Characterization of Recurrent Relevant Genes Reveals a Novel Role of RPL36A in Radioresistant Oral Squamous Cell Carcinoma
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Populations and Clinical Specimens
2.2. Identification of Specific Radioresistance-Associated Genes for OSCC from the TCGA Dataset
2.3. Core Molecular Pathways and Gene Ontology (GO) Analysis
2.4. Identification of Differentially Expressed Genes (DEGs) in OSCC from the TCGA Dataset
2.5. Analysis of the Relationship between RPL36A and Other Genes in the TCGA-OSCC Dataset
2.6. Cell Culture
2.7. Gene Knockdown of RPL36A Using Small Interfering RNA
2.8. RNA Extraction and qPCR
2.9. Flow Cytometry Analysis
2.10. Clonogenic Survival Assay
2.11. Statistical Analysis
3. Results
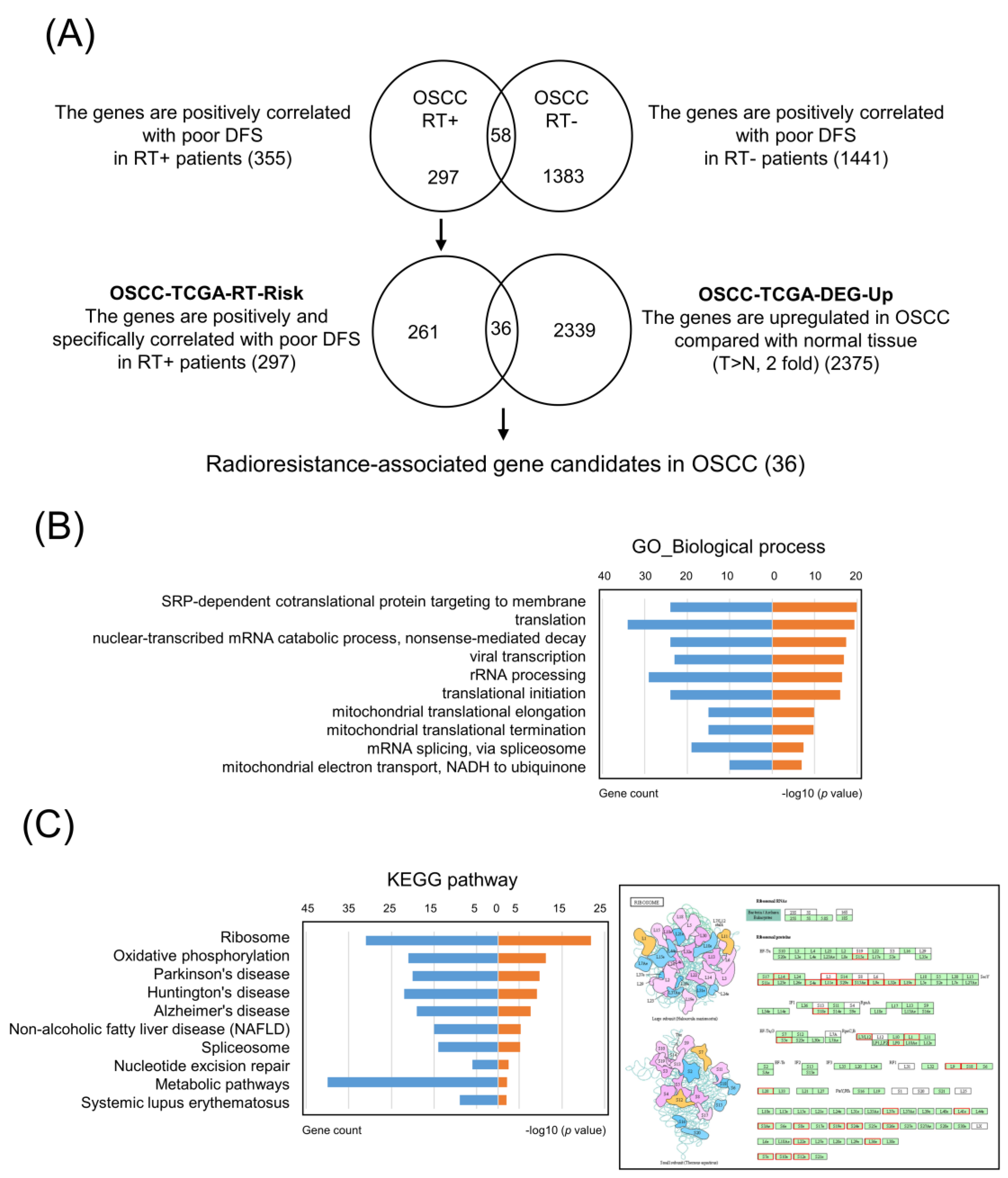
3.1. Identification of Specific Radioresistance-Associated Genes in OSCC from the TCGA Dataset
3.2. Core Molecular Pathway and Gene Ontology Analyses in OSCC-TCGA-RT-Risk
3.3. Selection of Upregulated Radioresistance-Associated Genes in OSCC Tumors
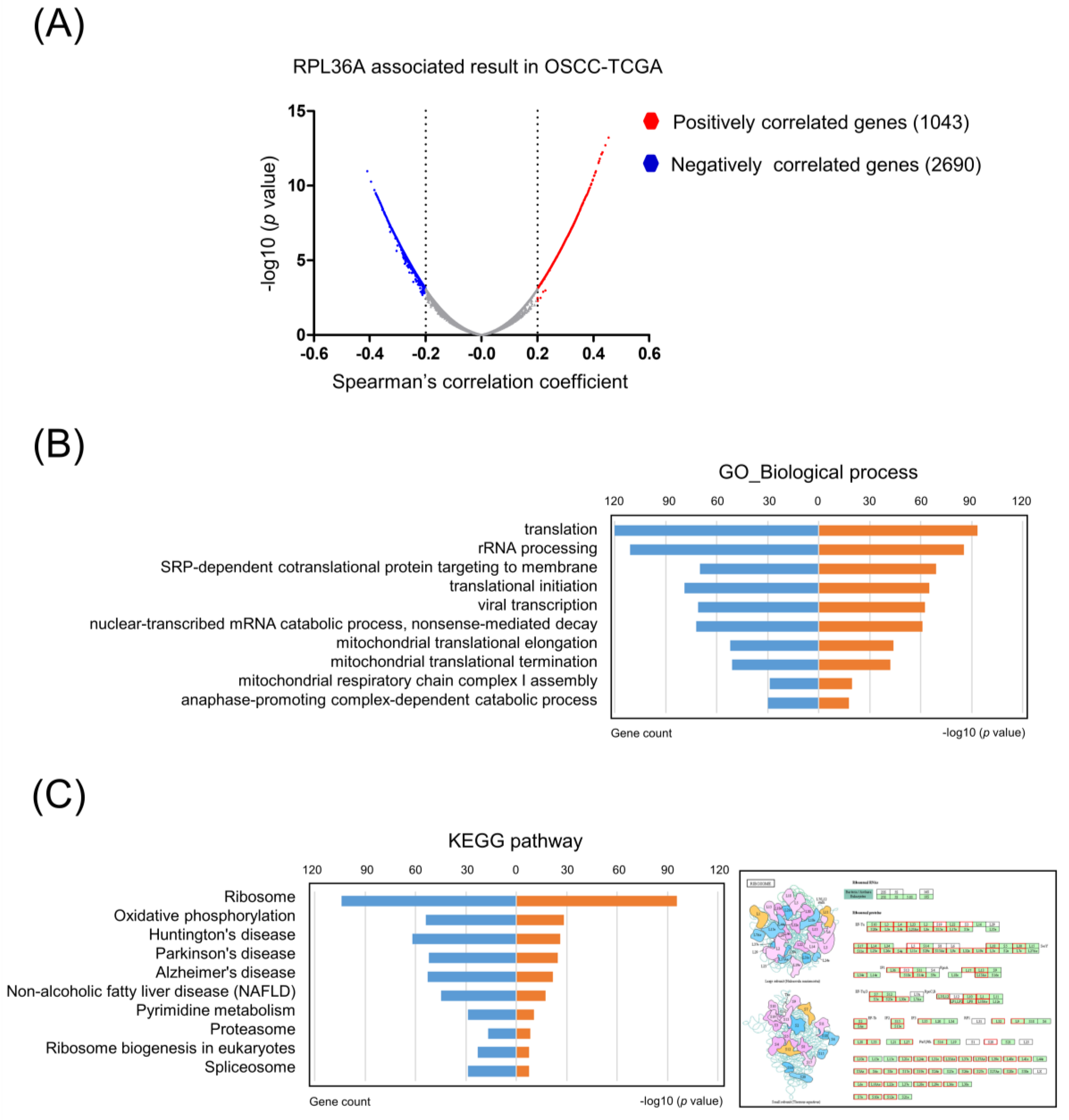
3.4. High RPL36A Expression Predicts a Poor Prognosis and Radioresistance in OSCC Patients
3.5. Core Molecular Pathways and Gene Ontology Are Similar between Radioresistance- and RPL36A-Associated Genes
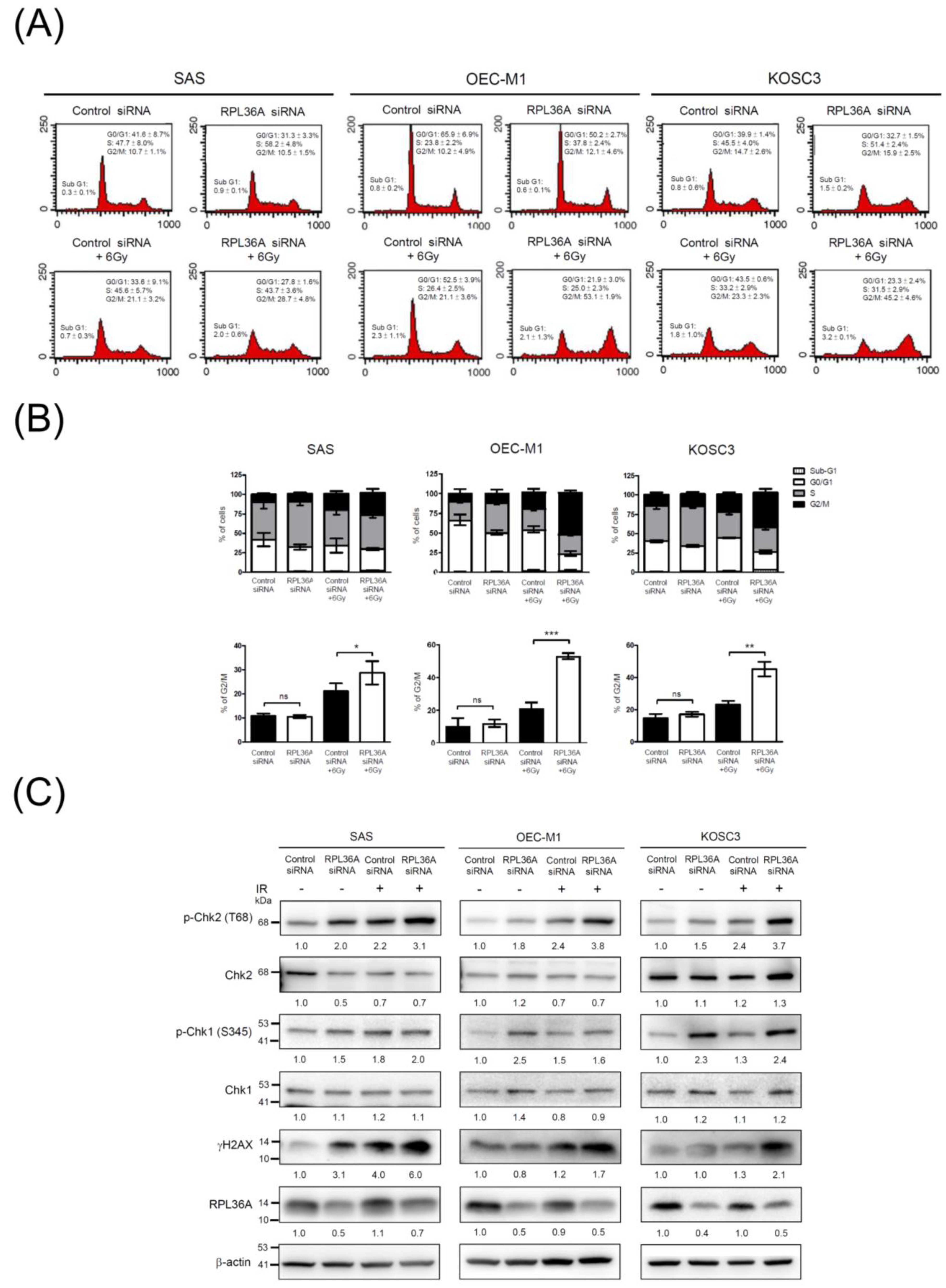
3.6. RPL36A Depletion Sensitizes Cell to DNA Damage and Promotes G2/M Cell Cycle Arrest in Response to Irradiation
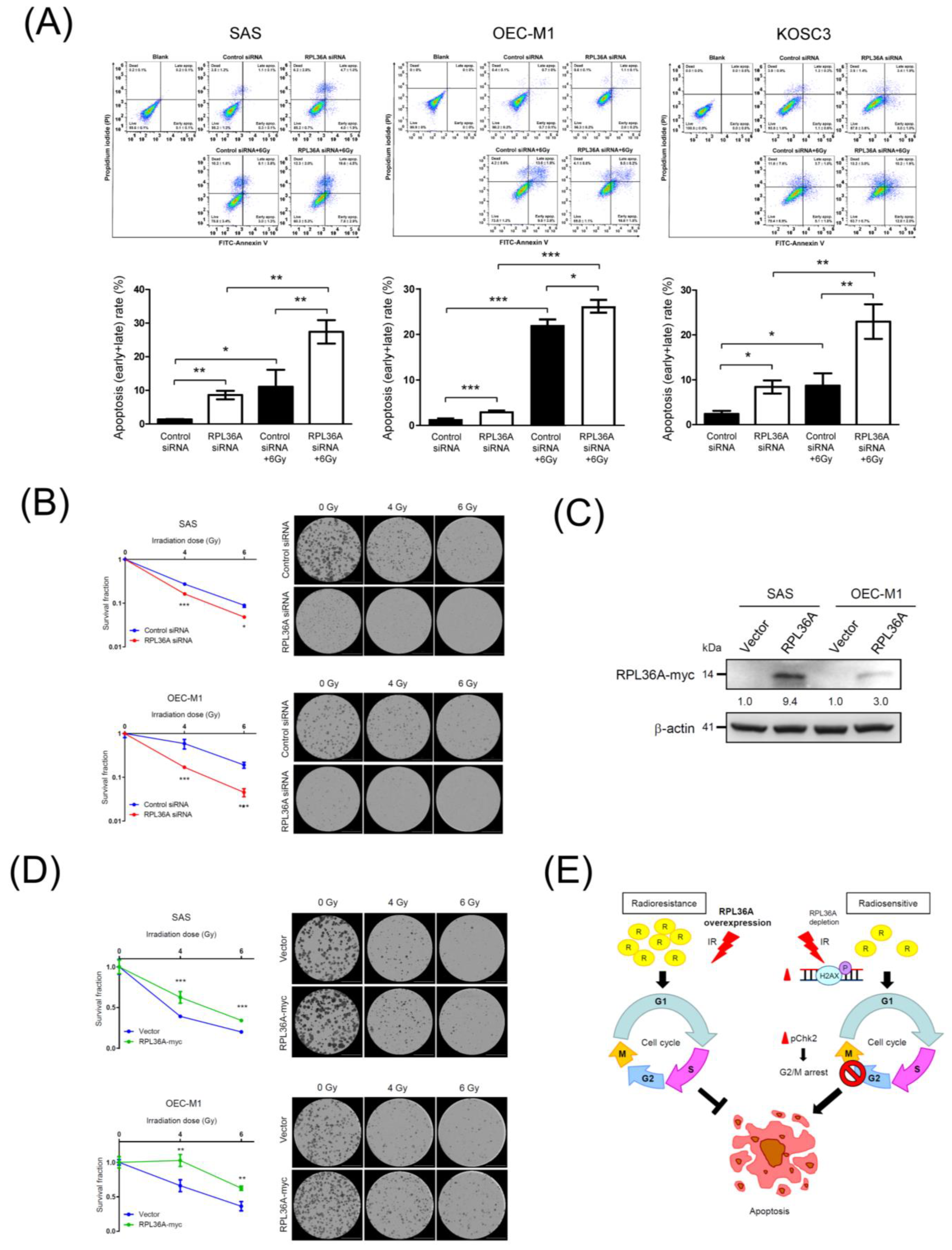
3.7. RPL36A Depletion Contributes to Radiosensitivity by Augmenting Irradiation-Induced Apoptosis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Parkin, D.M.; Bray, F.; Ferlay, J.; Pisani, P. Global cancer statistics, 2002. CA Cancer J. Clin. 2005, 55, 74–108. [Google Scholar] [CrossRef]
- Higgins, G.S.; O'Cathail, S.M.; Muschel, R.J.; McKenna, W.G. Drug radiotherapy combinations: Review of previous failures and reasons for future optimism. Cancer Treat. Rev. 2015, 41, 105–113. [Google Scholar] [CrossRef]
- Silverman, S., Jr. Demographics and occurrence of oral and pharyngeal cancers. The outcomes, the trends, the challenge. J. Am. Dent. Assoc. 2001, 132, 7S–11S. [Google Scholar] [CrossRef]
- Bennardo, L.; Bennardo, F.; Giudice, A.; Passante, M.; Dastoli, S.; Morrone, P.; Provenzano, E.; Patruno, C.; Nistico, S.P. Local Chemotherapy as an Adjuvant Treatment in Unresectable Squamous Cell Carcinoma: What Do We Know So Far? Curr. Oncol. 2021, 28, 2317–2325. [Google Scholar] [CrossRef] [PubMed]
- Pentangelo, G.; Nistico, S.P.; Provenzano, E.; Cisale, G.Y.; Bennardo, L. Topical 5% Imiquimod Sequential to Surgery for HPV-Related Squamous Cell Carcinoma of the Lip. Medicina 2021, 57, 563. [Google Scholar] [CrossRef]
- Camisasca, D.R.; Silami, M.A.; Honorato, J.; Dias, F.L.; de Faria, P.A.; Lourenco Sde, Q. Oral squamous cell carcinoma: Clinicopathological features in patients with and without recurrence. ORL J. Otorhinolaryngol. Relat. Spec. 2011, 73, 170–176. [Google Scholar] [CrossRef] [PubMed]
- Tsai, L.L.; Yu, C.C.; Chang, Y.C.; Yu, C.H.; Chou, M.Y. Markedly increased Oct4 and Nanog expression correlates with cisplatin resistance in oral squamous cell carcinoma. J. Oral Pathol. Med. 2011, 40, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Park, H.R.; Cho, N.H.; Choi, Y.P.; Rha, S.Y.; Park, S.W.; Kim, S.H. Identifying genes related to radiation resistance in oral squamous cell carcinoma cell lines. Int. J. Oral Maxillofac. Surg. 2013, 42, 169–176. [Google Scholar] [CrossRef]
- Wong, Y.F.; Sahota, D.S.; Cheung, T.H.; Lo, K.W.; Yim, S.F.; Chung, T.K.; Chang, A.M.; Smith, D.I. Gene expression pattern associated with radiotherapy sensitivity in cervical cancer. Cancer J. 2006, 12, 189–193. [Google Scholar] [CrossRef]
- Chen, Y.; Bao, C.; Zhang, X.; Lin, X.; Fu, Y. Knockdown of LINC00662 represses AK4 and attenuates radioresistance of oral squamous cell carcinoma. Cancer Cell Int. 2020, 20, 244. [Google Scholar] [CrossRef]
- Lin, C.Y.; Lin, T.Y.; Wang, H.M.; Huang, S.F.; Fan, K.H.; Liao, C.T.; Chen, I.H.; Lee, L.Y.; Li, Y.L.; Chen, Y.J.; et al. GP96 is over-expressed in oral cavity cancer and is a poor prognostic indicator for patients receiving radiotherapy. Radiat. Oncol. 2011, 6, 136. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.L.; Chang, J.T.; Lee, L.Y.; Fan, K.H.; Lu, Y.C.; Li, Y.C.; Chiang, C.H.; You, G.R.; Chen, H.Y.; Cheng, A.J. GDF15 contributes to radioresistance and cancer stemness of head and neck cancer by regulating cellular reactive oxygen species via a SMAD-associated signaling pathway. Oncotarget 2017, 8, 1508–1528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheu, J.J.; Hua, C.H.; Wan, L.; Lin, Y.J.; Lai, M.T.; Tseng, H.C.; Jinawath, N.; Tsai, M.H.; Chang, N.W.; Lin, C.F.; et al. Functional genomic analysis identified epidermal growth factor receptor activation as the most common genetic event in oral squamous cell carcinoma. Cancer Res. 2009, 69, 2568–2576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kundu, S.K.; Nestor, M. Targeted therapy in head and neck cancer. Tumour. Biol. 2012, 33, 707–721. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Erjala, K.; Kulmala, J.; Qiu, X.; Sundvall, M.; Elenius, K.; Grenman, R. Concurrent cetuximab, cisplatin, and radiation for squamous cell carcinoma of the head and neck in vitro. Radiother. Oncol. 2009, 92, 388–392. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 2011, 12, 323. [Google Scholar] [CrossRef] [Green Version]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Li, S.; Shi, J.; Gao, H.; Yuan, Y.; Chen, Q.; Zhao, Z.; Wang, X.; Li, B.; Ming, L.; Zhong, J.; et al. Identification of a gene signature associated with radiotherapy and prognosis in gliomas. Oncotarget 2017, 8, 88974–88987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mirisola, V.; Mora, R.; Esposito, A.I.; Guastini, L.; Tabacchiera, F.; Paleari, L.; Amaro, A.; Angelini, G.; Dellepiane, M.; Pfeffer, U.; et al. A prognostic multigene classifier for squamous cell carcinomas of the larynx. Cancer Lett. 2011, 307, 37–46. [Google Scholar] [CrossRef]
- Gao, C.B.; Wang, D.; Wang, Y.; Liu, Y.H. Artemin promotes proliferation and metastasis in human laryngeal squamous cell carcinoma. Int. J. Clin. Exp. Pathol. 2017, 10, 10413–10418. [Google Scholar] [PubMed]
- Tong, Q.; Wang, X.L.; Li, S.B.; Yang, G.L.; Jin, S.; Gao, Z.Y.; Liu, X.B. Combined detection of IL-6 and IL-8 is beneficial to the diagnosis of early stage esophageal squamous cell cancer: A preliminary study based on the screening of serum markers using protein chips. Onco Targets Ther. 2018, 11, 5777–5787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, W.T.; Yu, C.P.; Wu, D.H.; Zhang, L.; Xu, L.H.; Weng, G.X.; Zeng, M.S.; Song, L.B.; Li, J.S. Upregulation of CENP-H in tongue cancer correlates with poor prognosis and progression. J. Exp. Clin. Cancer Res. 2009, 28, 74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, W.; Liao, L.; Lai, H.; Yi, X.; Wang, D. Identification of core biomarkers associated with pathogenesis and prognostic outcomes of laryngeal squamous-cell cancer using bioinformatics analysis. Eur. Arch. Otorhinolaryngol. 2020, 277, 1397–1408. [Google Scholar] [CrossRef]
- Shen, L.; Zhao, K.; Li, H.; Ning, B.; Wang, W.; Liu, R.; Zhang, Y.; Zhang, A. Downregulation of UBE2T can enhance the radiosensitivity of osteosarcoma in vitro and in vivo. Epigenomics 2019, 11, 1283–1305. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, F.; Kasamatsu, A.; Endo-Sakamoto, Y.; Eizuka, K.; Hiroshima, K.; Kita, A.; Saito, T.; Koike, K.; Tanzawa, H.; Uzawa, K. Increased expression of tripartite motif (TRIM) like 2 promotes tumoral growth in human oral cancer. Biochem. Biophys. Res. Commun. 2019, 508, 1133–1138. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.S.; Zhang, E.X.; Sun, Q.F.; Ye, Z.J.; Liu, J.W.; Zhou, D.H.; Tang, Y. Integrated analysis of lncRNA-miRNA-mRNA ceRNA network in squamous cell carcinoma of tongue. BMC Cancer 2019, 19, 779. [Google Scholar] [CrossRef] [PubMed]
- Kudo, I.; Esumi, M.; Kusumi, Y.; Furusaka, T.; Oshima, T. Particular gene upregulation and p53 heterogeneous expression in TP53-mutated maxillary carcinoma. Oncol. Lett. 2017, 14, 4633–4640. [Google Scholar] [CrossRef] [Green Version]
- Snyder, E.R.; Ricker, J.L.; Chen, Z.; Waes, C.V. Variation in cisplatinum sensitivity is not associated with Fanconi Anemia/BRCA pathway inactivation in head and neck squamous cell carcinoma cell lines. Cancer Lett. 2007, 245, 75–80. [Google Scholar] [CrossRef]
- Turke, C.; Horn, S.; Petto, C.; Labudde, D.; Lauer, G.; Wittenburg, G. Loss of heterozygosity in FANCG, FANCF and BRIP1 from head and neck squamous cell carcinoma of the oral cavity. Int. J. Oncol. 2017, 50, 2207–2220. [Google Scholar] [CrossRef] [PubMed]
- Young, A.; Berry, R.; Holloway, A.F.; Blackburn, N.B.; Dickinson, J.L.; Skala, M.; Phillips, J.L.; Brettingham-Moore, K.H. RNA-seq profiling of a radiation resistant and radiation sensitive prostate cancer cell line highlights opposing regulation of DNA repair and targets for radiosensitization. BMC Cancer 2014, 14, 808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MacGregor, T.P.; Carter, R.; Gillies, R.S.; Findlay, J.M.; Kartsonaki, C.; Castro-Giner, F.; Sahgal, N.; Wang, L.M.; Chetty, R.; Maynard, N.D.; et al. Translational study identifies XPF and MUS81 as predictive biomarkers for oxaliplatin-based peri-operative chemotherapy in patients with esophageal adenocarcinoma. Sci. Rep. 2018, 8, 7265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carter, R.; Westhorpe, A.; Romero, M.J.; Habtemariam, A.; Gallevo, C.R.; Bark, Y.; Menezes, N.; Sadler, P.J.; Sharma, R.A. Radiosensitisation of human colorectal cancer cells by ruthenium(II) arene anticancer complexes. Sci. Rep. 2016, 6, 20596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, M.H.; Ueda, K.; Nakamura, Y.; Daigo, Y. Identification of a novel oncogene, MMS22L, involved in lung and esophageal carcinogenesis. Int. J. Oncol. 2012, 41, 1285–1296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, Y.; Zhang, L.; Piao, S.; Li, L.; Li, J.; Xia, Y.; Saiyin, W. NUDT1: A potential independent predictor for the prognosis of patients with oral squamous cell carcinoma. J. Oral Pathol. Med. 2020, 49, 210–218. [Google Scholar] [CrossRef] [PubMed]
- Bialkowski, K.; Szpila, A.; Kasprzak, K.S. Up-regulation of 8-oxo-dGTPase activity of MTH1 protein in the brain, testes and kidneys of mice exposed to (137)Cs gamma radiation. Radiat. Res. 2009, 172, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Mohanta, S.; Sekhar Khora, S.; Suresh, A. Cancer Stem Cell based molecular predictors of tumor recurrence in Oral squamous cell carcinoma. Arch. Oral Biol. 2019, 99, 92–106. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zheng, W.; Yao, D.; Chen, Q.; Zhu, L.; Zhang, J.; Pan, Y.; Shao, C. Upregulation of DNA Metabolism-Related Genes Contributes to Radioresistance of Glioblastoma. Hum. Gene Ther. Clin. Dev. 2019, 30, 74–87. [Google Scholar] [CrossRef]
- Li, J.N.; Feng, C.J.; Lu, Y.J.; Li, H.J.; Tu, Z.; Liao, G.Q.; Liang, C. mRNA expression of the DNA replication-initiation proteins in epithelial dysplasia and squamous cell carcinoma of the tongue. BMC Cancer 2008, 8, 395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Supiot, S.; Gouraud, W.; Campion, L.; Jezequel, P.; Buecher, B.; Charrier, J.; Heymann, M.F.; Mahe, M.A.; Rio, E.; Cherel, M. Early dynamic transcriptomic changes during preoperative radiotherapy in patients with rectal cancer: A feasibility study. World J. Gastroenterol. 2013, 19, 3249–3254. [Google Scholar] [CrossRef] [PubMed]
- Stumpf, C.R.; Moreno, M.V.; Olshen, A.B.; Taylor, B.S.; Ruggero, D. The translational landscape of the mammalian cell cycle. Mol. Cell 2013, 52, 574–582. [Google Scholar] [CrossRef] [Green Version]
- Sak, A.; Wurm, R.; Elo, B.; Grehl, S.; Pottgen, C.; Stuben, G.; Sinn, B.; Wolf, G.; Budach, V.; Stuschke, M. Increased radiation-induced apoptosis and altered cell cycle progression of human lung cancer cell lines by antisense oligodeoxynucleotides targeting p53 and p21(WAF1/CIP1). Cancer Gene Ther. 2003, 10, 926–934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, B.M.; Hong, Y.; Lee, S.; Liu, P.; Lim, J.H.; Lee, Y.H.; Lee, T.H.; Chang, K.T.; Hong, Y. Therapeutic Implications for Overcoming Radiation Resistance in Cancer Therapy. Int. J. Mol. Sci. 2015, 16, 26880–26913. [Google Scholar] [CrossRef] [Green Version]
- Wedekind, H.; Walz, K.; Buchbender, M.; Rieckmann, T.; Strasser, E.; Grottker, F.; Fietkau, R.; Frey, B.; Gaipl, U.S.; Ruckert, M. Head and neck tumor cells treated with hypofractionated irradiation die via apoptosis and are better taken up by M1-like macrophages. Strahlenther. Onkol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Li, M.Y.; Liu, J.Q.; Chen, D.P.; Li, Z.Y.; Qi, B.; He, L.; Yu, Y.; Yin, W.J.; Wang, M.Y.; Lin, L. Radiotherapy induces cell cycle arrest and cell apoptosis in nasopharyngeal carcinoma via the ATM and Smad pathways. Cancer Biol. Ther. 2017, 18, 681–693. [Google Scholar] [CrossRef]
- Ho, S.Y.; Wu, W.S.; Lin, L.C.; Wu, Y.H.; Chiu, H.W.; Yeh, Y.L.; Huang, B.M.; Wang, Y.J. Cordycepin Enhances Radiosensitivity in Oral Squamous Carcinoma Cells by Inducing Autophagy and Apoptosis Through Cell Cycle Arrest. Int. J. Mol. Sci. 2019, 20, 5366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costa, L.G.; de Laat, R.; Dao, K.; Pellacani, C.; Cole, T.B.; Furlong, C.E. Paraoxonase-2 (PON2) in brain and its potential role in neuroprotection. Neurotoxicology 2014, 43, 3–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.H.; You, K.R.; Kim, I.H.; Cho, B.H.; Kim, C.Y.; Kim, D.G. Over-expression of the ribosomal protein L36a gene is associated with cellular proliferation in hepatocellular carcinoma. Hepatology 2004, 39, 129–138. [Google Scholar] [CrossRef]
- Li, S.; Shi, J.; Gao, H.; Yuan, Y.; Chen, O.; Zhao, Z.; Wang, X.; Li, B.; Ming, L.; Zhong, J.; et al. Identification of a gene signature associated with radiotherapy and prognosis in gliomas. Oncotarget 2017, 8, 88974–88987. [Google Scholar] [CrossRef] [Green Version]
- Roberts, E.; Sethi, A.; Montoya, J.; Woese, C.R.; Luthey-Schulten, Z. Molecular signatures of ribosomal evolution. Proc. Natl. Acad. Sci. USA 2008, 105, 13953–13958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.Y.; Fan, L.N.; Han, D.H.; Yu, Z.; Ma, J.; Liu, Y.X.; Li, P.F.; Zhao, D.H.; Chai, J.; Jiang, L.; et al. Ribosomal S6 protein kinase 4 promotes radioresistance in esophageal squamous cell carcinoma. J. Clin. Investig. 2020, 130, 4301–4319. [Google Scholar] [CrossRef]
- Ma, J.; Lu, Y.; Zhang, S.; Li, Y.; Huang, J.; Yin, Z.; Ren, J.; Huang, K.; Liu, L.; Yang, K.; et al. beta-Trcp ubiquitin ligase and RSK2 kinase-mediated degradation of FOXN2 promotes tumorigenesis and radioresistance in lung cancer. Cell Death Differ. 2018, 25, 1473–1485. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Youn, H.; Lee, S.; Kim, E.; Kim, D.; Sub Lee, J.; Lee, J.M.; Youn, B. RNF138-mediated ubiquitination of rpS3 is required for resistance of glioblastoma cells to radiation-induced apoptosis. Exp. Mol. Med. 2018, 50, e434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernhard, E.J.; Maity, A.; Muschel, R.J.; McKenna, W.G. Effects of ionizing radiation on cell cycle progression. A review. Radiat. Environ. Biophys. 1995, 34, 79–83. [Google Scholar] [CrossRef] [PubMed]
- Norbury, C.J.; Hickson, I.D. Cellular responses to DNA damage. Annu. Rev. Pharmacol. Toxicol. 2001, 41, 367–401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Nie, J.; Wang, R.; Mao, W. The Cell Cycle G2/M Block Is an Indicator of Cellular Radiosensitivity. Dose Response 2019, 17. [Google Scholar] [CrossRef] [PubMed]
- Speidel, D. The role of DNA damage responses in p53 biology. Arch. Toxicol. 2015, 89, 501–517. [Google Scholar] [CrossRef] [PubMed]
- Knauf, J.A.; Ouyang, B.; Knudsen, E.S.; Fukasawa, K.; Babcock, G.; Fagin, J.A. Oncogenic RAS induces accelerated transition through G2/M and promotes defects in the G2 DNA damage and mitotic spindle checkpoints. J. Biol. Chem. 2006, 281, 3800–3809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gogineni, V.R.; Nalla, A.K.; Gupta, R.; Dinh, D.H.; Klopfenstein, J.D.; Rao, J.S. Chk2-mediated G2/M cell cycle arrest maintains radiation resistance in malignant meningioma cells. Cancer Lett. 2011, 313, 64–75. [Google Scholar] [CrossRef] [Green Version]

| Gene Name | DFS (p-Value) | Dysregulated in HNSCC (Ref.) | Function Related with Radioresistance (Ref.) |
|---|---|---|---|
| HIST1H2BH | 1.81 × 10−4 | No | No |
| HIST1H4I | 6.64 × 10−4 | No | No |
| RPL36A | 9.47 × 10−4 | No | Yes [18] |
| HIST1H3A | 1.34 × 10−3 | No | No |
| HIST1H3F | 1.41 × 10−3 | Yes [19] | No |
| ARTN | 1.94 × 10−3 | Yes [20,21] | No |
| PSMC3IP | 4.82 × 10−3 | No | No |
| CCDC78 | 9.17 × 10−3 | No | No |
| FBXL6 | 9.17 × 10−3 | No | No |
| CENPH | 9.29 × 10−3 | Yes [22] | No |
| ACOT7 | 1.07 × 10−2 | No | No |
| UBE2T | 1.10 × 10−2 | Yes [23] | Yes [24] |
| FAM72B | 1.26 × 10−2 | No | No |
| TRIML2 | 1.34 × 10−2 | Yes [25] | No |
| HIST1H2BI | 1.59 × 10−2 | No | No |
| OIP5 | 1.63 × 10−2 | No | No |
| ZWINT | 1.75 × 10−2 | No | No |
| NAGS | 2.01 × 10−2 | Yes [26] | No |
| EPPK1 | 2.30 × 10−2 | Yes [27] | No |
| HAUS8 | 2.43 × 10−2 | No | No |
| TMSB10 | 2.54 × 10−2 | No | No |
| FANCG | 2.69 × 10−2 | Yes [28,29] | Yes [30] |
| HIST1H3I | 2.71 × 10−2 | No | No |
| EME1 | 3.04 × 10−2 | Yes [31] | Yes [32] |
| GSDMB | 3.32 × 10−2 | No | No |
| RANBP1 | 3.51 × 10−2 | No | No |
| NFKBIL2 | 3.66 × 10−2 | Yes [33] | No |
| POC1A | 3.76 × 10−2 | No | No |
| NUDT1 | 4.15 × 10−2 | Yes [34] | Yes [35] |
| GINS2 | 4.20 × 10−2 | Yes [36] | Yes [37] |
| CDC45 | 4.37 × 10−2 | Yes [38] | No |
| PABPC1L | 4.56 × 10−2 | No | Yes [39] |
| GOLGA8B | 4.76 × 10−2 | No | No |
| ATHL1 | 479 × 10−2 | No | No |
| SPC25 | 4.83 × 10−2 | No | No |
| CKS2 | 4.85 × 10−2 | Yes [26] | No |
| Univariate | Multivariate | |||||
|---|---|---|---|---|---|---|
| Variables | Hazard Ratio | 95% CI | p-Value | Hazard Ratio | 95% CI | p-Value |
| Age, years a | 1.005 | 0.985–1.025 | 0.6130 | 1.014 | 0.990–1.038 | 0.2647 |
| Sex | ||||||
| Male | Reference | Reference | ||||
| Female | 1.014 | 0.611–1.684 | 0.9570 | 0.811 | 0.466–1.410 | 0.4579 |
| Smoking | ||||||
| No | Reference | Reference | ||||
| Yes | 0.969 | 0.591–1.589 | 0.9010 | 0.867 | 0.515–1.458 | 0.5899 |
| Overall TNM stage | ||||||
| I–II | Reference | Reference | ||||
| III–IV | 1.912 | 0.825–4.429 | 0.1310 | 1.500 | 0.608–3.706 | 0.3790 |
| Node classification | ||||||
| n ≤ 1 | Reference | Reference | ||||
| n > 1 | 2.032 | 1.271–3.246 | 0.0030 b | 2.055 | 1.237–3.415 | 0.0054 b |
| RPL36A | ||||||
| Low | Reference | Reference | ||||
| High | 2.144 | 1.328–3.462 | 0.0018 b | 1.980 | 1.216–3.224 | 0.0060 b |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, T.-W.; Chang, K.-P.; Cheng, C.-C.; Chen, C.-Y.; Hong, S.-W.; Sie, Z.-L.; Cheng, H.-W.; Yen, W.-C.; Huang, Y.; Liu, S.-C.; et al. Characterization of Recurrent Relevant Genes Reveals a Novel Role of RPL36A in Radioresistant Oral Squamous Cell Carcinoma. Cancers 2021, 13, 5623. https://doi.org/10.3390/cancers13225623
Chen T-W, Chang K-P, Cheng C-C, Chen C-Y, Hong S-W, Sie Z-L, Cheng H-W, Yen W-C, Huang Y, Liu S-C, et al. Characterization of Recurrent Relevant Genes Reveals a Novel Role of RPL36A in Radioresistant Oral Squamous Cell Carcinoma. Cancers. 2021; 13(22):5623. https://doi.org/10.3390/cancers13225623
Chicago/Turabian StyleChen, Ting-Wen, Kai-Ping Chang, Chun-Chia Cheng, Cheng-Yi Chen, Shu-Wen Hong, Zong-Lin Sie, Hsing-Wen Cheng, Wei-Chen Yen, Yenlin Huang, Shu-Chen Liu, and et al. 2021. "Characterization of Recurrent Relevant Genes Reveals a Novel Role of RPL36A in Radioresistant Oral Squamous Cell Carcinoma" Cancers 13, no. 22: 5623. https://doi.org/10.3390/cancers13225623







