inflammatory Responses
3.3. Inhibition of CD95L Exerts Synergistic Effects on the Gemcitabine Induced Cell Death of Pancreatic Tumor Cells
3.4. Effects of Death Ligands on the Cell Proliferation of PDAC Cells
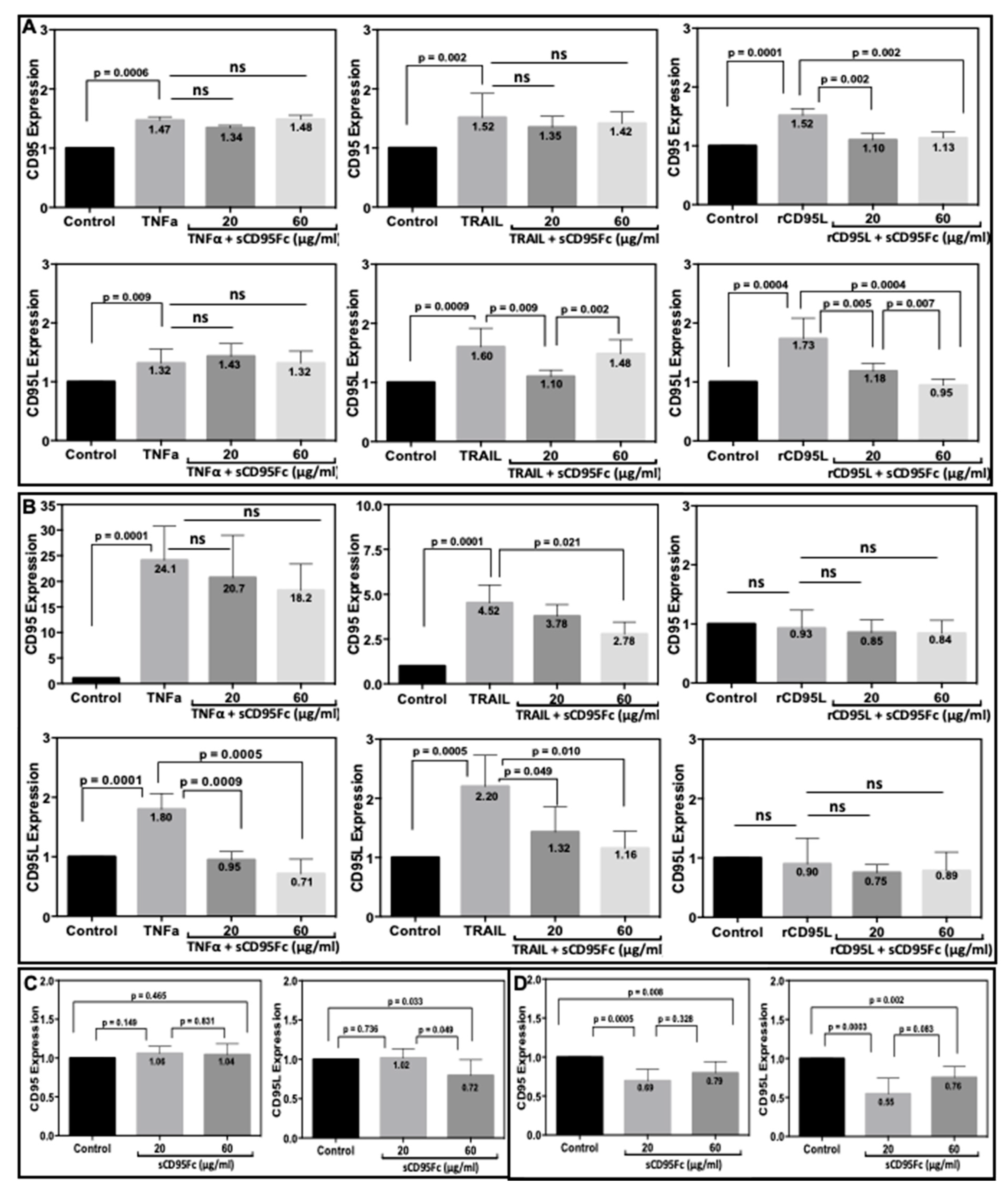
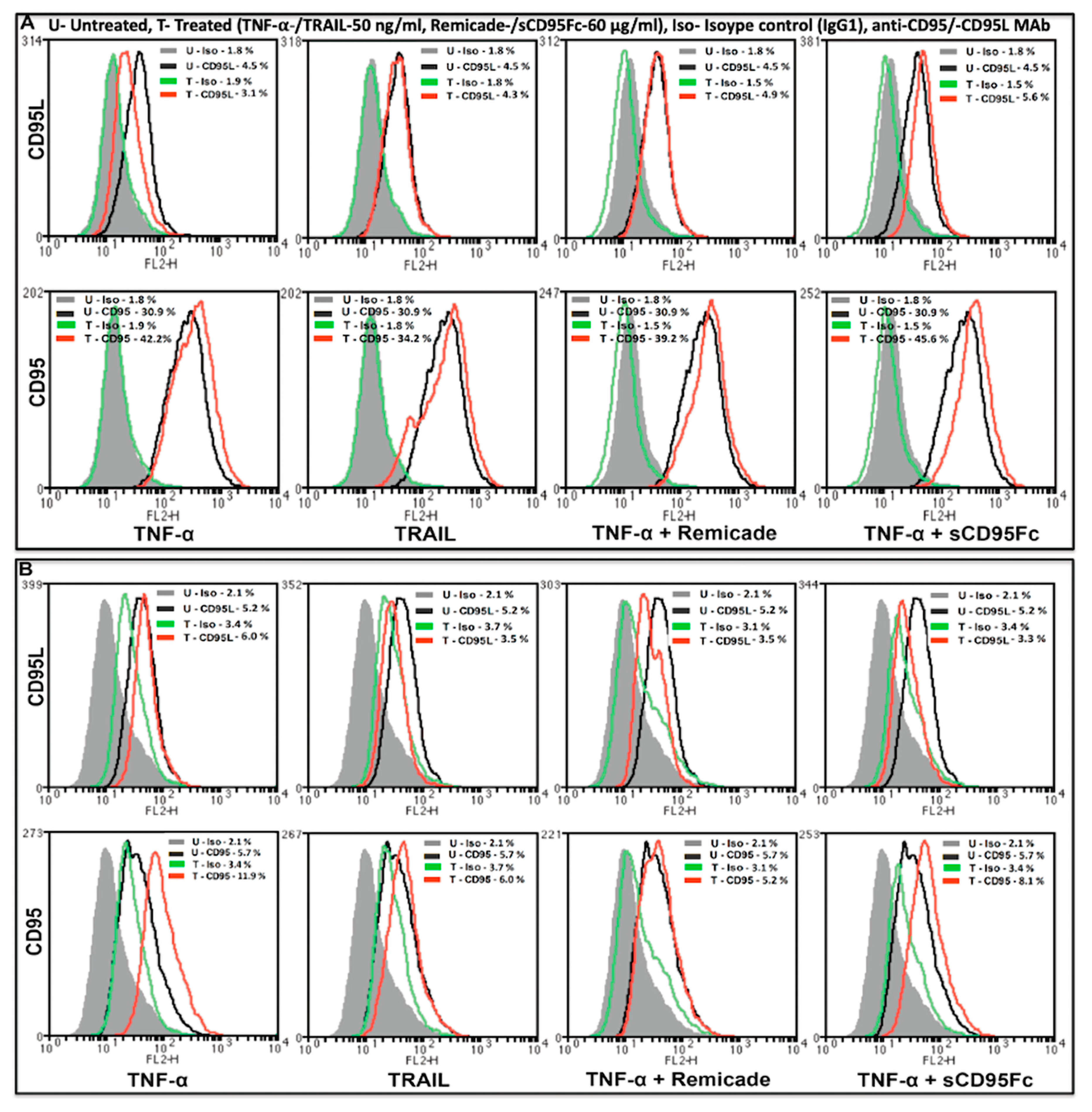
3.5. Effect of CD95L Inhibition on the Expression of CD95 and CD95L Induced upon TNF-α, TRAIL and rCD95L Stimulation
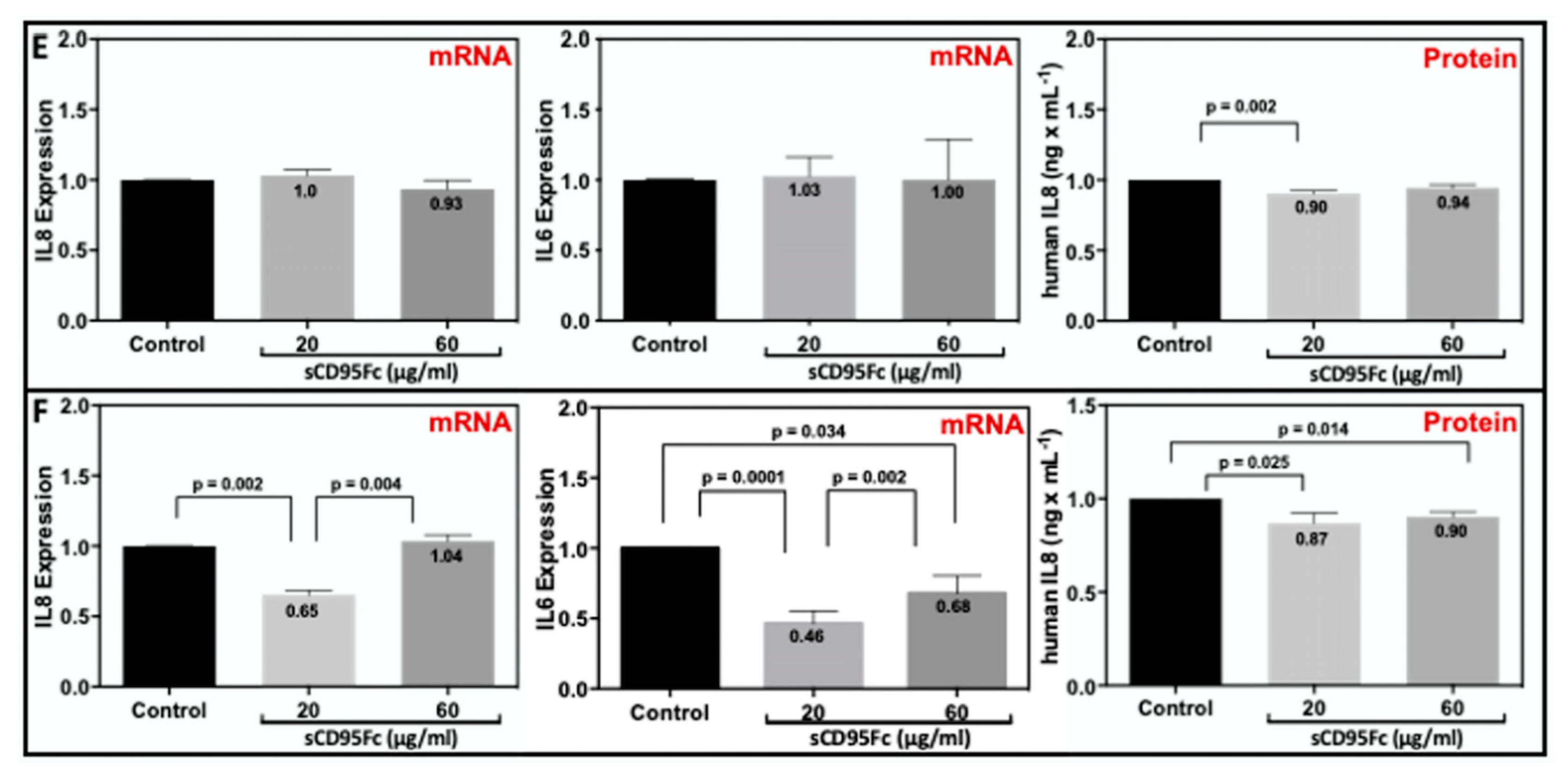
3.6. Effect of CD95L Inhibition on the Expression of IL8 and IL6 Induced upon TNF-α, TRAIL and CD95L Stimulation
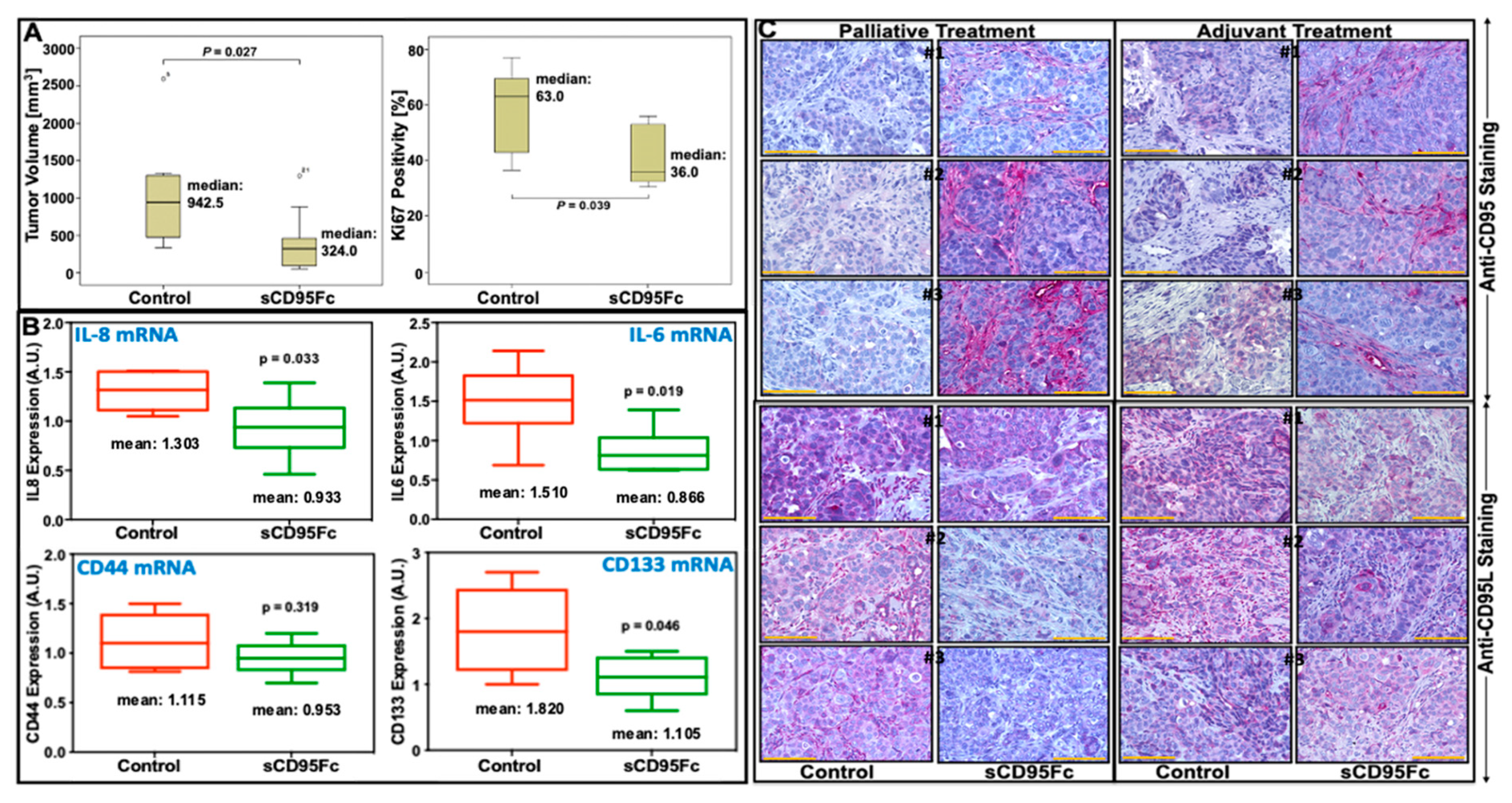
3.7. In Vivo Effects of CD95L Inhibition in Pancreatic Xenotransplant Models
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McGuigan, A.; Kelly, P.; Turkington, R.C.; Jones, C.; Coleman, H.G.; McCain, R.S. Pancreatic cancer: A review of clinical diagnosis, epidemiology, treatment and outcomes. World J. Gastroenterol. 2018, 24, 4846–4861. [Google Scholar] [CrossRef] [PubMed]
- Ling, Q.; Kalthoff, H. Transportome Malfunctions and the Hallmarks of Pancreatic Cancer. In Reviews of Physiology, Biochemistry and Pharmacology; Springer: Berlin/Heidelberg, Germany, 2020. [Google Scholar] [CrossRef]
- Ellenrieder, V.; Konig, A.; Seufferlein, T. Current Standard and Future Perspectives in First- and Second-Line Treatment of Metastatic Pancreatic Adenocarcinoma. Digestion 2016, 94, 44–49. [Google Scholar] [CrossRef] [PubMed]
- Binenbaum, Y.; Na’ara, S.; Gil, Z. Gemcitabine resistance in pancreatic ductal adenocarcinoma. Drug Resist. Updates 2015, 23, 55–68. [Google Scholar] [CrossRef]
- Fujiwara, Y.; Shiba, H.; Iwase, R.; Haruki, K.; Furukawa, K.; Uwagawa, T.; Misawa, T.; Ohashi, T.; Yanaga, K. Inhibition of nuclear factor kappa-B enhances the antitumor effect of combination treatment with tumor necrosis factor-alpha gene therapy and gemcitabine for pancreatic cancer in mice. J. Am. Coll. Surg. 2013, 216, 320–332.e3. [Google Scholar] [CrossRef]
- Harwood, F.G.; Kasibhatla, S.; Petak, I.; Vernes, R.; Green, D.R.; Houghton, J.A. Regulation of FasL by NF-kappaB and AP-1 in Fas-dependent thymineless death of human colon carcinoma cells. J. Biol. Chem. 2000, 275, 10023–10029. [Google Scholar] [CrossRef]
- Friesen, C.; Fulda, S.; Debatin, K.M. Cytotoxic drugs and the CD95 pathway. Leukemia 1999, 13, 1854–1858. [Google Scholar] [CrossRef] [PubMed]
- Quint, K.; Tonigold, M.; Di Fazio, P.; Montalbano, R.; Lingelbach, S.; Ruckert, F.; Alinger, B.; Ocker, M.; Neureiter, D. Pancreatic cancer cells surviving gemcitabine treatment express markers of stem cell differentiation and epithelial-mesenchymal transition. Int. J. Oncol. 2012, 41, 2093–2102. [Google Scholar] [CrossRef]
- Song, Y.; Baba, T.; Li, Y.Y.; Furukawa, K.; Tanabe, Y.; Matsugo, S.; Sasaki, S.; Mukaida, N. Gemcitabine-induced CXCL8 expression counteracts its actions by inducing tumor neovascularization. Biochem. Biophys. Res. Commun. 2015, 458, 341–346. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, Y.; Wang, W.; Zhu, Y.; Chen, Y.; Tian, B. Gemcitabine treatment induces endoplasmic reticular (ER) stress and subsequently upregulates urokinase plasminogen activator (uPA) to block mitochondrial-dependent apoptosis in Panc-1 cancer stem-like cells (CSCs). PLoS ONE 2017, 12, e0184110. [Google Scholar] [CrossRef]
- Rübe, C.E.; Wilfert, F.; Uthe, D.; König, J.; Liu, L.; Schuck, A.; Willich, N.; Remberger, K.; Rübe, C. Increased expression of pro-inflammatory cytokines as a cause of lung toxicity after combined treatment with gemcitabine and thoracic irradiation. Radiother. Oncol. 2004, 72, 231–241. [Google Scholar] [CrossRef]
- Adamska, A.; Elaskalani, O.; Emmanouilidi, A.; Kim, M.; Abdol Razak, N.B.; Metharom, P.; Falasca, M. Molecular and cellular mechanisms of chemoresistance in pancreatic cancer. Adv. Biol. Regul. 2018, 68, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Quinonero, F.; Mesas, C.; Doello, K.; Cabeza, L.; Perazzoli, G.; Jimenez-Luna, C.; Rama, A.R.; Melguizo, C.; Prados, J. The challenge of drug resistance in pancreatic ductal adenocarcinoma: A current overview. Cancer Biol. Med. 2019, 16, 688–699. [Google Scholar] [CrossRef]
- Trauth, B.C.; Klas, C.; Peters, A.M.; Matzku, S.; Moller, P.; Falk, W.; Debatin, K.M.; Krammer, P.H. Monoclonal antibody-mediated tumor regression by induction of apoptosis. Science 1989, 245, 301–305. [Google Scholar] [CrossRef]
- Choi, C.; Xu, X.; Oh, J.W.; Lee, S.J.; Gillespie, G.Y.; Park, H.; Jo, H.; Benveniste, E.N. Fas-induced expression of chemokines in human glioma cells: Involvement of extracellular signal-regulated kinase 1/2 and p38 mitogen-activated protein kinase. Cancer Res. 2001, 61, 3084–3091. [Google Scholar] [PubMed]
- Desbarats, J.; Birge, R.B.; Mimouni-Rongy, M.; Weinstein, D.E.; Palerme, J.-S.; Newell, M.K. Fas engagement induces neurite growth through ERK activation and p35 upregulation. Nat. Cell Biol. 2003, 5, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Kleber, S.; Sancho-Martinez, I.; Wiestler, B.; Beisel, A.; Gieffers, C.; Hill, O.; Thiemann, M.; Mueller, W.; Sykora, J.; Kuhn, A.; et al. Yes and PI3K bind CD95 to signal invasion of glioblastoma. Cancer Cell 2008, 13, 235–248. [Google Scholar] [CrossRef] [PubMed]
- Ponton, A.; Clement, M.V.; Stamenkovic, I. The CD95 (APO-1/Fas) receptor activates NF-kappaB independently of its cytotoxic function. J. Biol. Chem. 1996, 271, 8991–8995. [Google Scholar] [CrossRef] [PubMed]
- Steller, E.J.; Ritsma, L.; Raats, D.A.; Hoogwater, F.J.; Emmink, B.L.; Govaert, K.M.; Laoukili, J.; Rinkes, I.H.; van Rheenen, J.; Kranenburg, O. The death receptor CD95 activates the cofilin pathway to stimulate tumour cell invasion. EMBO Rep. 2011, 12, 931–937. [Google Scholar] [CrossRef]
- Toyoshima, F.; Moriguchi, T.; Nishida, E. Fas induces cytoplasmic apoptotic responses and activation of the MKK7-JNK/SAPK and MKK6-p38 pathways independent of CPP32-like proteases. J. Cell Biol. 1997, 139, 1005–1015. [Google Scholar] [CrossRef]
- Trauzold, A.; Roder, C.; Sipos, B.; Karsten, K.; Arlt, A.; Jiang, P.; Martin-Subero, J.I.; Siegmund, D.; Muerkoster, S.; Pagerols-Raluy, L.; et al. CD95 and TRAF2 promote invasiveness of pancreatic cancer cells. FASEB J. 2005, 19, 620–622. [Google Scholar] [CrossRef]
- Ceppi, P.; Hadji, A.; Kohlhapp, F.J.; Pattanayak, A.; Hau, A.; Liu, X.; Liu, H.; Murmann, A.E.; Peter, M.E. CD95 and CD95L promote and protect cancer stem cells. Nat. Commun. 2014, 5, 5238. [Google Scholar] [CrossRef]
- Egberts, J.H.; Cloosters, V.; Noack, A.; Schniewind, B.; Thon, L.; Klose, S.; Kettler, B.; von Forstner, C.; Kneitz, C.; Tepel, J.; et al. Anti-tumor necrosis factor therapy inhibits pancreatic tumor growth and metastasis. Cancer Res. 2008, 68, 1443–1450. [Google Scholar] [CrossRef]
- Ishimura, N.; Isomoto, H.; Bronk, S.F.; Gores, G.J. Trail induces cell migration and invasion in apoptosis-resistant cholangiocarcinoma cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2006, 290, G129–G136. [Google Scholar] [CrossRef] [PubMed]
- Leibovich, S.J.; Polverini, P.J.; Shepard, H.M.; Wiseman, D.M.; Shively, V.; Nuseir, N. Macrophage-induced angiogenesis is mediated by tumour necrosis factor-α. Nature 1987, 329, 630–632. [Google Scholar] [CrossRef]
- Trauzold, A.; Siegmund, D.; Schniewind, B.; Sipos, B.; Egberts, J.; Zorenkov, D.; Emme, D.; Roder, C.; Kalthoff, H.; Wajant, H. TRAIL promotes metastasis of human pancreatic ductal adenocarcinoma. Oncogene 2006, 25, 7434–7439. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.H.; Trauzold, A.; Roder, C.; Pan, G.; Zheng, C.; Kalthoff, H. The potential molecular mechanism of overexpression of uPA, IL-8, MMP-7 and MMP-9 induced by TRAIL in pancreatic cancer cell. Hepatobiliary Pancreat. Dis. Int. 2008, 7, 201–209. [Google Scholar]
- Zhou, D.H.; Yang, L.N.; Roder, C.; Kalthoff, H.; Trauzold, A. TRAIL-induced expression of uPA and IL-8 strongly enhanced by overexpression of TRAF2 and Bcl-xL in pancreatic ductal adenocarcinoma cells. Hepatobiliary Pancreat. Dis. Int. 2013, 12, 94–98. [Google Scholar] [CrossRef]
- Pacifico, F.; Leonardi, A. NF-kappaB in solid tumors. Biochem. Pharm. 2006, 72, 1142–1152. [Google Scholar] [CrossRef]
- Rayet, B.; Gelinas, C. Aberrant rel/nfkb genes and activity in human cancer. Oncogene 1999, 18, 6938–6947. [Google Scholar] [CrossRef] [PubMed]
- Karin, M.; Lin, A. NF-κB at the crossroads of life and death. Nat. Immunol. 2002, 3, 221–227. [Google Scholar] [CrossRef] [PubMed]
- RM, P.E.P.; Stein, I.; Bramovitch, R.; Amit, S.; Kasem, S.; Gutkovich-Pyest, E.; Urieli-Shoval, S.; Galun, E.; Ben-Neriah, Y. NF-kappaB functions as a tumour promoter in inflammation-associated cancer. Nature 2004, 431, 461–466. [Google Scholar]
- Kalthoff, H.; Roeder, C.; Humburg, I.; Thiele, H.; Greten, H.; Schmiegel, W. Modulation of platelet-derived growth factor A-and B-chain/c-sis mRNA by tumor necrosis factor and other agents in adenocarcinoma cells. Oncogene 1991, 6, 1015–1021. [Google Scholar] [PubMed]
- Tepel, J.; Kruse, M.-L.; March, C.; Fiedler, A.; Kapischke, M.; Ketterer, T.; Sipos, B.; Kremer, B.; Kalthoff, H. Terminally modified oligodeoxynucleotides directed against p53 in an orthotopic xenograft model: A novel adjuvant treatment strategy for pancreatic ductal carcinoma. Pancreas 2004, 28, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Kettler, B.; Trauzold, A.; Röder, C.; Egberts, J.-H.; Kalthoff, H. Topology impacts TRAIL therapy: Differences in primary cancer growth and liver metastasis between orthotopic and subcutaneous xenotransplants of pancreatic ductal adenocarcinoma cells. Hepatobiliary Pancreat. Dis. Int. 2021, 20, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Egberts, J.-H.; Schniewind, B.; Pätzold, M.; Kettler, B.; Tepel, J.; Kalthoff, H.; Trauzold, A. Dexamethasone reduces tumor recurrence and metastasis after pancreatic tumor resection in SCID mice. Cancer Biol. Ther. 2008, 7, 1044–1050. [Google Scholar] [CrossRef] [PubMed]
- Egberts, J.-H.; Schniewind, B.; Sipos, B.; Sipos, B.; Kalthoff, H.; Tepel, J. Superiority of extended neoadjuvant chemotherapy with gemcitabine in pancreatic cancer: A comparative analysis in a clinically adapted orthotopic xenotransplantation model in SCID beige mice. Cancer Biol. Ther. 2007, 6, 1238–1243. [Google Scholar] [CrossRef]
- Ungefroren, H.; Voss, M.; Jansen, M.; Roeder, C.; Henne-Bruns, D.; Kremer, B.; Kalthoff, H. Human pancreatic adenocarcinomas express Fas and Fas ligand yet are resistant to Fas-mediated apoptosis. Cancer Res. 1998, 58, 1741–1749. [Google Scholar] [PubMed]
- Trauzold, A.; Schmiedel, S.; Roder, C.; Tams, C.; Christgen, M.; Oestern, S.; Arlt, A.; Westphal, S.; Kapischke, M.; Ungefroren, H.; et al. Multiple and synergistic deregulations of apoptosis-controlling genes in pancreatic carcinoma cells. Br. J. Cancer 2003, 89, 1714–1721. [Google Scholar] [CrossRef]
- Tepel, J.; March, C.; Ketterer, T.; Kapischke, M.; Arlt, A.; Kremer, B.; Kalthoff, H.; Kruse, M.-L. A modified random oligonucleotide-based combination therapy for adjuvant treatment of pancreatic ductal adenocarcinoma. Int. J. Oncol. 2006, 28, 1105–1112. [Google Scholar] [CrossRef][Green Version]
- Olempska, M.; Eisenach, P.A.; Ammerpohl, O.; Ungefroren, H.; Fandrich, F.; Kalthoff, H. Detection of tumor stem cell markers in pancreatic carcinoma cell lines. Hepatobiliary Pancreat. Dis. Int. 2007, 6, 92–97. [Google Scholar]
- Hsu, C.-P.; Lee, L.-Y.; Hsu, J.-T.; Hsu, Y.-P.; Wu, Y.-T.; Wang, S.-Y.; Yeh, C.-N.; Chen, T.-C.; Hwang, T.-L. CD44 predicts early recurrence in pancreatic cancer patients undergoing radical surgery. In Vivo 2018, 32, 1533–1540. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Chen, C.; Chang, K.; Karnad, A.; Jagirdar, J.; Kumar, A.P.; Freeman, J.W. CD44 expression level and isoform contributes to pancreatic cancer cell plasticity, invasiveness, and response to therapy. Clin. Cancer Res. 2016, 22, 5592–5604. [Google Scholar] [CrossRef]
- Qadir, A.S.; Ceppi, P.; Brockway, S.; Law, C.; Mu, L.; Khodarev, N.N.; Kim, J.; Zhao, J.C.; Putzbach, W.; Murmann, A.E.; et al. CD95/Fas Increases Stemness in Cancer Cells by Inducing a STAT1-Dependent Type I Interferon Response. Cell Rep. 2017, 18, 2373–2386. [Google Scholar] [CrossRef]
- Teodorczyk, M.; Kleber, S.; Wollny, D.; Sefrin, J.P.; Aykut, B.; Mateos, A.; Herhaus, P.; Sancho-Martinez, I.; Hill, O.; Gieffers, C.; et al. CD95 promotes metastatic spread via Sck in pancreatic ductal adenocarcinoma. Cell Death Differ. 2015, 22, 1192–1202. [Google Scholar] [CrossRef] [PubMed]
- Bernstorff, W.v.; Spanjaard, R.A.; Chan, A.K.; Lockhart, D.C.; Sadanaga, N.; Wood, I.; Peiper, M.; Goedegebuure, P.S.; Eberlein, T.J. Pancreatic cancer cells can evade immune surveillance via nonfunctional Fas (APO-1/CD95) receptors and aberrant expression of functional Fas ligand. Surgery 1999, 125, 73–84. [Google Scholar] [CrossRef]
- Burris, H.r.; Moore, M.J.; Andersen, J.; Green, M.R.; Rothenberg, M.L.; Modiano, M.R.; Christine Cripps, M.; Portenoy, R.K.; Storniolo, A.M.; Tarassoff, P. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: A randomized trial. J. Clin. Oncol. 1997, 15, 2403–2413. [Google Scholar] [CrossRef]
- Singh, S.; Srivastava, S.; Bhardwaj, A.; Owen, L.; Singh, A. CXCL12–CXCR4 signalling axis confers gemcitabine resistance to pancreatic cancer cells: A novel target for therapy. Br. J. Cancer 2010, 103, 1671–1679. [Google Scholar] [CrossRef]
- Arora, S.; Bhardwaj, A.; Singh, S.; Srivastava, S.K.; McClellan, S.; Grizzle, W.E.; Owen, L.B.; Singh, A.P. An undesired effect of chemotherapy: Gemcitabine promotes pancreatic cancer cell invasiveness through upregulation of CXCR4. Cancer Res. 2013, 73, 4794. [Google Scholar] [CrossRef]
- Gaertner, F.; Krüger, S.; Röder, C.; Trauzold, A.; Röcken, C.; Kalthoff, H. The expression of death receptor systems TRAIL-R1/-R2/-R4, CD95 and TNF-R1 and their cognate ligands in pancreatic ductal adenocarcinoma. Histol. Histopathol. 2018, 34, 491–501. [Google Scholar]
- Almendro Navarro, V.; Ametller, E.; Garcia Recio, S.; Collazo, O.; Casas, I.; Augé Fradera, J.M.; Maurel Santasusana, J.; Gascón, P. The Role of MMP7 and its cross-talk with the FAS/FASL system during the acquisition of chemoresistance to oxaliplatin. PLoS ONE 2009, 4, 4728. [Google Scholar] [CrossRef]
- Mitsiades, N.; Yu, W.H.; Poulaki, V.; Tsokos, M.; Stamenkovic, I. Matrix metalloproteinase-7-mediated cleavage of Fas ligand protects tumor cells from chemotherapeutic drug cytotoxicity. Cancer Res. 2001, 61, 577–581. [Google Scholar] [PubMed]
- Villunger, A.; Egle, A.; Kos, M.; Hartmann, B.L.; Geley, S.; Kofler, R.; Greil, R. Drug-induced apoptosis is associated with enhanced Fas (Apo-1/CD95) ligand expression but occurs independently of Fas (Apo-1/CD95) signaling in human T-acute lymphatic leukemia cells. Cancer Res. 1997, 57, 3331–3334. [Google Scholar] [PubMed]
- Wesselborg, S.; Engels, I.H.; Rossmann, E.; Los, M.; Schulze-Osthoff, K. Anticancer drugs induce caspase-8/FLICE activation and apoptosis in the absence of CD95 receptor/ligand interaction. Blood 1999, 93, 3053–3063. [Google Scholar] [CrossRef] [PubMed]






| Cell Line | TNF-α | TRAIL | CD95L | Gemcitabine |
|---|---|---|---|---|
| PancTuI-luc | + + | + + | − | − |
| A818-4 | − | − − | + + | + |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rashid, K.; Röder, C.; Goumas, F.; Egberts, J.-H.; Kalthoff, H. CD95L Inhibition Impacts Gemcitabine-Mediated Effects and Non-Apoptotic Signaling of TNF-α and TRAIL in Pancreatic Tumor Cells. Cancers 2021, 13, 5458. https://doi.org/10.3390/cancers13215458
Rashid K, Röder C, Goumas F, Egberts J-H, Kalthoff H. CD95L Inhibition Impacts Gemcitabine-Mediated Effects and Non-Apoptotic Signaling of TNF-α and TRAIL in Pancreatic Tumor Cells. Cancers. 2021; 13(21):5458. https://doi.org/10.3390/cancers13215458
Chicago/Turabian StyleRashid, Khalid, Christian Röder, Freya Goumas, Jan-Hendrik Egberts, and Holger Kalthoff. 2021. "CD95L Inhibition Impacts Gemcitabine-Mediated Effects and Non-Apoptotic Signaling of TNF-α and TRAIL in Pancreatic Tumor Cells" Cancers 13, no. 21: 5458. https://doi.org/10.3390/cancers13215458
APA StyleRashid, K., Röder, C., Goumas, F., Egberts, J.-H., & Kalthoff, H. (2021). CD95L Inhibition Impacts Gemcitabine-Mediated Effects and Non-Apoptotic Signaling of TNF-α and TRAIL in Pancreatic Tumor Cells. Cancers, 13(21), 5458. https://doi.org/10.3390/cancers13215458





