Mitotic Centromere-Associated Kinesin (MCAK/KIF2C) Regulates Cell Migration and Invasion by Modulating Microtubule Dynamics and Focal Adhesion Turnover
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. CRISPRi/A Cell Line Generation, Cell Culture and siRNA Transfection
2.2. Western Blot and Immunofluorescence (IF) Staining
2.3. RNA Extraction and Real-Time PCR
2.4. Cell Cycle, Cell Viability, Motility, Migration and Invasion
2.5. Cell Spreading/Adhesion, Actin Repolymerization and Nocodazole Washout Assay
2.6. FA Turnover and EB3 (End Binding Protein 3) Tracking
2.7. Statistical Analysis
3. Results
3.1. Characterization of HeLa/RPE CRISPR/dCas9 sgMCAK Knockdown (CRISPRi) and Overexpression (CRISPRa) Cell Lines
3.2. Interfering with the Expression of MCAK Triggers Mitotic Defects
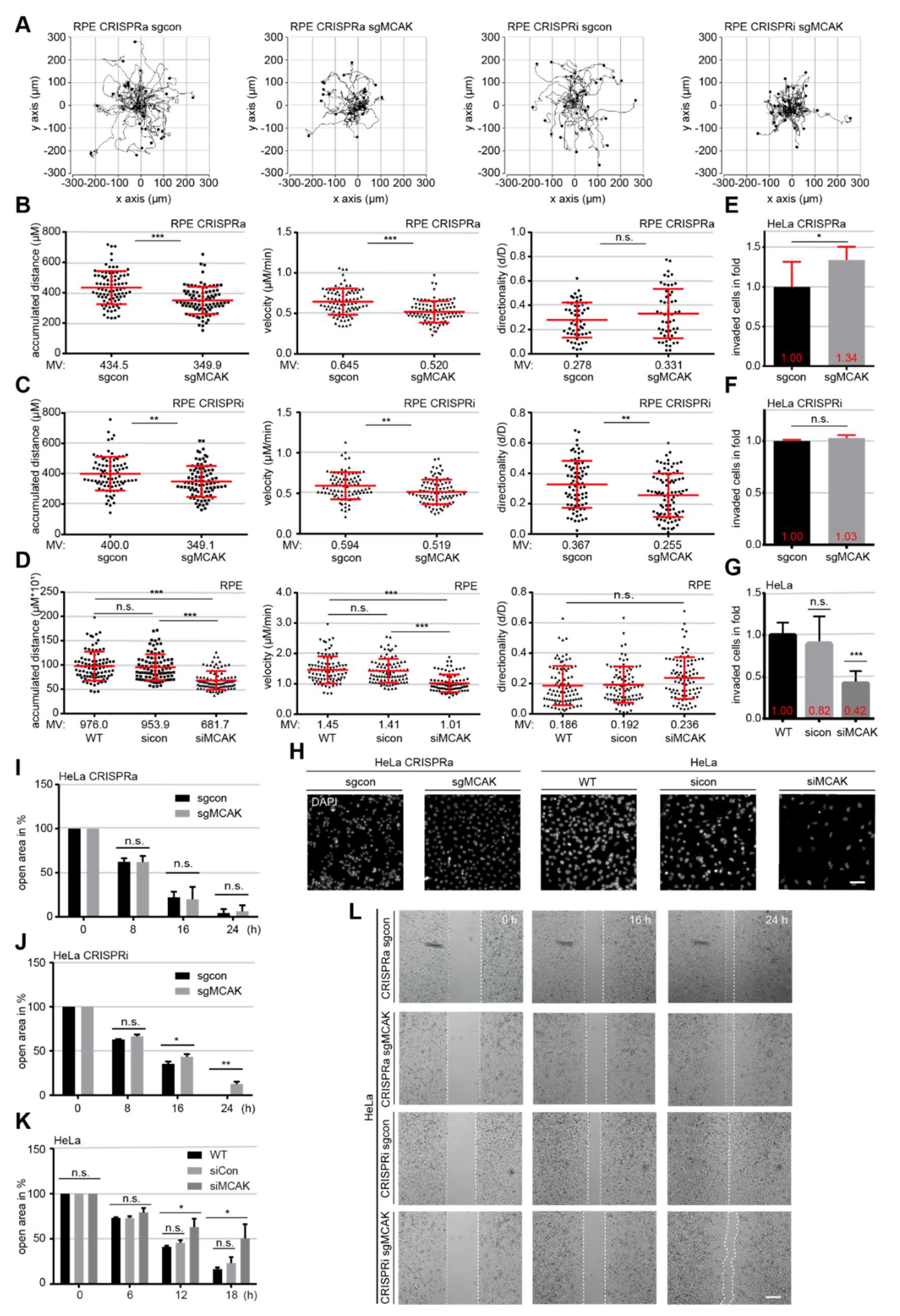
3.3. MCAK Overexpression or Downregulation Impairs Cell Motility and Migration
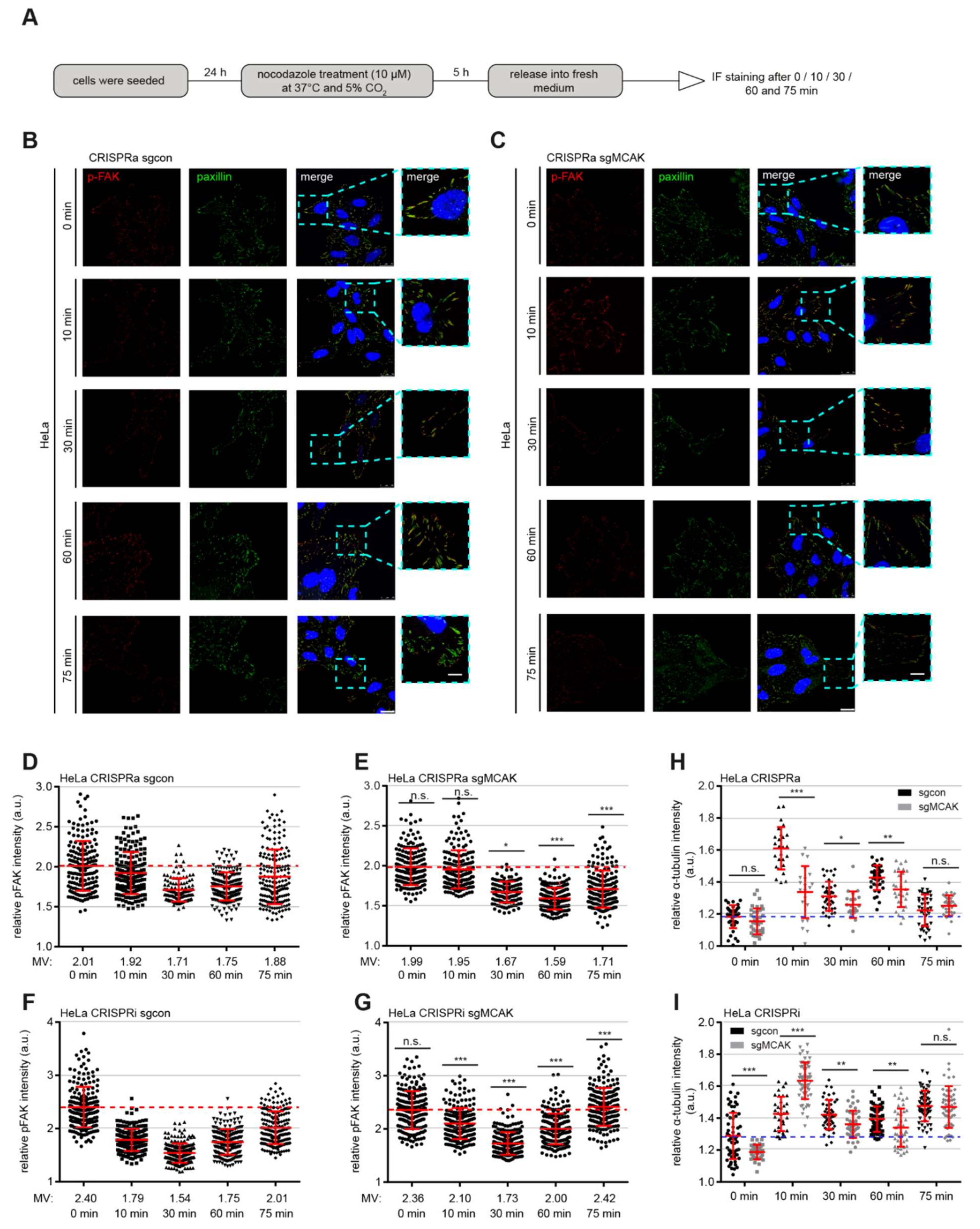
3.4. Interfering with the Expression of MCAK Impacts the Timely Accumulation of Paxillin, FAK, and Their Phosphorylated Forms in FAs
3.5. Deregulation of MCAK Delays CELL Spreading and Adhesion
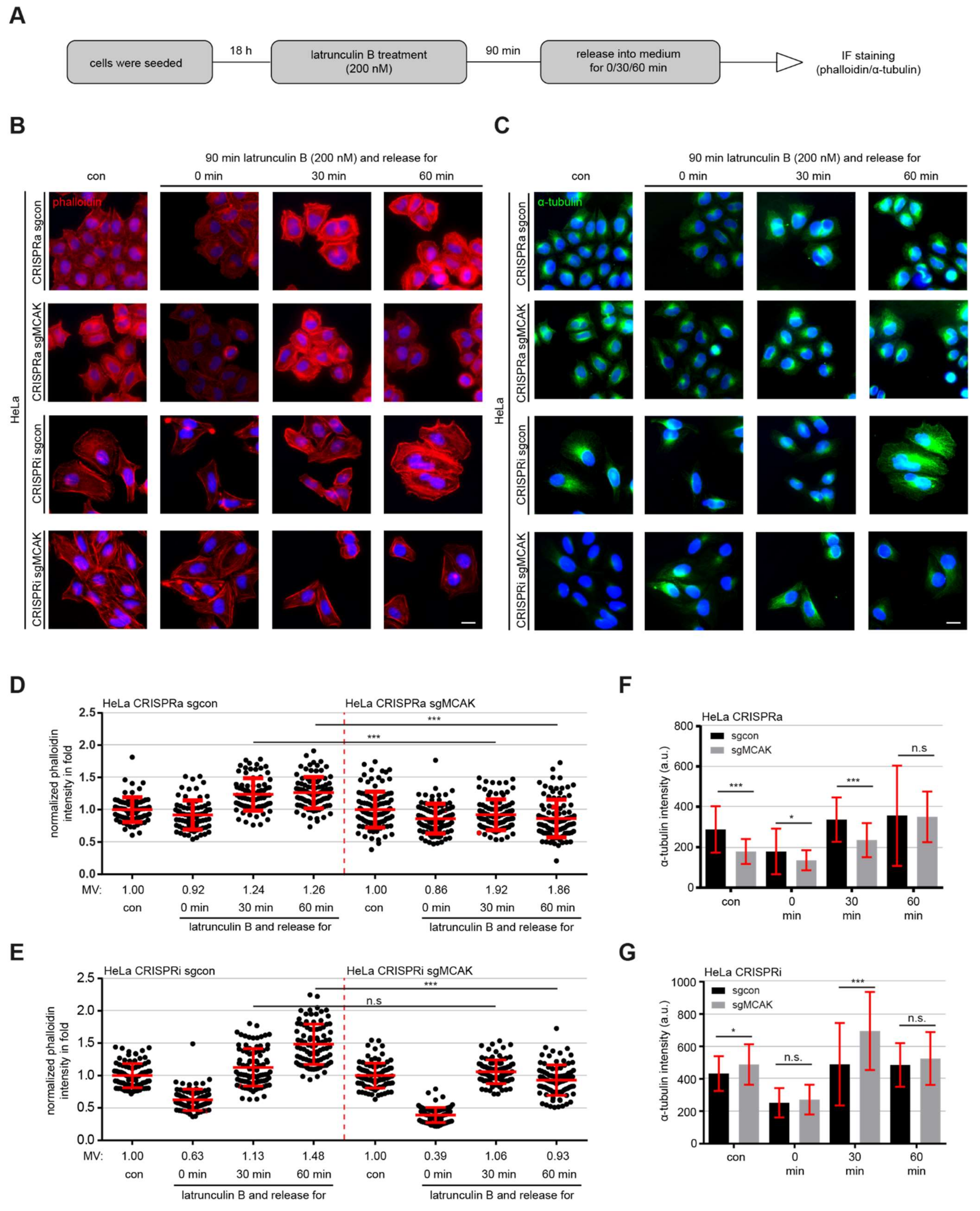
3.6. MCAK Modulates Actin- and MT Dynamics
3.7. Overexpression or Suppression of MCAK Perturbs MT-Induced FA Turnover
3.8. MCAK Is Involved in FA Lifetime and MT Plus-Tip Dynamics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- SenGupta, S.; Parent, C.A.; Bear, J.E. The principles of directed cell migration. Nat. Rev. Mol. Cell Biol. 2021, 22, 529–547. [Google Scholar] [CrossRef]
- Bravo-Cordero, J.J.; Hodgson, L.; Condeelis, J. Directed cell invasion and migration during metastasis. Curr. Opin. Cell Biol. 2012, 24, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Akhshi, T.K.; Wernike, D.; Piekny, A. Microtubules and actin crosstalk in cell migration and division. Cytoskeleton 2014, 71, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Seetharaman, S.; Etienne-Manneville, S. Cytoskeletal Crosstalk in Cell Migration. Trends Cell Biol. 2020, 30, 720–735. [Google Scholar] [CrossRef] [PubMed]
- Garcin, C.; Straube, A. Microtubules in cell migration. Essays Biochem. 2019, 63, 509–520. [Google Scholar] [CrossRef] [PubMed]
- Wehrle-Haller, B.; Imhof, B.A. Actin, microtubules and focal adhesion dynamics during cell migration. Int. J. Biochem. Cell Biol. 2003, 35, 39–50. [Google Scholar] [CrossRef]
- Schaks, M.; Giannone, G.; Rottner, K. Actin dynamics in cell migration. Essays Biochem. 2019, 63, 483–495. [Google Scholar] [CrossRef]
- Seetharaman, S.; Etienne-Manneville, S. Microtubules at focal adhesions—A double-edged sword. J. Cell Sci. 2019, 132, jcs232843. [Google Scholar] [CrossRef] [PubMed]
- Müntjes, K.; Devan, S.K.; Reichert, A.S.; Feldbrügge, M. Linking transport and translation of mRNAs with endosomes and mitochondria. EMBO Rep. 2021, 22, e52445. [Google Scholar] [CrossRef] [PubMed]
- Rooney, C.; White, G.; Nazgiewicz, A.; Woodcock, S.A.; Anderson, K.I.; Ballestrem, C.; Malliri, A. The Rac activator STEF (Tiam2) regulates cell migration by microtubule-mediated focal adhesion disassembly. EMBO Rep. 2010, 11, 292–298. [Google Scholar] [CrossRef] [PubMed]
- Paradzik, M.; Humphries, J.D.; Stojanovic, N.; Nestic, D.; Majhen, D.; Dekanic, A.; Samarzija, I.; Sedda, D.; Weber, I.; Humphries, M.J.; et al. KANK2 Links alphaVbeta5 Focal Adhesions to Microtubules and Regulates Sensitivity to Microtubule Poisons and Cell Migration. Front. Cell Dev. Biol. 2020, 8, 125. [Google Scholar] [CrossRef]
- Humphries, J.D.; Chastney, M.R.; Askari, J.A.; Humphries, M.J. Signal transduction via integrin adhesion complexes. Curr. Opin. Cell Biol. 2019, 56, 14–21. [Google Scholar] [CrossRef]
- Lopez-Colome, A.M.; Lee-Rivera, I.; Benavides-Hidalgo, R.; Lopez, E. Paxillin: A crossroad in pathological cell migration. J. Hematol. Oncol. 2017, 10, 50. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Ruest, P.J.; Hanks, S.K. FAK regulates tyrosine phosphorylation of CAS, paxillin, and PYK2 in cells expressing v-Src, but is not a critical determinant of v-Src transformation. J. Cell. Biochem. 2002, 84, 377–388. [Google Scholar] [CrossRef] [PubMed]
- Ems-McClung, S.C.; Walczak, C.E. Kinesin-13s in mitosis: Key players in the spatial and temporal organization of spindle microtubules. Semin. Cell Dev. Biol. 2010, 21, 276–282. [Google Scholar] [CrossRef] [PubMed]
- Ritter, A.; Kreis, N.N.; Louwen, F.; Wordeman, L.; Yuan, J. Molecular insight into the regulation and function of MCAK. Crit. Rev. Biochem. Mol. Biol. 2015, 51, 228–245. [Google Scholar] [CrossRef]
- Steblyanko, Y.; Rajendraprasad, G.; Osswald, M.; Eibes, S.; Jacome, A.; Geley, S.; Pereira, A.J.; Maiato, H.; Barisic, M. Microtubule poleward flux in human cells is driven by the coordinated action of four kinesins. EMBO J. 2020, 39, e105432. [Google Scholar] [CrossRef] [PubMed]
- Sanhaji, M.; Friel, C.T.; Wordeman, L.; Louwen, F.; Yuan, J. Mitotic centromere-associated kinesin (MCAK): A potential cancer drug target. Oncotarget 2011, 2, 935–947. [Google Scholar] [CrossRef]
- Ritter, A.; Sanhaji, M.; Friemel, A.; Roth, S.; Rolle, U.; Louwen, F.; Yuan, J. Functional analysis of phosphorylation of the mitotic centromere-associated kinesin by Aurora B kinase in human tumor cells. Cell Cycle 2015, 14, 3755–3767. [Google Scholar] [CrossRef] [PubMed]
- Braun, A.; Dang, K.; Buslig, F.; Baird, M.A.; Davidson, M.W.; Waterman, C.M.; Myers, K.A. Rac1 and Aurora A regulate MCAK to polarize microtubule growth in migrating endothelial cells. J. Cell Biol. 2014, 206, 97–112. [Google Scholar] [CrossRef] [PubMed]
- Zong, H.; Hazelbaker, M.; Moe, C.; Ems-McClung, S.C.; Hu, K.; Walczak, C.E. Spatial regulation of MCAK promotes cell polarization and focal adhesion turnover to drive robust cell migration. Mol. Biol. Cell 2021, 32, 590–604. [Google Scholar] [CrossRef] [PubMed]
- Jost, M.; Chen, Y.; Gilbert, L.A.; Horlbeck, M.A.; Krenning, L.; Menchon, G.; Rai, A.; Cho, M.Y.; Stern, J.J.; Prota, A.E.; et al. Combined CRISPRi/a-Based Chemical Genetic Screens Reveal that Rigosertib Is a Microtubule-Destabilizing Agent. Mol. Cell 2017, 68, 210–223e6. [Google Scholar] [CrossRef] [PubMed]
- Sanhaji, M.; Ritter, A.; Belsham, H.R.; Friel, C.T.; Roth, S.; Louwen, F.; Yuan, J. Polo-like kinase 1 regulates the stability of the mitotic centromere-associated kinesin in mitosis. Oncotarget 2014, 5, 3130–3144. [Google Scholar] [CrossRef]
- Ritter, A.; Sanhaji, M.; Steinhauser, K.; Roth, S.; Louwen, F.; Yuan, J. The activity regulation of the mitotic centromere-associated kinesin by Polo-like kinase 1. Oncotarget 2015, 6, 6641–6655. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Muschol-Steinmetz, C.; Friemel, A.; Kreis, N.N.; Reinhard, J.; Yuan, J.; Louwen, F. Function of survivin in trophoblastic cells of the placenta. PLoS ONE 2013, 8, e73337. [Google Scholar] [CrossRef] [PubMed]
- Kreis, N.N.; Sommer, K.; Sanhaji, M.; Kramer, A.; Matthess, Y.; Kaufmann, M.; Strebhardt, K.; Yuan, J. Long-term downregulation of Polo-like kinase 1 increases the cyclin-dependent kinase inhibitor p21(WAF1/CIP1). Cell Cycle 2009, 8, 460–472. [Google Scholar] [CrossRef]
- Kreis, N.N.; Louwen, F.; Zimmer, B.; Yuan, J. Loss of p21Cip1/CDKN1A renders cancer cells susceptible to Polo-like kinase 1 inhibition. Oncotarget 2015, 6, 6611–6626. [Google Scholar] [CrossRef]
- Ritter, A.; Roth, S.; Kreis, N.N.; Friemel, A.; Hoock, S.C.; Souto, A.S.; Eichbaum, C.; Neuhoff, A.; Chen, Q.; Solbach, C.; et al. Primary Cilia in Trophoblastic Cells: Potential Involvement in Preeclampsia. Hypertension 2020, 76, 1491–1505. [Google Scholar] [CrossRef]
- Applegate, K.T.; Besson, S.; Matov, A.; Bagonis, M.H.; Jaqaman, K.; Danuser, G. plusTipTracker: Quantitative image analysis software for the measurement of microtubule dynamics. J. Struct. Biol. 2011, 176, 168–184. [Google Scholar] [CrossRef]
- Gilbert, L.A.; Larson, M.H.; Morsut, L.; Liu, Z.; Brar, G.A.; Torres, S.E.; Stern-Ginossar, N.; Brandman, O.; Whitehead, E.H.; Doudna, J.A.; et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 2013, 154, 442–451. [Google Scholar] [CrossRef]
- Jones, M.C.; Zha, J.Z.; Humphries, M.J. Connections between the cell cycle, cell adhesion and the cytoskeleton. Philos. Trans. R. Soc. B 2019, 374, 20180227. [Google Scholar] [CrossRef]
- Vicente, J.J.; Wordeman, L. The quantification and regulation of microtubule dynamics in the mitotic spindle. Curr. Opin. Cell Biol. 2019, 60, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Qi, Q.; Hou, S.; Chen, Z.; Jiang, N.; Zhang, L.; Lin, C. Activation of the TGF-β1/Smad signaling by KIF2C contributes to the malignant phenotype of thyroid carcinoma cells. Tissue Cell 2021, 73, 101655. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Jia, H.; Yu, F.; Guo, H.; Li, B. KIF2C promotes the proliferation of hepatocellular carcinoma cells in vitro and in vivo. Exp. Ther. Med. 2021, 22, 1094. [Google Scholar] [CrossRef]
- Eichenlaub-Ritter, U.; Staubach, N.; Trapphoff, T. Chromosomal and cytoplasmic context determines predisposition to maternal age-related aneuploidy: Brief overview and update on MCAK in mammalian oocytes. Biochem. Soc. Trans. 2010, 38, 1681–1686. [Google Scholar] [CrossRef] [PubMed]
- Ruggiero, C.; Lalli, E. Targeting the cytoskeleton against metastatic dissemination. Cancer Metastasis Rev. 2021, 40, 89–140. [Google Scholar] [CrossRef]
- Ritter, A.; Friemel, A.; Kreis, N.N.; Hoock, S.C.; Roth, S.; Kielland-Kaisen, U.; Bruggmann, D.; Solbach, C.; Louwen, F.; Yuan, J. Primary Cilia Are Dysfunctional in Obese Adipose-Derived Mesenchymal Stem Cells. Stem Cell Rep. 2018, 10, 583–599. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Dai, M.; Zhang, C.; Teng, K.; Wang, F.; Li, H.; Sun, W.; Feng, Z.; Kang, T.; Guan, X.; et al. KIF2C: A novel link between Wnt/β-catenin and mTORC1 signaling in the pathogenesis of hepatocellular carcinoma. Protein Cell 2020, 12, 788–809. [Google Scholar] [CrossRef] [PubMed]
- Zuo, X.; Meng, P.; Bao, Y.; Tao, C.; Wang, Y.; Liu, X.; Bu, Y.; Zhu, J. Cell cycle dysregulation with overexpression of KIF2C/MCAK is a critical event in nasopharyngeal carcinoma. Genes Dis. 2021, in press. [Google Scholar] [CrossRef]
- LaFlamme, S.E.; Mathew-Steiner, S.; Singh, N.; Colello-Borges, D.; Nieves, B. Integrin and microtubule crosstalk in the regulation of cellular processes. Cell. Mol. Life Sci. 2018, 75, 4177–4185. [Google Scholar] [CrossRef]
- Kim, D.-H.; Wirtz, D. Focal adhesion size uniquely predicts cell migration. FASEB J. 2013, 27, 1351–1361. [Google Scholar] [CrossRef]
- Nagano, M.; Hoshino, D.; Koshikawa, N.; Akizawa, T.; Seiki, M. Turnover of focal adhesions and cancer cell migration. Int. J. Cell Biol. 2012, 2012, 310616. [Google Scholar] [CrossRef] [PubMed]
- Hoock, S.C.; Ritter, A.; Steinhauser, K.; Roth, S.; Behrends, C.; Oswald, F.; Solbach, C.; Louwen, F.; Kreis, N.N.; Yuan, J. RITA modulates cell migration and invasion by affecting focal adhesion dynamics. Mol. Oncol. 2019, 13, 2121–2141. [Google Scholar] [CrossRef]
- Ritter, A.; Safdar, B.K.; Jasmer, B.; Kreis, N.N.; Friemel, A.; Roth, S.; Solbach, C.; Louwen, F.; Yuan, J.P. The Function of Oncogene B-Cell Lymphoma 6 in the Regulation of the Migration and Invasion of Trophoblastic Cells. Int. J. Mol. Sci. 2020, 21, 8393. [Google Scholar] [CrossRef]
- Zhao, B.; Li, L.; Wang, L.; Wang, C.Y.; Yu, J.; Guan, K.L. Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes Dev. 2012, 26, 54–68. [Google Scholar] [CrossRef] [PubMed]
- Piper, M.; Lee, A.C.; van Horck, F.P.; McNeilly, H.; Lu, T.B.; Harris, W.A.; Holt, C.E. Differential requirement of F-actin and microtubule cytoskeleton in cue-induced local protein synthesis in axonal growth cones. Neural Dev. 2015, 10, 3. [Google Scholar] [CrossRef]
- Magliocca, V.; Petrini, S.; Franchin, T.; Borghi, R.; Niceforo, A.; Abbaszadeh, Z.; Bertini, E.; Compagnucci, C. Identifying the dynamics of actin and tubulin polymerization in iPSCs and in iPSC-derived neurons. Oncotarget 2017, 8, 111096–111109. [Google Scholar] [CrossRef]
- Colin, A.; Singaravelu, P.; Thery, M.; Blanchoin, L.; Gueroui, Z. Actin-Network Architecture Regulates Microtubule Dynamics. Curr. Biol. 2018, 28, 2647–2656.e4. [Google Scholar] [CrossRef] [PubMed]
- Waterman-Storer, C.M.; Worthylake, R.A.; Liu, B.P.; Burridge, K.; Salmon, E.D. Microtubule growth activates Rac1 to promote lamellipodial profusion in fibroblasts. Nat. Cell Biol. 1999, 1, 45–50. [Google Scholar] [CrossRef]
- Zieve, G.W.; Turnbull, D.; Mullins, J.M.; McIntosh, J.R. Production of large numbers of mitotic mammalian cells by use of the reversible microtubule inhibitor nocodazole: Nocodazole accumulated mitotic cells. Exp. Cell Res. 1980, 126, 397–405. [Google Scholar] [CrossRef]
- Hu, Y.L.; Lu, S.Y.; Szeto, K.W.; Sun, J.; Wang, Y.X.; Lasheras, J.C.; Chien, S. FAK and paxillin dynamics at focal adhesions in the protrusions of migrating cells. Sci. Rep. 2014, 4, 6024. [Google Scholar] [CrossRef] [PubMed]
- Roll-Mecak, A. The Tubulin Code in Microtubule Dynamics and Information Encoding. Dev. Cell 2020, 54, 7–20. [Google Scholar] [CrossRef] [PubMed]
- Matov, A.; Applegate, K.; Kumar, P.; Thoma, C.; Krek, W.; Danuser, G.; Wittmann, T. Analysis of microtubule dynamic instability using a plus-end growth marker. Nat. Methods 2010, 7, 761–768. [Google Scholar] [CrossRef] [PubMed]
- Gan, H.; Lin, L.; Hu, N.; Yang, Y.; Gao, Y.; Pei, Y.; Chen, K.; Sun, B. KIF2C exerts an oncogenic role in nonsmall cell lung cancer and is negatively regulated by miR-325-3p. Cell Biochem. Funct. 2019, 37, 424–431. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Xiong, L.; Zhu, M.; Yang, Z.; Zhao, J.; Tang, H. Co-expression network analysis identified KIF2C in association with progression and prognosis in lung adenocarcinoma. Cancer Biomark. 2019, 24, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Bie, L.; Zhao, G.; Wang, Y.P.; Zhang, B. Kinesin family member 2C (KIF2C/MCAK) is a novel marker for prognosis in human gliomas. Clin. Neurol. Neurosurg. 2012, 114, 356–360. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Tanaka, F.; Haraguchi, N.; Mimori, K.; Matsumoto, T.; Inoue, H.; Yanaga, K.; Mori, M. Clinicopathological and biological significance of mitotic centromere-associated kinesin overexpression in human gastric cancer. Br. J. Cancer 2007, 97, 543–549. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Wang, C.; Xue, L.; Zhang, Q.; Luh, F.; Wang, J.; Lin, T.G.; Yen, Y.; Liu, X. Human Mitotic Centromere-Associated Kinesin Is Targeted by MicroRNA 485-5p/181c and Prognosticates Poor Survivability of Breast Cancer. J. Oncol. 2019, 2019, 2316237. [Google Scholar] [CrossRef]
- Zinn, A.; Goicoechea, S.M.; Kreider-Letterman, G.; Maity, D.; Awadia, S.; Cedeno-Rosario, L.; Chen, Y.; Garcia-Mata, R. The small GTPase RhoG regulates microtubule-mediated focal adhesion disassembly. Sci. Rep. 2019, 9, 5163. [Google Scholar] [CrossRef]
- Efimov, A.; Schiefermeier, N.; Grigoriev, I.; Brown, M.C.; Turner, C.E.; Small, J.V.; Kaverina, I. Paxillin-dependent stimulation of microtubule catastrophes at focal adhesion sites. J. Cell Sci. 2008, 121, 196–204. [Google Scholar] [CrossRef]
- Xie, T.; Li, X.; Ye, F.; Lu, C.; Huang, H.; Wang, F.; Cao, X.; Zhong, C. High KIF2A expression promotes proliferation, migration and predicts poor prognosis in lung adenocarcinoma. Biochem. Biophys. Res. Commun. 2018, 497, 65–72. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, Y.; Liu, X.; Zhao, A.; Yang, Z.; Kong, F.; Sun, L.; Yu, Y.; Jiang, L. KIF2A promotes the progression via AKT signaling pathway and is upregulated by transcription factor ETV4 in human gastric cancer. Biomed. Pharmacother. 2020, 125, 109840. [Google Scholar] [CrossRef]
- Wang, J.; Ma, S.; Ma, R.; Qu, X.; Liu, W.; Lv, C.; Zhao, S.; Gong, Y. KIF2A silencing inhibits the proliferation and migration of breast cancer cells and correlates with unfavorable prognosis in breast cancer. BMC Cancer 2014, 14, 461. [Google Scholar] [CrossRef]
- Mitra, S.K.; Schlaepfer, D.D. Integrin-regulated FAK–Src signaling in normal and cancer cells. Curr. Opin. Cell Biol. 2006, 18, 516–523. [Google Scholar] [CrossRef] [PubMed]
- Kanteti, R.; Batra, S.K.; Lennon, F.E.; Salgia, R. FAK and paxillin, two potential targets in pancreatic cancer. Oncotarget 2016, 7, 31586–31601. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, M.; Kukkurainen, S.; Hytonen, V.P.; Wehrle-Haller, B. Cell Adhesion by Integrins. Physiol. Rev. 2019, 99, 1655–1699. [Google Scholar] [CrossRef]
- Paul, N.R.; Jacquemet, G.; Caswell, P.T. Endocytic Trafficking of Integrins in Cell Migration. Curr. Biol. 2015, 25, R1092–R1105. [Google Scholar] [CrossRef]
- Hohmann, T.; Dehghani, F. The Cytoskeleton-A Complex Interacting Meshwork. Cells 2019, 8, 362. [Google Scholar] [CrossRef] [PubMed]
- Ezratty, E.J.; Partridge, M.A.; Gundersen, G.G. Microtubule-induced focal adhesion disassembly is mediated by dynamin and focal adhesion kinase. Nat. Cell Biol. 2005, 7, 581–590. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Luo, Y.; Lyu, R.; Chen, J.; Liu, R.; Li, D.; Liu, M.; Zhou, J. Proto-Oncogenic Src Phosphorylates EB1 to Regulate the Microtubule-Focal Adhesion Crosstalk and Stimulate Cell Migration. Theranostics 2016, 6, 2129–2140. [Google Scholar] [CrossRef] [PubMed]
- Schlaepfer, D.D.; Hunter, T. Integrin signalling and tyrosine phosphorylation: Just the FAKs? Trends Cell Biol. 1998, 8, 151–157. [Google Scholar] [CrossRef]
- Gahmberg, C.G.; Gronholm, M.; Madhavan, S.; Jahan, F.; Mikkola, E.; Viazmina, L.; Koivunen, E. Regulation of cell adhesion: A collaborative effort of integrins, their ligands, cytoplasmic actors, and phosphorylation. Q. Rev. Biophys. 2019, 52, e10. [Google Scholar] [CrossRef]
- Li, J.X.H.; Tang, V.W.; Brieher, W.M. Actin protrusions push at apical junctions to maintain E-cadherin adhesion. Proc. Natl. Acad. Sci. USA 2020, 117, 432–438. [Google Scholar] [CrossRef] [PubMed]
- Vasioukhin, V.; Bauer, C.; Yin, M.; Fuchs, E. Directed actin polymerization is the driving force for epithelial cell-cell adhesion. Cell 2000, 100, 209–219. [Google Scholar] [CrossRef]
- Machesky, L.M.; Hall, A. Role of Actin Polymerization and Adhesion to Extracellular Matrix in Rac- and Rho-induced Cytoskeletal Reorganization. J. Cell Biol. 1997, 138, 913–926. [Google Scholar] [CrossRef]
- Stehbens, S.; Wittmann, T. Targeting and transport: How microtubules control focal adhesion dynamics. J. Cell Biol. 2012, 198, 481–489. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moon, H.H.; Kreis, N.-N.; Friemel, A.; Roth, S.; Schulte, D.; Solbach, C.; Louwen, F.; Yuan, J.; Ritter, A. Mitotic Centromere-Associated Kinesin (MCAK/KIF2C) Regulates Cell Migration and Invasion by Modulating Microtubule Dynamics and Focal Adhesion Turnover. Cancers 2021, 13, 5673. https://doi.org/10.3390/cancers13225673
Moon HH, Kreis N-N, Friemel A, Roth S, Schulte D, Solbach C, Louwen F, Yuan J, Ritter A. Mitotic Centromere-Associated Kinesin (MCAK/KIF2C) Regulates Cell Migration and Invasion by Modulating Microtubule Dynamics and Focal Adhesion Turnover. Cancers. 2021; 13(22):5673. https://doi.org/10.3390/cancers13225673
Chicago/Turabian StyleMoon, Ha Hyung, Nina-Naomi Kreis, Alexandra Friemel, Susanne Roth, Dorothea Schulte, Christine Solbach, Frank Louwen, Juping Yuan, and Andreas Ritter. 2021. "Mitotic Centromere-Associated Kinesin (MCAK/KIF2C) Regulates Cell Migration and Invasion by Modulating Microtubule Dynamics and Focal Adhesion Turnover" Cancers 13, no. 22: 5673. https://doi.org/10.3390/cancers13225673
APA StyleMoon, H. H., Kreis, N.-N., Friemel, A., Roth, S., Schulte, D., Solbach, C., Louwen, F., Yuan, J., & Ritter, A. (2021). Mitotic Centromere-Associated Kinesin (MCAK/KIF2C) Regulates Cell Migration and Invasion by Modulating Microtubule Dynamics and Focal Adhesion Turnover. Cancers, 13(22), 5673. https://doi.org/10.3390/cancers13225673






