Dysregulation of Cytoskeleton Remodeling Drives Invasive Leading Cells Detachment
Abstract
:Simple Summary
Abstract
1. Introduction
2. Results
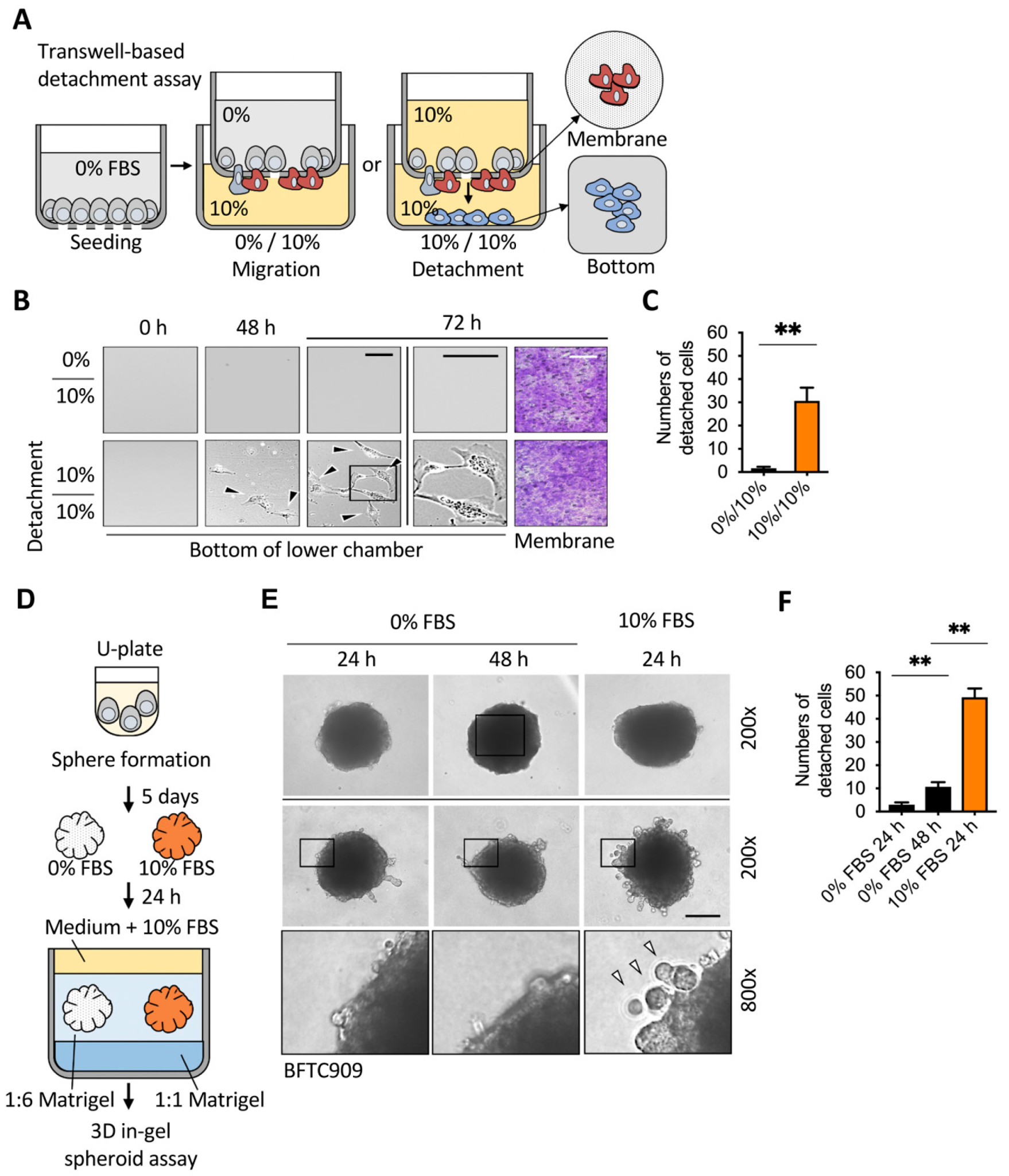
2.1. Sufficient Nutrients Induced the Detachment of Invasive Leading Cells
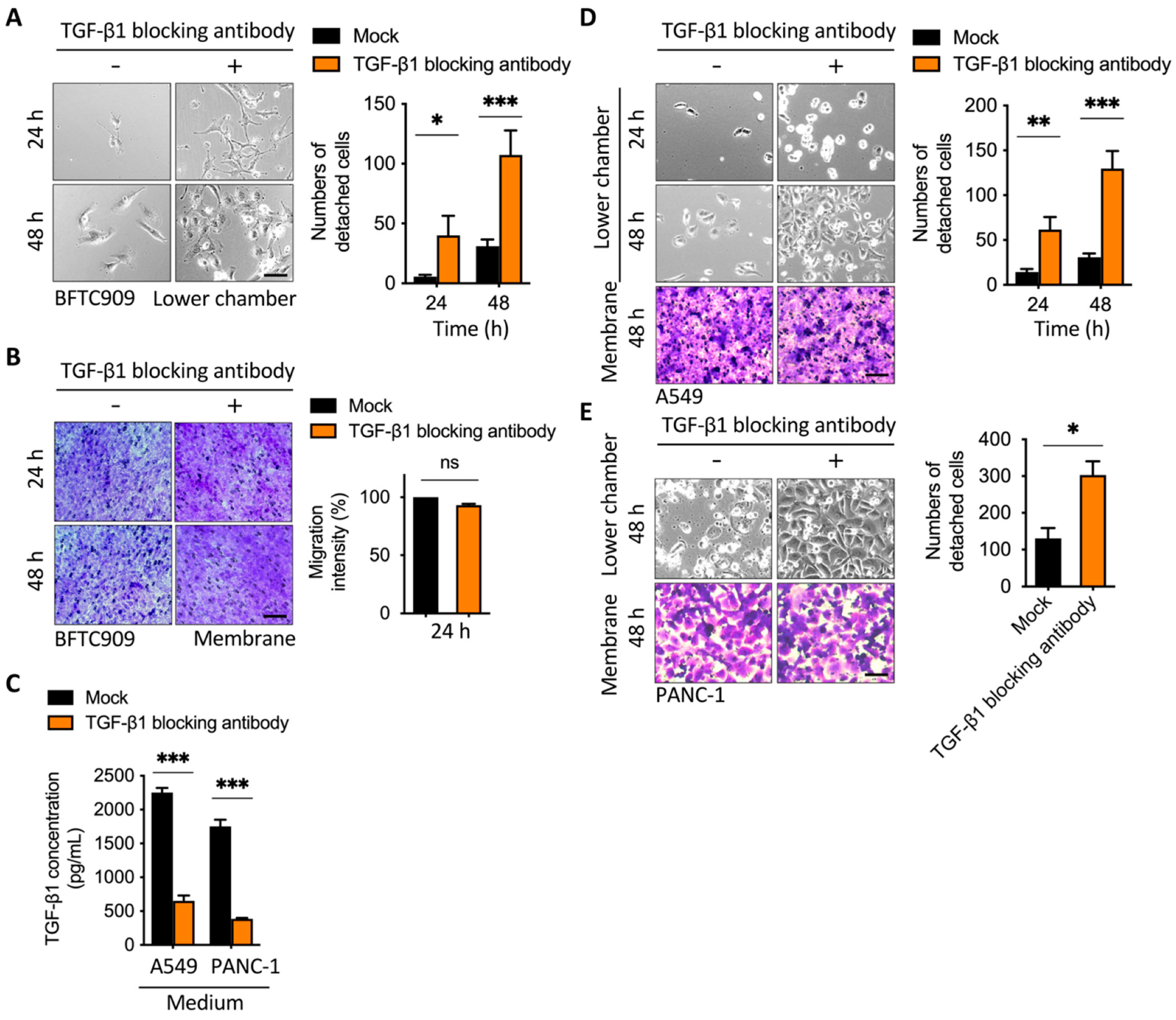
2.2. Blockage of TGF-β1 Enhanced Cell Detachment after Transmembrane Migration
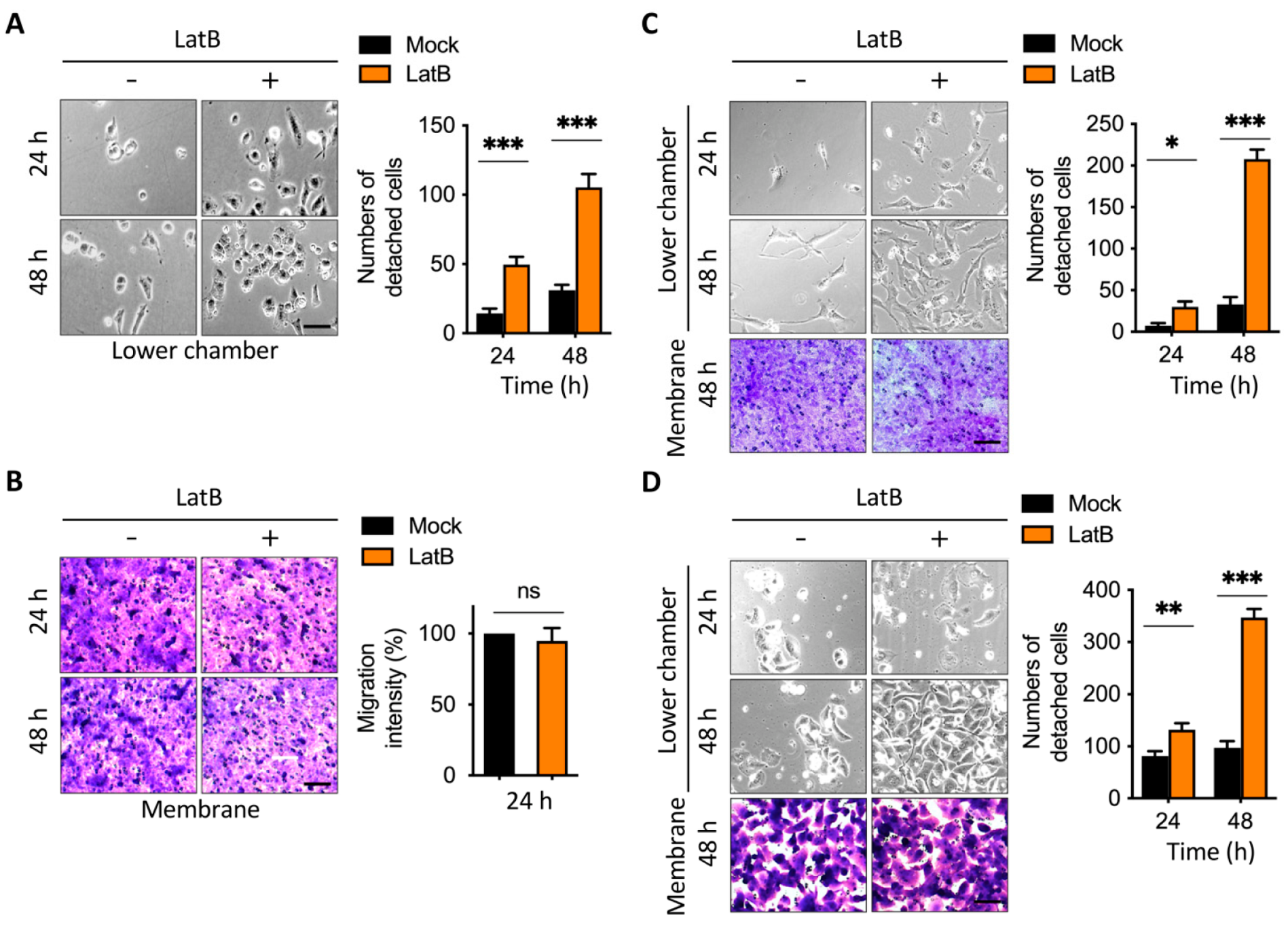
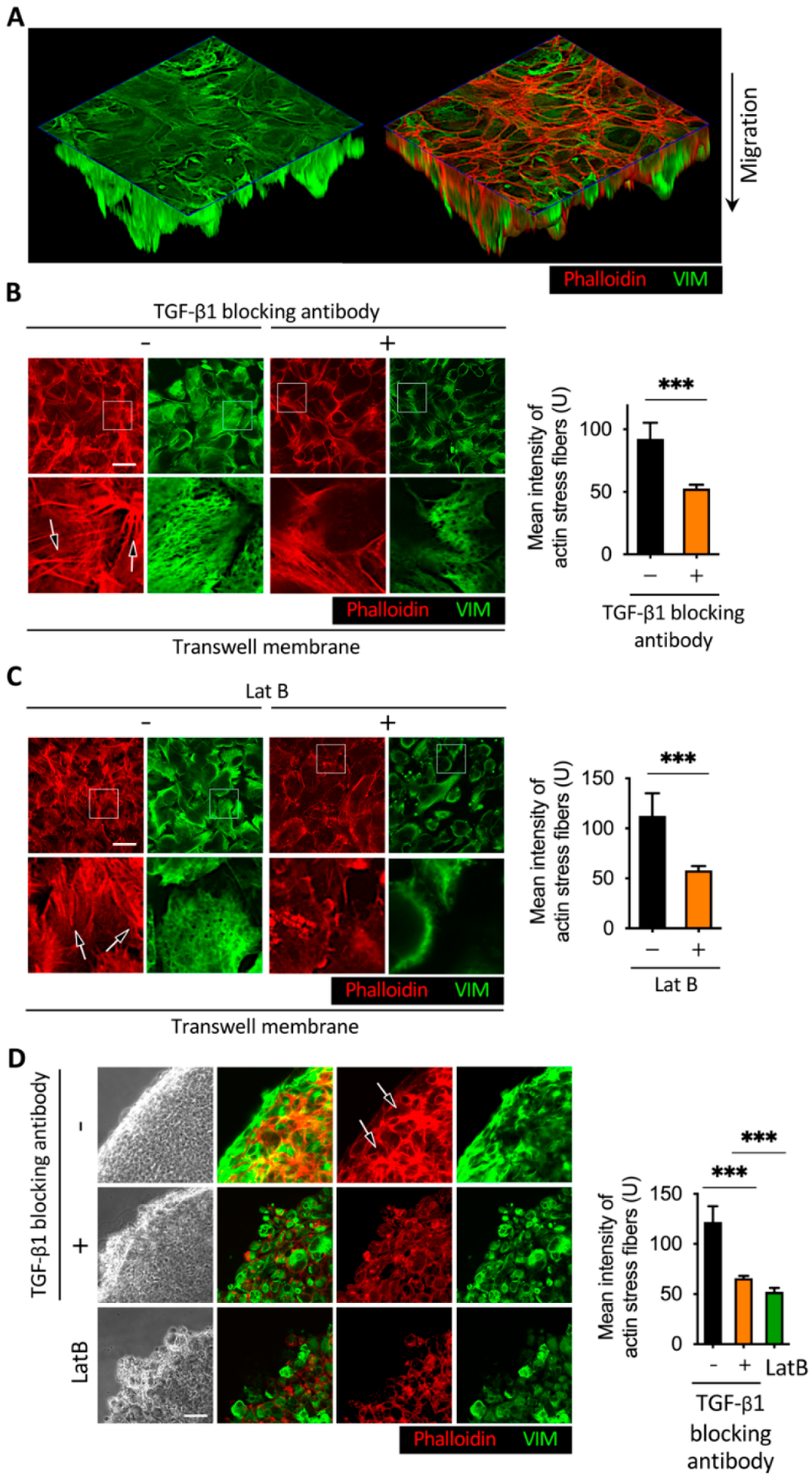
2.3. Disruption of Actin Cytoskeleton Remodeling Led to Reduction in TGF-β1-Induced Transmembrane Anchoring
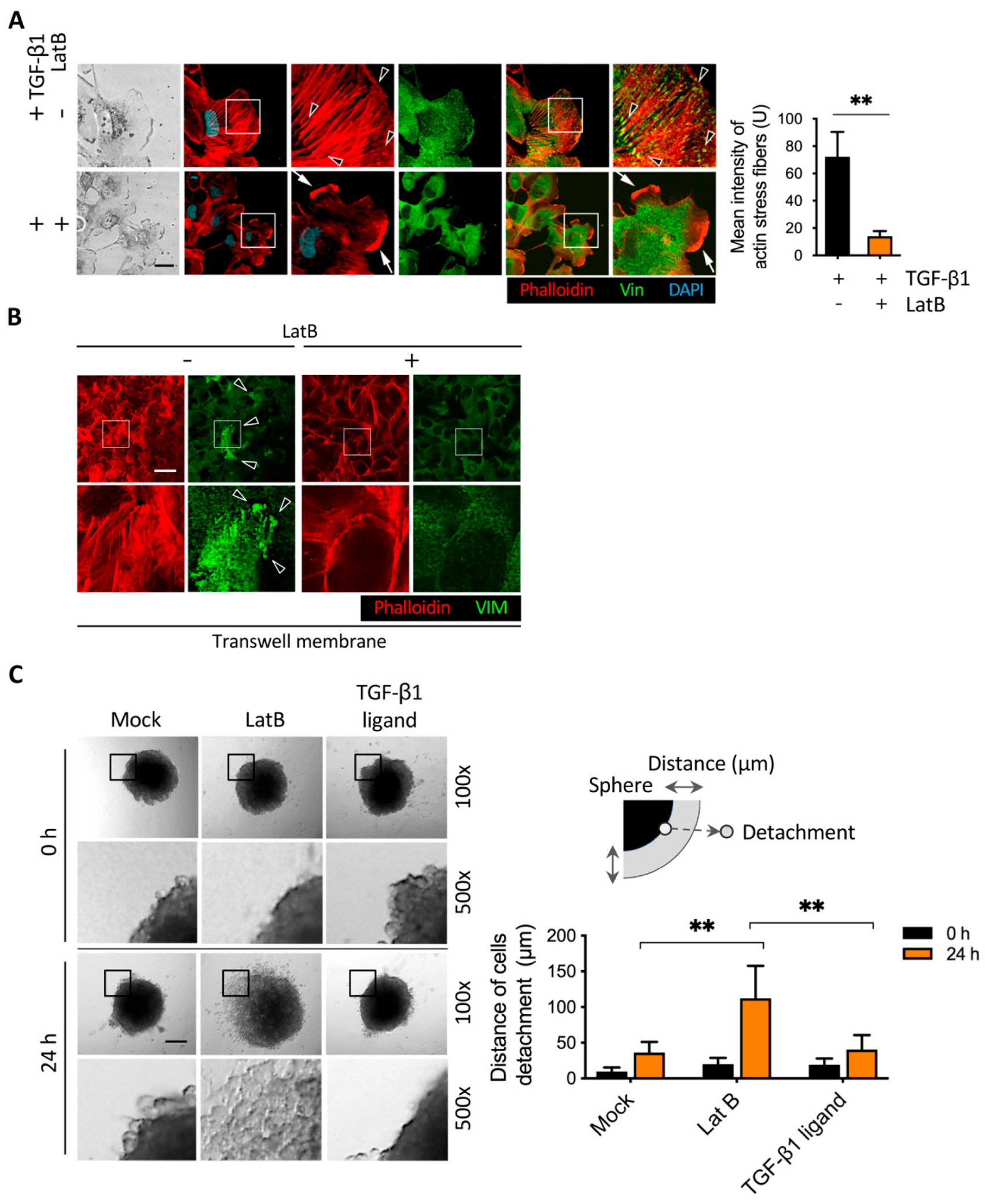
2.4. Latrunculin B Disrupted TGF-β1-Mediated Actin Stress Fibers and Vimentin Intermediate Filament Networks
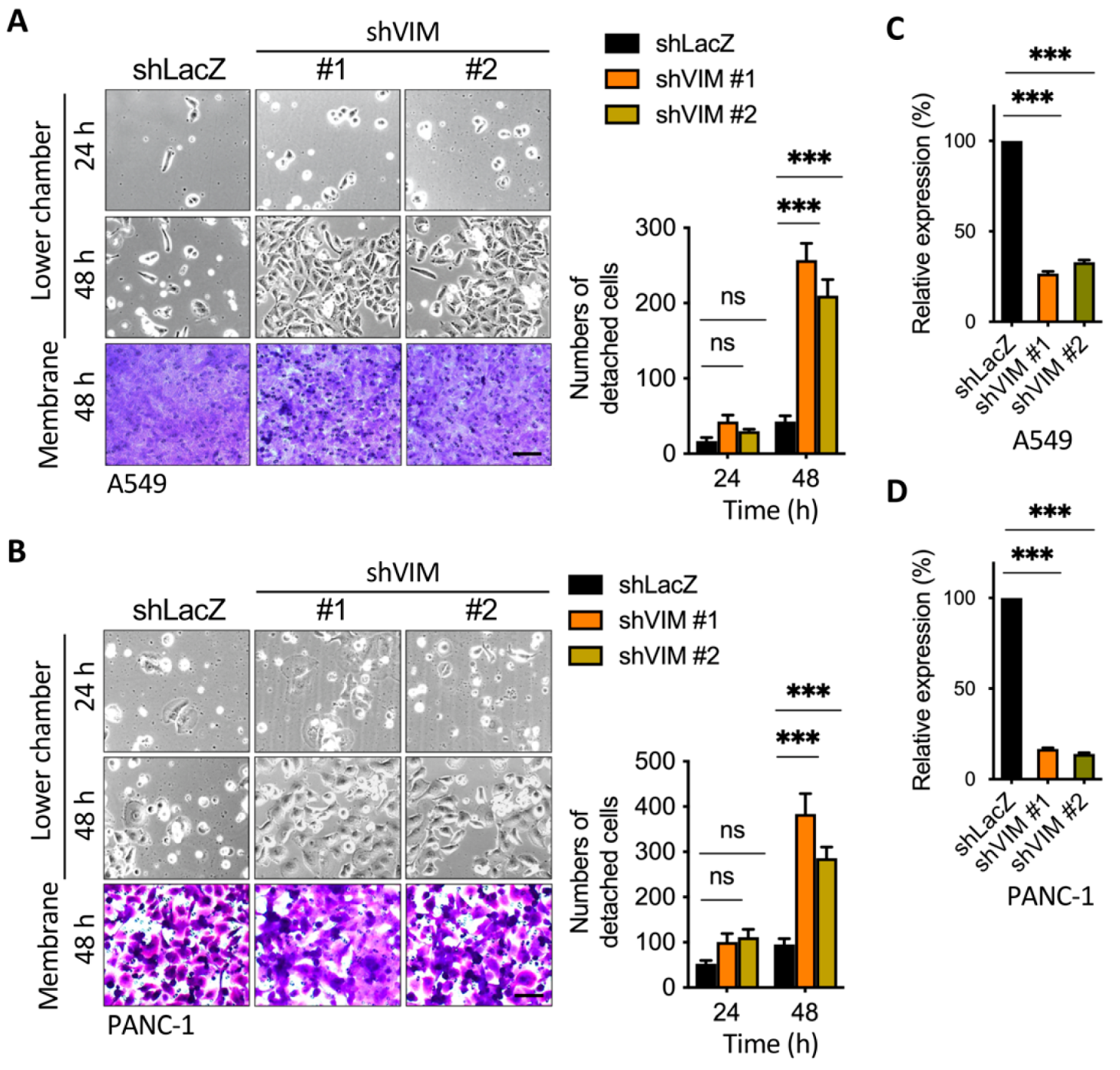
2.5. Silencing of Vimentin Led to a Reduction in Membrane Anchoring in the Invasive Leading Cells
2.6. Latrunculin B-Induced Focal Adhesion Proteins Translocation and Cell Detachment from Tumor Spheres
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Materials
4.2. shRNA, Lentivirus Infection, and Quantitative Real-Time PCR
4.3. Boyden Chamber Migration and Detachment Assay
4.4. Three-Dimensional In-Gel Spheroid Detachment Assay
4.5. Wound Healing Assay
4.6. Immunofluorescence Assay
4.7. ELISA of Secreted TGF-β1
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fletcher, D.A.; Mullins, R.D. Cell mechanics and the cytoskeleton. Nature 2010, 463, 485–492. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Wang, J. Mechanical tumor microenvironment and transduction: Cytoskeleton mediates cancer cell invasion and metastasis. Int. J. Biol. Sci. 2020, 16, 2014–2028. [Google Scholar] [CrossRef]
- Fife, C.M.; McCarroll, J.A.; Kavallaris, M. Movers and shakers: Cell cytoskeleton in cancer metastasis. Br. J. Pharmacol. 2014, 171, 5507–5523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marjoram, R.; Lessey, E.; Burridge, K. Regulation of RhoA Activity by Adhesion Molecules and Mechanotransduction. Curr. Mol. Med. 2014, 14, 199–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hinz, B. The extracellular matrix and transforming growth factor-β1: Tale of a strained relationship. Matrix Biol. 2015, 47, 54–65. [Google Scholar] [CrossRef] [PubMed]
- Hoot, K.E.; Lighthall, J.; Han, G.; Lu, S.L.; Li, A.; Ju, W.; Kulesz-Martin, M.; Bottinger, E.; Wang, X.J. Keratinocyte-specific Smad2 ablation results in increased epithelial-mesenchymal transition during skin cancer formation and progression. J. Clin. Investig. 2008, 118, 2722–2732. [Google Scholar] [CrossRef] [Green Version]
- Morita, T.; Mayanagi, T.; Sobue, K. Dual roles of myocardin-related transcription factors in epithelial–mesenchymal transition via slug induction and actin remodeling. J. Cell Biol. 2007, 179, 1027–1042. [Google Scholar] [CrossRef]
- Nishimura, G.; Manabe, I.; Tsushima, K.; Fujiu, K.; Oishi, Y.; Imai, Y.; Maemura, K.; Miyagishi, M.; Higashi, Y.; Kondoh, H.; et al. δEF1 Mediates TGF-β Signaling in Vascular Smooth Muscle Cell Differentiation. Dev. Cell 2006, 11, 93–104. [Google Scholar] [CrossRef] [Green Version]
- Xie, L.; Law, B.K.; Chytil, A.M.; Brown, K.A.; Aakre, M.E.; Moses, H.L. Activation of the Erk Pathway Is Required for TGF-β1-Induced EMT In Vitro. Neoplasia 2004, 6, 603–610. [Google Scholar] [CrossRef] [Green Version]
- Sousa-Squiavinato, A.C.M.; Rocha, M.R.; Barcellos-de-Souza, P.; de Souza, W.F.; Morgado-Diaz, J.A. Cofilin-1 signaling mediates epithelial-mesenchymal transition by promoting actin cytoskeleton reorganization and cell-cell adhesion regulation in colorectal cancer cells. Biochim. Et Biophys. Acta (BBA) Mol. Cell Res. 2019, 1866, 418–429. [Google Scholar] [CrossRef]
- Ridley, A. Life at the Leading Edge. Cell 2011, 145, 1012–1022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Derynck, R.; Zhang, Y.E. Smad-dependent and Smad-independent pathways in TGF-β family signalling. Nature 2003, 425, 577–584. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.E. Non-Smad Signaling Pathways of the TGF-β Family. Cold Spring Harb. Perspect. Biol. 2016, 9, a022129. [Google Scholar] [CrossRef] [PubMed]
- Pardoll, D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 2012, 12, 252–264. [Google Scholar] [CrossRef] [Green Version]
- Mariathasan, S.; Turley, S.J.; Nickles, D.; Castiglioni, A.; Yuen, K.; Wang, Y.; Kadel III, E.E.; Koeppen, H.; Astarita, J.L.; Cubas, R.; et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 2018, 554, 544–548. [Google Scholar] [CrossRef] [PubMed]
- Esue, O.; Carson, A.A.; Tseng, Y.; Wirtz, D. A Direct Interaction between Actin and Vimentin Filaments Mediated by the Tail Domain of Vimentin. J. Biol. Chem. 2006, 281, 30393–30399. [Google Scholar] [CrossRef] [Green Version]
- Tavares, S.; Vieira, A.F.; Taubenberger, A.V.; Araújo, M.; Martins, N.P.; Brás-Pereira, C.; Polónia, A.; Herbig, M.; Barreto, C.; Otto, O.; et al. Actin stress fiber organization promotes cell stiffening and proliferation of pre-invasive breast cancer cells. Nat. Commun. 2017, 8, 15237. [Google Scholar] [CrossRef]
- Havel, L.S.; Kline, E.R.; Salgueiro, A.M.; Marcus, A.I. Vimentin regulates lung cancer cell adhesion through a VAV2–Rac1 pathway to control focal adhesion kinase activity. Oncogene 2014, 34, 1979–1990. [Google Scholar] [CrossRef] [Green Version]
- Vuoriluoto, K.; Haugen, H.; Kiviluoto, S.; Mpindi, J.P.; Nevo, J.; Gjerdrum, C.; Tiron, C.; Lorens, J.B.; Ivaska, J. Vimentin regulates EMT induction by Slug and oncogenic H-Ras and migration by governing Axl expression in breast cancer. Oncogene 2010, 30, 1436–1448. [Google Scholar] [CrossRef] [Green Version]
- Sulzmaier, F.J.; Jean, C.; Schlaepfer, D.D. FAK in cancer: Mechanistic findings and clinical applications. Nat. Rev. Cancer 2014, 14, 598–610. [Google Scholar] [CrossRef] [Green Version]
- Serrels, A.; Lund, T.; Serrels, B.; Byron, A.; McPherson, R.; von Kriegsheim, A.; Gómez-Cuadrado, L.; Canel, M.; Muir, M.; Ring, J.; et al. Nuclear FAK Controls Chemokine Transcription, Tregs, and Evasion of Anti-tumor Immunity. Cell 2015, 163, 160–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, H.; Hegde, S.; Knolhoff, B.L.; Zhu, Y.; Herndon, J.M.; Meyer, M.A.; Nywening, T.M.; Hawkins, W.G.; Shapiro, I.M.; Weaver, D.T.; et al. Targeting focal adhesion kinase renders pancreatic cancers responsive to checkpoint immunotherapy. Nat. Med. 2016, 22, 851–860. [Google Scholar] [CrossRef] [PubMed]
- Orgaz, J.L.; Crosas-Molist, E.; Sadok, A.; Perdrix-Rosell, A.; Maiques, O.; Rodriguez-Hernandez, I.; Monger, J.; Mele, S.; Georgouli, M.; Bridgeman, V.; et al. Myosin II Reactivation and Cytoskeletal Remodeling as a Hallmark and a Vulnerability in Melanoma Therapy Resistance. Cancer Cell 2020, 37, 85–103.e9. [Google Scholar] [CrossRef] [PubMed]
- Jiu, Y.; Peränen, J.; Schaible, N.; Cheng, F.; Eriksson, J.E.; Krishnan, R.; Lappalainen, P. Vimentin intermediate filaments control actin stress fiber assembly through GEF-H1 and RhoA. J. Cell Sci. 2017, 130, 892–902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ostrowska-Podhorodecka, Z.; McCulloch, C.A. Vimentin regulates the assembly and function of matrix adhesions. Wound Repair Regen. 2021, 29, 602–612. [Google Scholar] [CrossRef]
- Van Zijl, F.; Krupitza, G.; Mikulits, W. Initial steps of metastasis: Cell invasion and endothelial transmigration. Mutat. Res./Rev. Mutat. Res. 2011, 728, 23–34. [Google Scholar] [CrossRef]
- Ungefroren, H. Autocrine TGF-β in Cancer: Review of the Literature and Caveats in Experimental Analysis. Int. J. Mol. Sci. 2021, 22, 977. [Google Scholar] [CrossRef]
- Ungefroren, H.; Otterbein, H.; Wellner, U.F.; Keck, T.; Lehnert, H.; Marquardt, J.U. RAC1B Regulation of TGFB1 Reveals an Unexpected Role of Autocrine TGFβ1 in the Suppression of Cell Motility. Cancers 2020, 12, 3570. [Google Scholar] [CrossRef]
- Principe, D.R.; Doll, J.A.; Bauer, J.; Jung, B.; Munshi, H.G.; Bartholin, L.; Pasche, B.; Lee, C.; Grippo, P.J. TGF-β: Duality of Function Between Tumor Prevention and Carcinogenesis. J. Natl. Cancer Inst. 2014, 106, djt369. [Google Scholar] [CrossRef]
- Peng, J.M.; Bera, R.; Chiou, C.Y.; Yu, M.C.; Chen, T.C.; Chen, C.W.; Wang, T.R.; Chiang, W.L.; Chai, S.P.; Wei, Y.; et al. Actin cytoskeleton remodeling drives epithelial-mesenchymal transition for hepatoma invasion and metastasis in mice. Hepatology 2018, 67, 2226–2243. [Google Scholar] [CrossRef] [Green Version]
- Hubchak, S.C.; Runyan, C.E.; Kreisberg, J.I.; Schnaper, H.W. Cytoskeletal Rearrangement and Signal Transduction in TGF-β1–Stimulated Mesangial Cell Collagen Accumulation. J. Am. Soc. Nephrol. 2003, 14, 1969–1980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Itoh, T.; Erdmann, K.S.; Roux, A.; Habermann, B.; Werner, H.; de Camilli, P. Dynamin and the Actin Cytoskeleton Cooperatively Regulate Plasma Membrane Invagination by BAR and F-BAR Proteins. Dev. Cell 2005, 9, 791–804. [Google Scholar] [CrossRef] [PubMed]
- Jiu, Y.; Lehtimäki, J.; Tojkander, S.; Cheng, F.; Jäälinoja, H.; Liu, X.; Varjosalo, M.; Eriksson, J.; Lappalainen, P. Bidirectional Interplay between Vimentin Intermediate Filaments and Contractile Actin Stress Fibers. Cell Rep. 2015, 11, 1511–1518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edlund, S.; Landström, M.; Heldin, C.H.; Aspenström, P. Transforming Growth Factor-β–induced Mobilization of Actin Cytoskeleton Requires Signaling by Small GTPases Cdc42 and RhoA. Mol. Biol. Cell 2002, 13, 902–914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.N.; Koo, K.H.; Sung, J.Y.; Yun, U.J.; Kim, H. Anoikis Resistance: An Essential Prerequisite for Tumor Metastasis. Int. J. Cell Biol. 2012, 2012, 306879. [Google Scholar] [CrossRef] [Green Version]
- Chen, N.; Balasenthil, S.; Reuther, J.; Frayna, A.; Wang, Y.; Chandler, D.S.; Abruzzo, L.V.; Rashid, A.; Rodriguez, J.; Lozano, G.; et al. DEAR1 Is a Chromosome 1p35 Tumor Suppressor and Master Regulator of TGF-β–Driven Epithelial–Mesenchymal Transition. Cancer Discov. 2013, 3, 1172–1189. [Google Scholar] [CrossRef] [Green Version]
- Yan, X.; Zhou, R.; Ma, Z. Autophagy—Cell Survival and Death. Autophagy Biol. Dis. 2019, 667–696. [Google Scholar] [CrossRef]
- White, E. Deconvoluting the context-dependent role for autophagy in cancer. Nat. Rev. Cancer 2012, 12, 401–410. [Google Scholar] [CrossRef] [Green Version]
- Fung, C.; Lock, R.; Gao, S.; Salas, E.; Debnath, J. Induction of Autophagy during Extracellular Matrix Detachment Promotes Cell Survival. Mol. Biol. Cell 2008, 19, 797–806. [Google Scholar] [CrossRef] [Green Version]
- Avivar-Valderas, A.; Salas, E.; Bobrovnikova-Marjon, E.; Diehl, J.A.; Nagi, C.; Debnath, J.; Aguirre-Ghiso, J.A. PERK Integrates Autophagy and Oxidative Stress Responses to Promote Survival during Extracellular Matrix Detachment. Mol. Cell. Biol. 2011, 31, 3616–3629. [Google Scholar] [CrossRef] [Green Version]
- Avivar-Valderas, A.; Bobrovnikova-Marjon, E.; Alan Diehl, J.; Bardeesy, N.; Debnath, J.; Aguirre-Ghiso, J.A. Regulation of autophagy during ECM detachment is linked to a selective inhibition of mTORC1 by PERK. Oncogene 2012, 32, 4932–4940. [Google Scholar] [CrossRef] [Green Version]
- Chen, N.; Debnath, J. IκB kinase complex (IKK) triggers detachment-induced autophagy in mammary epithelial cells independently of the PI3K-AKT-MTORC1 pathway. Autophagy 2013, 9, 1214–1227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schafer, Z.T.; Grassian, A.R.; Song, L.; Jiang, Z.; Gerhart-Hines, Z.; Irie, H.Y.; Gao, S.; Puigserver, P.; Brugge, J.S. Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature 2009, 461, 109–113. [Google Scholar] [CrossRef] [Green Version]
- Davison, C.A.; Durbin, S.M.; Thau, M.R.; Zellmer, V.R.; Chapman, S.E.; Diener, J.; Wathen, C.; Leevy, W.M.; Schafer, Z.T. Antioxidant Enzymes Mediate Survival of Breast Cancer Cells Deprived of Extracellular Matrix. Cancer Res. 2013, 73, 3704–3715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grassian, A.R.; Metallo, C.M.; Coloff, J.L.; Stephanopoulos, G.; Brugge, J.S. Erk regulation of pyruvate dehydrogenase flux through PDK4 modulates cell proliferation. Genes Dev. 2011, 25, 1716–1733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeNicola, G.M.; Karreth, F.A.; Humpton, T.J.; Gopinathan, A.; Wei, C.; Frese, K.; Mangal, D.; Yu, K.H.; Yeo, C.J.; Calhoun, E.S.; et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature 2011, 475, 106–109. [Google Scholar] [CrossRef] [PubMed]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular mechanisms of epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196. [Google Scholar] [CrossRef] [Green Version]
- Derynck, R.; Turley, S.J.; Akhurst, R.J. TGFβ biology in cancer progression and immunotherapy. Nat. Rev. Clin. Oncol. 2020, 18, 9–34. [Google Scholar] [CrossRef]
- Tauriello, D.V.F.; Palomo-Ponce, S.; Stork, D.; Berenguer-Llergo, A.; Badia-Ramentol, J.; Iglesias, M.; Sevillano, M.; Ibiza, S.; Cañellas, A.; Hernando-Momblona, X.; et al. TGFβ drives immune evasion in genetically reconstituted colon cancer metastasis. Nature 2018, 554, 538–543. [Google Scholar] [CrossRef] [Green Version]
- Zhao, F.; Evans, K.; Xiao, C.; DeVito, N.; Theivanthiran, B.; Holtzhausen, A.; Siska, P.J.; Blobe, G.C.; Hanks, B.A. Stromal Fibroblasts Mediate Anti–PD-1 Resistance via MMP-9 and Dictate TGFβ Inhibitor Sequencing in Melanoma. Cancer Immunol. Res. 2018, 6, 1459–1471. [Google Scholar] [CrossRef]
- Martin, C.J.; Datta, A.; Littlefield, C.; Kalra, A.; Chapron, C.; Wawersik, S.; Dagbay, K.B.; Brueckner, C.T.; Nikiforov, A.; Danehy, F.T.; et al. Selective inhibition of TGFβ1 activation overcomes primary resistance to checkpoint blockade therapy by altering tumor immune landscape. Sci. Transl. Med. 2020, 12, eaay8456. [Google Scholar] [CrossRef]
- Dodagatta-Marri, E.; Meyer, D.S.; Reeves, M.Q.; Paniagua, R.; To, M.D.; Binnewies, M.; Broz, M.L.; Mori, H.; Wu, D.; Adoumie, M.; et al. α-PD-1 therapy elevates Treg/Th balance and increases tumor cell pSmad3 that are both targeted by α-TGFβ antibody to promote durable rejection and immunity in squamous cell carcinomas. J. ImmunoTherapy Cancer 2019, 7, 62. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Ye, W.L.; Zhang, R.N.; He, X.S.; Wang, J.R.; Liu, Y.X.; Wang, Y.; Yang, X.M.; Zhang, Y.J.; Gan, W.J. The Role of TGF-β Signaling Pathways in Cancer and Its Potential as a Therapeutic Target. Evid. Based Complementary Altern. Med. 2021, 2021, 6675208. [Google Scholar] [CrossRef] [PubMed]
- Sato, K.; Tsuchihara, K.; Fujii, S.; Sugiyama, M.; Goya, T.; Atomi, Y.; Ueno, T.; Ochiai, A.; Esumi, H. Autophagy Is Activated in Colorectal Cancer Cells and Contributes to the Tolerance to Nutrient Deprivation. Cancer Res. 2007, 67, 9677–9684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kato, K.; Ogura, T.; Kishimoto, A.; Minegishi, Y.; Nakajima, N.; Miyazaki, M.; Esumi, H. Critical roles of AMP-activated protein kinase in constitutive tolerance of cancer cells to nutrient deprivation and tumor formation. Oncogene 2002, 21, 6082–6090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, H.L.; Liu, H.Y.; Chang, Y.L.; Sung, M.T.; Chen, P.Y.; Su, Y.L.; Huang, C.C.; Peng, J.M. Hypomethylated RRBP1 Potentiates Tumor Malignancy and Chemoresistance in Upper Tract Urothelial Carcinoma. Int. J. Mol. Sci. 2021, 22, 8761. [Google Scholar] [CrossRef] [PubMed]
- Sherman, H.; Gitschier, H.J.; Rossi, A.E. A Novel Three-Dimensional Immune Oncology Model for High-Throughput Testing of Tumoricidal Activity. Front. Immunol. 2018, 9, 857. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, J.-M.; Chen, W.-Y.; Cheng, J.-H.; Luo, J.-W.; Tzeng, H.-T. Dysregulation of Cytoskeleton Remodeling Drives Invasive Leading Cells Detachment. Cancers 2021, 13, 5648. https://doi.org/10.3390/cancers13225648
Peng J-M, Chen W-Y, Cheng J-H, Luo J-W, Tzeng H-T. Dysregulation of Cytoskeleton Remodeling Drives Invasive Leading Cells Detachment. Cancers. 2021; 13(22):5648. https://doi.org/10.3390/cancers13225648
Chicago/Turabian StylePeng, Jei-Ming, Wei-Yu Chen, Jai-Hong Cheng, Jia-Wun Luo, and Hong-Tai Tzeng. 2021. "Dysregulation of Cytoskeleton Remodeling Drives Invasive Leading Cells Detachment" Cancers 13, no. 22: 5648. https://doi.org/10.3390/cancers13225648






