Modeling the Tumor Microenvironment of Ovarian Cancer: The Application of Self-Assembling Biomaterials
Abstract
:Simple Summary
Abstract
1. Introduction
2. Components of the Ovarian TME
2.1. Cellular Composition
2.2. Matrix Composition
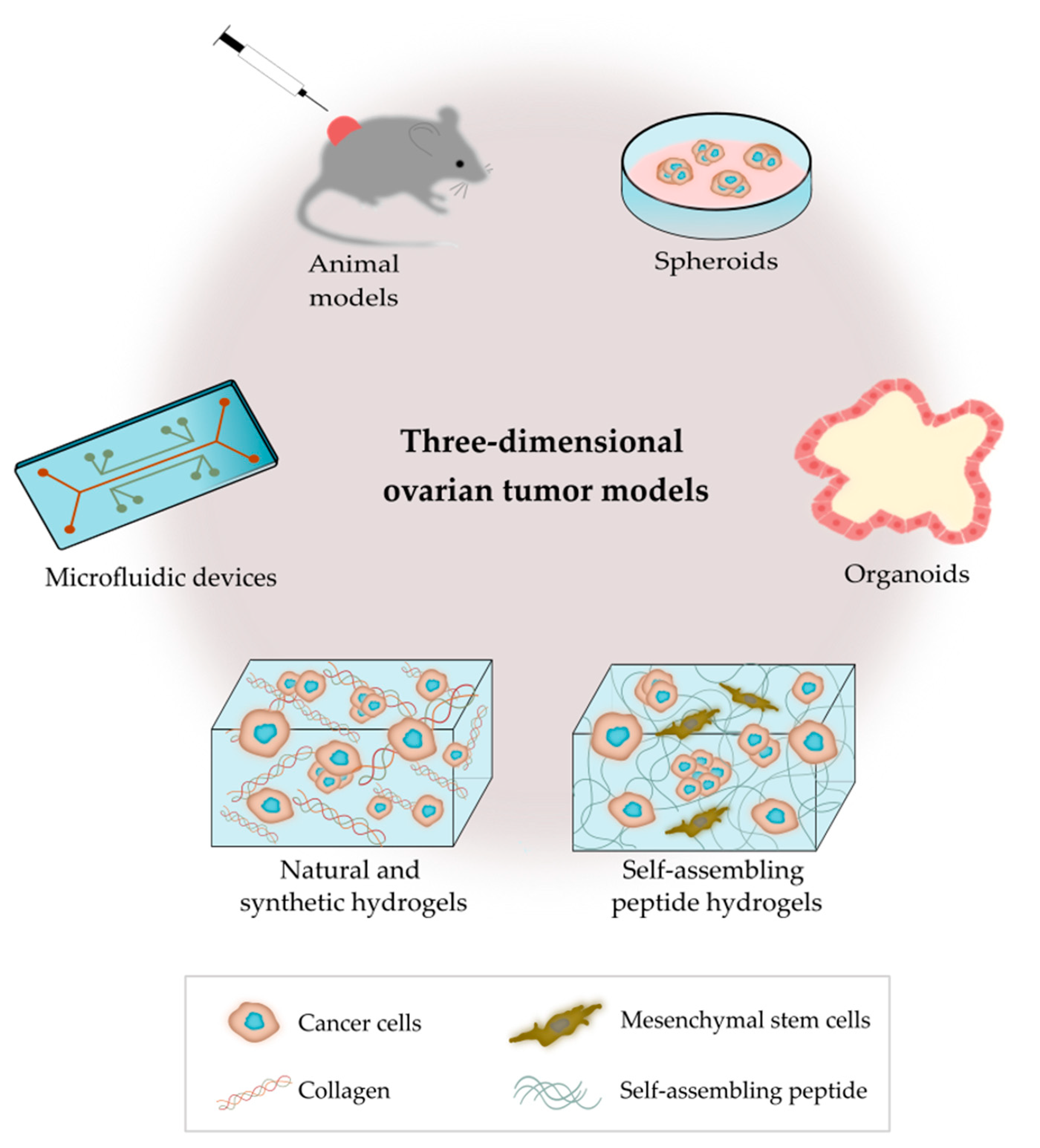
3. 3D Ovarian Cancer Models
3.1. Animal Models
3.1.1. Rodent Models
3.1.2. Laying Hen Model
3.2. 3D In Vitro Models
3.2.1. Spheroids
3.2.2. Organoids
3.2.3. Microfluidic Devices
3.2.4. Hydrogels Based on Polymer/Protein Networks
Natural Hydrogels
Synthetic Hydrogels
3.2.5. Hydrogels Based on Self-Assembled Peptide Networks
Ionic Complementary Self-Assembling Peptides
Peptide Amphiphiles
3.2.6. Mechanical Stimuli in OvCa Models
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef]
- Motohara, T.; Masuda, K.; Morotti, M.; Zheng, Y.; El-Sahhar, S.; Chong, K.Y.; Wietek, N.; Alsaadi, A.; KaramiNejadRanjbar, M.; Hu, Z.; et al. An evolving story of the metastatic voyage of ovarian cancer cells: Cellular and molecular orchestration of the adipose-rich metastatic microenvironment. Oncogene 2019, 38, 2885–2898. [Google Scholar] [CrossRef] [Green Version]
- Lane, D.; Matte, I.; Garde-Granger, P.; Laplante, C.; Carignan, A.; Rancourt, C.; Piché, A. Inflammation-regulating factors in ascites as predictive biomarkers of drug resistance and progression-free survival in serous epithelial ovarian cancers. BMC Cancer 2015, 15, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karnezis, A.N.; Cho, K.R.; Gilks, C.B.; Pearce, C.L.; Huntsman, D.G. The disparate origins of ovarian cancers: Pathogenesis and prevention strategies. Nat. Rev. Cancer 2017, 17, 65–74. [Google Scholar] [CrossRef]
- Kipps, E.; Tan, D.; Kaye, S.B. Meeting the challenge of ascites in ovarian cancer: New avenues for therapy and research. Nat. Rev. Cancer 2013, 13, 273–282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shield, K.; Ackland, L.; Ahmed, N.; Rice, G. Multicellular spheroids in ovarian cancer metastases: Biology and pathology. Gynecol. Oncol. 2009, 113, 143–148. [Google Scholar] [CrossRef]
- Ahmed, N.; Thompson, E.W.; Quinn, M. Epithelial–mesenchymal interconversions in normal ovarian surface epithelium and ovarian carcinomas: An exception to the norm. J. Cell. Physiol. 2007, 213, 581–588. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Xu, H.; Zhao, X. Designer Self-Assembling Peptide Hydrogels to Engineer 3D Cell Microenvironments for Cell Constructs Formation and Precise Oncology Remodeling in Ovarian Cancer. Adv. Sci. 2020, 7, 1903718. [Google Scholar] [CrossRef] [Green Version]
- Pape, J.; Emberton, M.; Cheema, U. 3D Cancer Models: The Need for a Complex Stroma, Compartmentalization and Stiffness. Front. Bioeng. Biotechnol. 2021, 9, 276. [Google Scholar] [CrossRef]
- Echo, A.; Howell, V.M.; Colvin, E.K. The Extracellular Matrix in Epithelial Ovarian Cancer—A Piece of a Puzzle. Front. Oncol. 2015, 5, 245. [Google Scholar] [CrossRef] [Green Version]
- Park, K.M.; Lewis, D.; Gerecht, S. Bioinspired Hydrogels to Engineer Cancer Microenvironments. Annu. Rev. Biomed. Eng. 2017, 19, 109–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, Y.; Hua, S.; Tanwar, P.S. Extracellular matrix-mediated regulation of cancer stem cells and chemoresistance. Int. J. Biochem. Cell Biol. 2019, 109, 90–104. [Google Scholar] [CrossRef]
- Shih, A.J.; Menzin, A.; Whyte, J.; Lovecchio, J.; Liew, A.; Khalili, H.; Bhuiya, T.; Gregersen, P.K.; Lee, A.T. Correction: Identification of grade and origin specific cell populations in serous epithelial ovarian cancer by single cell RNA-seq. PLoS ONE 2018, 13, e0208778. [Google Scholar] [CrossRef] [PubMed]
- Barbato, L.; Bocchetti, M.; Di Biase, A.; Regad, T. Cancer Stem Cells and Targeting Strategies. Cells 2019, 8, 926. [Google Scholar] [CrossRef] [Green Version]
- Yu, Z.; Pestell, T.G.; Lisanti, M.; Pestell, R.G. Cancer stem cells. Int. J. Biochem. Cell Biol. 2012, 44, 2144–2151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foster, R.; Buckanovich, R.J.; Rueda, B.R. Ovarian cancer stem cells: Working towards the root of stemness. Cancer Lett. 2013, 338, 147–157. [Google Scholar] [CrossRef]
- Zhao, J. Cancer stem cells and chemoresistance: The smartest survives the raid. Pharmacol. Ther. 2016, 160, 145–158. [Google Scholar] [CrossRef] [Green Version]
- Nowak, M.; Klink, M. The Role of Tumor-Associated Macrophages in the Progression and Chemoresistance of Ovarian Cancer. Cells 2020, 9, 1299. [Google Scholar] [CrossRef]
- Luo, Z.; Wang, Q.; Lau, W.B.; Lau, B.; Xu, L.; Zhao, L.; Yang, H.; Feng, M.; Xuan, Y.; Yang, Y.; et al. Tumor microenvironment: The culprit for ovarian cancer metastasis? Cancer Lett. 2016, 377, 174–182. [Google Scholar] [CrossRef]
- Ge, Z.; Ding, S. The Crosstalk Between Tumor-Associated Macrophages (TAMs) and Tumor Cells and the Corresponding Targeted Therapy. Front. Oncol. 2020, 10, 2404. [Google Scholar] [CrossRef]
- Hansen, J.M.; Coleman, R.L.; Sood, A.K. Targeting the tumour microenvironment in ovarian cancer. Eur. J. Cancer 2016, 56, 131–143. [Google Scholar] [CrossRef] [Green Version]
- Salas-Benito, D.; Vercher, E.; Conde, E.; Glez-Vaz, J.; Tamayo, I.; Hervas-Stubbs, S. Inflammation and immunity in ovarian cancer. Eur. J. Cancer Suppl. 2020, 15, 56–66. [Google Scholar] [CrossRef]
- Ostrand-Rosenberg, S.; Sinha, P.; Beury, D.W.; Clements, V.K. Cross-talk between myeloid-derived suppressor cells (MDSC), macrophages, and dendritic cells enhances tumor-induced immune suppression. Semin. Cancer Biol. 2012, 22, 275–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Liu, Y.; Zheng, S.; Zhang, T.; Wu, J.; Sun, Y.; Zhang, J.; Liu, G. Role of exosomes in the immune microenvironment of ovarian cancer (Review). Oncol. Lett. 2021, 21, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Dasari, S.; Fang, Y.; Mitra, A.K. Cancer Associated Fibroblasts: Naughty Neighbors That Drive Ovarian Cancer Progression. Cancers 2018, 10, 406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sahai, E.; Astsaturov, I.; Cukierman, E.; DeNardo, D.G.; Egeblad, M.; Evans, R.M.; Fearon, D.; Greten, F.R.; Hingorani, S.R.; Hunter, T.; et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat. Rev. Cancer 2020, 20, 174–186. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, G.M.; Galpin, K.J.C.; McCloskey, C.W.; Vanderhyden, B.C. The Tumor Microenvironment of Epithelial Ovarian Cancer and Its Influence on Response to Immunotherapy. Cancers 2018, 10, 242. [Google Scholar] [CrossRef] [Green Version]
- Han, Q.; Huang, B.; Huang, Z.; Cai, J.; Gong, L.; Zhang, Y.; Jiang, J.; Dong, W.; Wang, Z. Tumor cell-fibroblast heterotypic aggregates in malignant ascites of patients with ovarian cancer. Int. J. Mol. Med. 2019, 44, 2245–2255. [Google Scholar] [CrossRef] [Green Version]
- Coffman, L.G.; Choi, Y.-J.; McLean, K.; Allen, B.L.; di Magliano, M.P.; Buckanovich, R.J. Human carcinoma-associated mesenchymal stem cells promote ovarian cancer chemotherapy resistance via a BMP4/HH signaling loop. Oncotarget 2016, 7, 6916–6932. [Google Scholar] [CrossRef] [Green Version]
- McLean, K.; Gong, Y.; Choi, Y.; Deng, N.; Yang, K.; Bai, S.; Cabrera, L.; Keller, E.; McCauley, L.; Cho, K.; et al. Human ovarian carcinoma–associated mesenchymal stem cells regulate cancer stem cells and tumorigenesis via altered BMP production. J. Clin. Investig. 2011, 121, 3206–3219. [Google Scholar] [CrossRef] [Green Version]
- Coffman, L.G.; Pearson, A.; Frisbie, L.G.; Freeman, Z.; Christie, E.; Bowtell, D.D.; Buckanovich, R.J. Ovarian Carcinoma-Associated Mesenchymal Stem Cells Arise from Tissue-Specific Normal Stroma. STEM Cells 2018, 37, 257–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nieman, K.; Kenny, H.A.; Penicka, C.V.; Ladanyi, A.; Buell-Gutbrod, R.; Zillhardt, M.R.; Romero, I.; Carey, M.S.; Mills, G.B.; Hotamisligil, G.S.; et al. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat. Med. 2011, 17, 1498–1503. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Zaman, M.M.; Vlasakov, I.; Roy, R.; Huang, L.; Martin, C.R.; Freedman, S.D.; Serhan, C.N.; Moses, M.A. Adipocytes promote ovarian cancer chemoresistance. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Moghaddam, S.M.; Amini, A.; Morris, D.L.; Pourgholami, M.H. Significance of vascular endothelial growth factor in growth and peritoneal dissemination of ovarian cancer. Cancer Metastasis Rev. 2011, 31, 143–162. [Google Scholar] [CrossRef] [Green Version]
- McKenzie, A.J.; Hicks, S.R.; Svec, K.V.; Naughton, H.; Edmunds, Z.L.; Howe, A.K. The mechanical microenvironment regulates ovarian cancer cell morphology, migration, and spheroid disaggregation. Sci. Rep. 2018, 8, 1–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bregenzer, M.E.; Horst, E.N.; Mehta, P.; Novak, C.M.; Repetto, T. The Role of Cancer Stem Cells and Mechanical Forces in Ovarian Cancer Metastasis. Cancers 2019, 11, 1008. [Google Scholar] [CrossRef] [Green Version]
- Fan, Y.; Sun, Q.; Li, X.; Feng, J.; Ao, Z.; Li, X.; Wang, J. Substrate Stiffness Modulates the Growth, Phenotype, and Chemoresistance of Ovarian Cancer Cells. Front. Cell Dev. Biol. 2021, 9, 1–13. [Google Scholar] [CrossRef]
- Liu, M.; Zhang, X.; Long, C.; Xu, H.; Cheng, X.; Chang, J.; Zhang, C.; Zhang, C.; Wang, X. Collagen-based three-dimensional culture microenvironment promotes epithelial to mesenchymal transition and drug resistance of human ovarian cancerin vitro. RSC Adv. 2018, 8, 8910–8919. [Google Scholar] [CrossRef] [Green Version]
- Pearce, O.; Delaine-Smith, R.M.; Maniati, E.; Nichols, S.; Wang, J.; Böhm, S.; Rajeeve, V.; Ullah, D.; Chakravarty, P.; Jones, R.R.; et al. Deconstruction of a Metastatic Tumor Microenvironment Reveals a Common Matrix Response in Human Cancers. Cancer Discov. 2018, 8, 304–319. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.-H.; Chang, T.-H.; Huang, Y.-F.; Huang, H.-D.; Chou, C.-Y. COL11A1 promotes tumor progression and predicts poor clinical outcome in ovarian cancer. Oncogene 2014, 33, 3432–3440. [Google Scholar] [CrossRef]
- Ajeti, V.; Lara-Santiago, J.; Alkmin, S.; Campagnola, P.J. Ovarian and Breast Cancer Migration Dynamics on Laminin and Fibronectin Bi-directional Gradient Fibers Fabricated via Multiphoton Excited Photochemistry. Cell. Mol. Bioeng. 2017, 10, 295–311. [Google Scholar] [CrossRef]
- Bar, J.K.; Grelewski, P.; Popiela, A.; Noga, L.; Rabczyñski, J. Type IV collagen and CD44v6 expression in benign, malignant primary and metastatic ovarian tumors: Correlation with Ki-67 and p53 immunoreactivity. Gynecol. Oncol. 2004, 95, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Ricciardelli, C.; Rodgers, R.J. Extracellular Matrix of Ovarian Tumors. Semin. Reprod. Med. 2006, 24, 270–282. [Google Scholar] [CrossRef] [PubMed]
- Anttila, M.A.; Tammi, R.H.; Tammi, M.I.; Syrjänen, K.J.; Saarikoski, S.V.; Kosma, V.M. High levels of stromal hyaluronan predict poor disease outcome in epithelial ovarian cancer. Cancer Res. 2000, 60, 150–155. [Google Scholar]
- Chen, C.; Zhao, S.; Karnad, A.; Freeman, J.W. The biology and role of CD44 in cancer progression: Therapeutic implications. J. Hematol. Oncol. 2018, 11, 1–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kenny, H.A.; Chiang, C.-Y.; White, E.A.; Schryver, E.M.; Habis, M.; Romero, I.; Ladanyi, A.; Penicka, C.V.; George, J.; Matlin, K.; et al. Mesothelial cells promote early ovarian cancer metastasis through fibronectin secretion. J. Clin. Investig. 2014, 124, 4614–4628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Didem, T.; Faruk, T.; Senem, K.; Derya, D.; Murat, S.; Murat, G.; Oznur, K. Clinical significance of serum tenascin-c levels in epithelial ovarian cancer. Tumor Biol. 2014, 35, 6777–6782. [Google Scholar] [CrossRef] [PubMed]
- Kramer, M.; Pierredon, S.; Ribaux, P.; Tille, J.-C.; Petignat, P.; Cohen, M. Secretome Identifies Tenascin-X as a Potent Marker of Ovarian Cancer. BioMed Res. Int. 2015, 2015, 208017. [Google Scholar] [CrossRef] [Green Version]
- Ricci, C.; Moroni, L.; Danti, S. Cancer tissue engineering-new perspectives in understanding the biology of solid tumours—A critical review. OA Tissue Eng. 2013, 1, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Cavo, M.; Fato, M.M.; Peñuela, L.; Beltrame, F.; Raiteri, R.; Scaglione, S. Microenvironment complexity and matrix stiffness regulate breast cancer cell activity in a 3D in vitro model. Sci. Rep. 2016, 6, 35367. [Google Scholar] [CrossRef] [Green Version]
- Worthington, P.; Pochan, D.J.; Langhans, S.A. Peptide Hydrogels—Versatile Matrices for 3D Cell Culture in Cancer Medicine. Front. Oncol. 2015, 5, 92. [Google Scholar] [CrossRef] [Green Version]
- Nyga, A.; Cheema, U.; Loizidou, M. 3D tumour models: Novel in vitro approaches to cancer studies. J. Cell Commun. Signal. 2011, 5, 239–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kast, V.; Loessner, D. 3D Models for Ovarian Cancer. Adv. Exp. Med. Biol. 2021, 1330, 139–149. [Google Scholar] [CrossRef]
- Tudrej, P.; Kujawa, K.A.; Cortez, A.J.; Lisowska, K.M. Characteristics of in Vivo Model Systems for Ovarian Cancer Studies. Diagnostics 2019, 9, 120. [Google Scholar] [CrossRef] [Green Version]
- House, C.D.; Ehernandez, L.; Annunziata, C.M. Recent Technological Advances in Using Mouse Models to Study Ovarian Cancer. Front. Oncol. 2014, 4, 26. [Google Scholar] [CrossRef] [PubMed]
- Bleijs, M.; Van De Wetering, M.; Clevers, H.; Drost, J. Xenograft and organoid model systems in cancer research. EMBO J. 2019, 38, e101654. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Luo, L.; Zheng, X.; Yu, T. An Advanced Orthotopic Ovarian Cancer Model in Mice for Therapeutic Trials. BioMed Res. Int. 2016, 2016, 1–4. [Google Scholar] [CrossRef]
- Lee, M.W.; Miljanic, M.; Triplett, T.; Ramirez, C.; Aung, K.L.; Eckhardt, S.G.; Capasso, A. Current methods in translational cancer research. Cancer Metastasis Rev. 2021, 40, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Dobbin, Z.C.; Katre, A.A.; Steg, A.D.; Erickson, B.; Shah, M.M.; Alvarez, R.D.; Conner, M.G.; Schneider, D.; Chen, D.; Landen, C.N. Using heterogeneity of the patient-derived xenograft model to identify the chemoresistant population in ovarian cancer. Oncotarget 2014, 5, 8750–8764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morgan, K.M.; Riedlinger, G.M.; Rosenfeld, J.; Ganesan, S.; Pine, S.R. Patient-Derived Xenograft Models of Non-Small Cell Lung Cancer and Their Potential Utility in Personalized Medicine. Front. Oncol. 2017, 7, 2. [Google Scholar] [CrossRef] [Green Version]
- Topp, M.D.; Hartley, L.; Cook, M.; Heong, V.; Boehm, E.; McShane, L.; Pyman, J.; McNally, O.; Ananda, S.; Harrell, M.; et al. Molecular correlates of platinum response in human high-grade serous ovarian cancer patient-derived xenografts. Mol. Oncol. 2014, 8, 656–668. [Google Scholar] [CrossRef]
- Ricci, F.; Bizzaro, F.; Cesca, M.; Guffanti, F.; Ganzinelli, M.; Decio, A.; Ghilardi, C.; Perego, P.; Fruscio, R.; Buda, A.; et al. Patient-Derived Ovarian Tumor Xenografts Recapitulate Human Clinicopathology and Genetic Alterations. Cancer Res. 2014, 74, 6980–6990. [Google Scholar] [CrossRef] [Green Version]
- Weroha, S.J.; Becker, M.A.; Enderica-Gonzalez, S.; Harrington, S.C.; Oberg, A.L.; Maurer, M.; Perkins, S.E.; Al Hilli, M.; Butler, K.A.; McKinstry, S.; et al. Tumorgrafts as In Vivo Surrogates for Women with Ovarian Cancer. Clin. Cancer Res. 2014, 20, 1288–1297. [Google Scholar] [CrossRef] [Green Version]
- Heo, E.J.; Cho, Y.J.; Cho, W.C.; Hong, J.E.; Jeon, H.-K.; Oh, D.-Y.; Choi, Y.-L.; Song, S.Y.; Choi, J.-J.; Bae, D.-S.; et al. Patient-Derived Xenograft Models of Epithelial Ovarian Cancer for Preclinical Studies. Cancer Res. Treat. 2017, 49, 915–926. [Google Scholar] [CrossRef] [Green Version]
- Whittle, J.R.; Lewis, M.T.; Lindeman, G.J.; Visvader, J.E. Patient-derived xenograft models of breast cancer and their predictive power. Breast Cancer Res. 2015, 17, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Hidalgo, M.; Amant, F.; Biankin, A.; Budinská, E.; Byrne, A.; Caldas, C.; Clarke, R.; De Jong, S.; Jonkers, J.; Mælandsmo, G.M.; et al. Patient-Derived Xenograft Models: An Emerging Platform for Translational Cancer Research. Cancer Discov. 2014, 4, 998–1013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bobbs, A.S.; Cole, J.M.; Dahl, K.D.C. Emerging and Evolving Ovarian Cancer Animal Models. Cancer Growth Metastasis 2015, 8 (Suppl. 1), 29–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, Q.; Yan, L.; Xu, Y. Anoikis resistance is a critical feature of highly aggressive ovarian cancer cells. Oncogene 2015, 34, 3315–3324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCloskey, C.W.; Goldberg, R.L.; Carter, L.E.; Gamwell, L.F.; Al-Hujaily, E.M.; Collins, O.; Macdonald, E.A.; Garson, K.; Daneshmand, M.; Carmona, E.; et al. A New Spontaneously Transformed Syngeneic Model of High-Grade Serous Ovarian Cancer with a Tumor-Initiating Cell Population. Front. Oncol. 2014, 4, 53. [Google Scholar] [CrossRef] [Green Version]
- Gómez-Cuadrado, L.; Tracey, N.; Ma, R.; Qian, B.; Brunton, V.G. Mouse models of metastasis: Progress and prospects. Dis. Model. Mech. 2017, 10, 1061–1074. [Google Scholar] [CrossRef] [Green Version]
- Mullany, L.K.; Richards, J.S. Minireview: Animal Models and Mechanisms of Ovarian Cancer Development. Endocrinology 2012, 153, 1585–1592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Der Horst, P.H.; Van Der Zee, M.; Heijmans-Antonissen, C.; Jia, Y.; DeMayo, F.J.; Lydon, J.P.; Van Deurzen, C.H.; Ewing, P.C.; Burger, C.W.; Blok, L.J. A mouse model for endometrioid ovarian cancer arising from the distal oviduct. Int. J. Cancer 2014, 135, 1028–1037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perets, R.; Wyant, G.A.; Muto, K.W.; Bijron, J.G.; Poole, B.B.; Chin, K.T.; Chen, J.Y.H.; Ohman, A.; Stepule, C.D.; Kwak, S.; et al. Transformation of the Fallopian Tube Secretory Epithelium Leads to High-Grade Serous Ovarian Cancer in Brca;Tp53;Pten Models. Cancer Cell 2013, 24, 751–765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sherman-Baust, C.A.; Kuhn, E.; Valle, B.L.; Shih, I.-M.; Kurman, R.J.; Wang, T.-L.; Amano, T.; Ko, M.S.; Miyoshi, I.; Araki, Y.; et al. A genetically engineered ovarian cancer mouse model based on fallopian tube transformation mimics human high-grade serous carcinoma development. J. Pathol. 2014, 233, 228–237. [Google Scholar] [CrossRef]
- Zhai, Y.; Wu, R.; Kuick, R.; Sessine, M.S.; Schulman, S.; Green, M.; Fearon, E.R.; Cho, K.R. High-grade serous carcinomas arise in the mouse oviduct via defects linked to the human disease. J. Pathol. 2017, 243, 16–25. [Google Scholar] [CrossRef] [PubMed]
- Bernardo, A.D.M.; Thorsteinsdóttir, S.; Mummery, C.L. Advantages of the avian model for human ovarian cancer. Mol. Clin. Oncol. 2015, 3, 1191–1198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hasan, N.; Ohman, A.W.; Dinulescu, D.M. The promise and challenge of ovarian cancer models. Transl. Cancer Res. 2015, 4, 14–28. [Google Scholar]
- Giles, J.R.; Shivaprasad, H.; Johnson, P.A. Ovarian tumor expression of an oviductal protein in the hen: A model for human serous ovarian adenocarcinoma. Gynecol. Oncol. 2004, 95, 530–533. [Google Scholar] [CrossRef]
- Treviño, L.S.; Giles, J.R.; Wang, W.; Urick, M.E.; Johnson, P.A. Gene Expression Profiling Reveals Differentially Expressed Genes in Ovarian Cancer of the Hen: Support for Oviductal Origin? Horm. Cancer 2010, 1, 177–186. [Google Scholar] [CrossRef]
- Johnson, P.A.; Giles, J.R. The hen as a model of ovarian cancer. Nat. Rev. Cancer 2013, 13, 432–436. [Google Scholar] [CrossRef]
- Gao, Q.; Yang, Z.; Xu, S.; Li, X.; Yang, X.; Jin, P.; Liu, Y.; Zhou, X.; Zhang, T.; Gong, C.; et al. Heterotypic CAF-tumor spheroids promote early peritoneal metastasis of ovarian cancer. J. Exp. Med. 2019, 216, 688–703. [Google Scholar] [CrossRef] [Green Version]
- Raghavan, S.; Snyder, C.S.; Wang, A.; McLean, K.; Zamarin, D.; Buckanovich, R.J.; Mehta, G. Carcinoma-Associated Mesenchymal Stem Cells Promote Chemoresistance in Ovarian Cancer Stem Cells via PDGF Signaling. Cancers 2020, 12, 2063. [Google Scholar] [CrossRef] [PubMed]
- Ip, C.K.; Li, S.-S.; Tang, M.Y.H.; Sy, S.K.H.; Ren, Y.; Shum, H.C.; Wong, A.S.T. Stemness and chemoresistance in epithelial ovarian carcinoma cells under shear stress. Sci. Rep. 2016, 6, 26788. [Google Scholar] [CrossRef] [PubMed]
- Al Habyan, S.; Kalos, C.; Szymborski, J.; Mc Caffrey, L. Multicellular detachment generates metastatic spheroids during intra-abdominal dissemination in epithelial ovarian cancer. Oncogene 2018, 37, 5127–5135. [Google Scholar] [CrossRef]
- Gunay, G.; Kirit, H.A.; Kamatar, A.; Baghdasaryan, O.; Hamsici, S.; Acar, H. The effects of size and shape of the ovarian cancer spheroids on the drug resistance and migration. Gynecol. Oncol. 2020, 159, 563–572. [Google Scholar] [CrossRef] [PubMed]
- Boylan, K.L.; Manion, R.D.; Shah, H.; Skubitz, K.M.; Skubitz, A.P.N. Inhibition of Ovarian Cancer Cell Spheroid Formation by Synthetic Peptides Derived from Nectin-4. Int. J. Mol. Sci. 2020, 21, 4637. [Google Scholar] [CrossRef]
- L’Espérance, S.; Bachvarova, M.; Tetu, B.; Mes-Masson, A.-M.; Bachvarov, D. Global gene expression analysis of early response to chemotherapy treatment in ovarian cancer spheroids. BMC Genom. 2008, 9, 99. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Li, S.; Yang, L.; Zhang, D.; Zhao, Z.; Gao, J.; Liu, L. CDC25A Facilitates Chemo-resistance in Ovarian Cancer Multicellular Spheroids by Promoting E-cadherin Expression and Arresting Cell Cycles. J. Cancer 2019, 10, 2874–2884. [Google Scholar] [CrossRef] [Green Version]
- Lupia, M.; Angiolini, F.; Bertalot, G.; Freddi, S.; Sachsenmeier, K.F.; Chisci, E.; Kutryb-Zajac, B.; Confalonieri, S.; Smolenski, R.; Giovannoni, R.; et al. CD73 Regulates Stemness and Epithelial-Mesenchymal Transition in Ovarian Cancer-Initiating Cells. Stem Cell Rep. 2018, 10, 1412–1425. [Google Scholar] [CrossRef] [Green Version]
- Kwon, A.-Y.; Kim, G.-I.; Jeong, J.-Y.; Song, J.-Y.; Kwack, K.-B.; Lee, C.; Kang, H.-Y.; Kim, T.-H.; Heo, J.-H.; An, H.J. VAV3 Overexpressed in Cancer Stem Cells Is a Poor Prognostic Indicator in Ovarian Cancer Patients. Stem Cells Dev. 2015, 24, 1521–1535. [Google Scholar] [CrossRef] [Green Version]
- Park, H.; Hwang, S.; Jeong, J.-Y.; Jung, S.G.; Choi, M.C.; Joo, W.D.; Song, S.H.; Lee, C.; An, H.J. Integrative analysis of transcription factors and microRNAs in ovarian cancer cell spheroids. J. Ovarian Res. 2020, 13, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Ishiguro, T.; Sato, A.; Ohata, H.; Ikarashi, Y.; Takahashi, R.-U.; Ochiya, T.; Yoshida, M.; Tsuda, H.; Onda, T.; Kato, T.; et al. Establishment and Characterization of an In Vitro Model of Ovarian Cancer Stem-like Cells with an Enhanced Proliferative Capacity. Cancer Res. 2016, 76, 150–160. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, K.; Berger, H.; Kulbe, H.; Thillainadarasan, S.; Mollenkopf, H.; Zemojtel, T.; Taube, E.; Darb-Esfahani, S.; Mangler, M.; Sehouli, J.; et al. Stable expansion of high-grade serous ovarian cancer organoids requires a low-Wnt environment. EMBO J. 2020, 39, e104013. [Google Scholar] [CrossRef] [PubMed]
- Kopper, O.; de Witte, C.J.; Lõhmussaar, K.; Valle-Inclan, J.E.; Hami, N.; Kester, L.; Balgobind, A.V.; Korving, J.; Proost, N.; Begthel, H.; et al. An organoid platform for ovarian cancer captures intra- and interpatient heterogeneity. Nat. Med. 2019, 25, 838–849. [Google Scholar] [CrossRef]
- Maru, Y.; Tanaka, N.; Itami, M.; Hippo, Y. Efficient use of patient-derived organoids as a preclinical model for gynecologic tumors. Gynecol. Oncol. 2019, 154, 189–198. [Google Scholar] [CrossRef]
- Nanki, Y.; Chiyoda, T.; Hirasawa, A.; Ookubo, A.; Itoh, M.; Ueno, M.; Akahane, T.; Kameyama, K.; Yamagami, W.; Kataoka, F.; et al. Patient-derived ovarian cancer organoids capture the genomic profiles of primary tumours applicable for drug sensitivity and resistance testing. Sci. Rep. 2020, 10, 12581. [Google Scholar] [CrossRef]
- Chen, H.; Gotimer, K.; De Souza, C.; Tepper, C.G.; Karnezis, A.N.; Leiserowitz, G.S.; Chien, J.; Smith, L.H. Short-term organoid culture for drug sensitivity testing of high-grade serous carcinoma. Gynecol. Oncol. 2020, 157, 783–792. [Google Scholar] [CrossRef]
- Hill, S.J.; Decker, B.; Roberts, E.A.; Horowitz, N.S.; Muto, M.G.; Worley, M.J.; Feltmate, C.M.; Nucci, M.R.; Swisher, E.M.; Nguyen, H.; et al. Prediction of DNA Repair Inhibitor Response in Short-Term Patient-Derived Ovarian Cancer Organoids. Cancer Discov. 2018, 8, 1404–1421. [Google Scholar] [CrossRef] [Green Version]
- de Witte, C.J.; Valle-Inclan, J.E.; Hami, N.; Lõhmussaar, K.; Kopper, O.; Vreuls, C.P.H.; Jonges, G.N.; van Diest, P.; Nguyen, L.; Clevers, H.; et al. Patient-Derived Ovarian Cancer Organoids Mimic Clinical Response and Exhibit Heterogeneous Inter- and Intrapatient Drug Responses. Cell Rep. 2020, 31, 107762. [Google Scholar] [CrossRef]
- Jabs, J.; Zickgraf, F.M.; Park, J.; Wagner, S.; Jiang, X.; Jechow, K.; Kleinheinz, K.; Toprak, U.H.; Schneider, M.A.; Meister, M.; et al. Screening drug effects in patient-derived cancer cells links organoid responses to genome alterations. Mol. Syst. Biol. 2017, 13, 955. [Google Scholar] [CrossRef]
- Maenhoudt, N.; Defraye, C.; Boretto, M.; Jan, Z.; Heremans, R.; Boeckx, B.; Hermans, F.; Arijs, I.; Cox, B.; Van Nieuwenhuysen, E.; et al. Developing Organoids from Ovarian Cancer as Experimental and Preclinical Models. Stem Cell Rep. 2020, 14, 717–729. [Google Scholar] [CrossRef]
- Li, S.-S.; Ip, C.K.; Tang, M.Y.H.; Sy, S.K.H.; Yung, S.; Chan, T.-M.; Yang, M.; Shum, H.C.; Wong, A.S. Modeling Ovarian Cancer Multicellular Spheroid Behavior in a Dynamic 3D Peritoneal Microdevice. J. Vis. Exp. 2017, e55337. [Google Scholar] [CrossRef] [Green Version]
- Rizvi, I.; Gurkan, U.; Tasoglu, S.; Alagic, N.; Celli, J.P.; Mensah, L.B.; Mai, Z.; Demirci, U.; Hasan, T. Flow induces epithelial-mesenchymal transition, cellular heterogeneity and biomarker modulation in 3D ovarian cancer nodules. Proc. Natl. Acad. Sci. USA 2013, 110, E1974–E1983. [Google Scholar] [CrossRef] [Green Version]
- Dadgar, N.; Gonzalez-Suarez, A.M.; Fattahi, P.; Hou, X.; Weroha, J.S.; Gaspar-Maia, A.; Stybayeva, G.; Revzin, A. A microfluidic platform for cultivating ovarian cancer spheroids and testing their responses to chemotherapies. Microsyst. Nanoeng. 2020, 6, 1–12. [Google Scholar] [CrossRef]
- Flont, M.; Jastrzębska, E.; Brzózka, Z. Synergistic effect of the combination therapy on ovarian cancer cells under microfluidic conditions. Anal. Chim. Acta 2020, 1100, 138–148. [Google Scholar] [CrossRef]
- Ding, Y.; Li, J.; Xiao, W.; Xiao, K.; Lee, J.; Bhardwaj, U.; Zhu, Z.; Digiglio, P.; Yang, G.; Lam, K.S.; et al. Microfluidic-Enabled Print-to-Screen Platform for High-Throughput Screening of Combinatorial Chemotherapy. Anal. Chem. 2015, 87, 10166–10171. [Google Scholar] [CrossRef]
- Marimuthu, M.; Rousset, N.; St-Georges-Robillard, A.; Lateef, M.A.; Ferland, M.; Mes-Masson, A.-M.; Gervais, T. Multi-size spheroid formation using microfluidic funnels. Lab Chip 2018, 18, 304–314. [Google Scholar] [CrossRef]
- Amatangelo, M.D.; Garipov, A.; Li, H.; Conejo-Garcia, J.R.; Speicher, D.W.; Zhang, R. Three-dimensional culture sensitizes epithelial ovarian cancer cells to EZH2 methyltransferase inhibition. Cell Cycle 2013, 12, 2113–2119. [Google Scholar] [CrossRef] [Green Version]
- Novak, C.; Horst, E.; Mehta, G. Review: Mechanotransduction in ovarian cancer: Shearing into the unknown. APL Bioeng. 2018, 2, 031701. [Google Scholar] [CrossRef] [Green Version]
- Klymenko, Y.; Kim, O.; Loughran, E.; Yang, J.; Lombard, R.; Alber, M.; Stack, M.S. Cadherin composition and multicellular aggregate invasion in organotypic models of epithelial ovarian cancer intraperitoneal metastasis. Oncogene 2017, 36, 5840–5851. [Google Scholar] [CrossRef] [Green Version]
- Paradiso, F.; Fitzgerald, J.; Yao, S.; Barry, F.; Taraballi, F.; Gonzalez, D.; Conlan, R.S.; Francis, L. Marine Collagen Substrates for 2D and 3D Ovarian Cancer Cell Systems. Front. Bioeng. Biotechnol. 2019, 7, 343. [Google Scholar] [CrossRef] [Green Version]
- Shin, S.; Ikram, M.; Subhan, F.; Kang, H.Y.; Lim, Y.; Lee, R.; Jin, S.; Jeong, Y.H.; Kwak, J.-Y.; Na, Y.-J.; et al. Alginate–marine collagen–agarose composite hydrogels as matrices for biomimetic 3D cell spheroid formation. RSC Adv. 2016, 6, 46952–46965. [Google Scholar] [CrossRef]
- Loessner, D.; Stok, K.S.; Lutolf, M.P.; Hutmacher, D.W.; Clements, J.A.; Rizzi, S.C. Bioengineered 3D platform to explore cell–ECM interactions and drug resistance of epithelial ovarian cancer cells. Biomaterials 2010, 31, 8494–8506. [Google Scholar] [CrossRef] [Green Version]
- Guo, Z.; Zhang, T.; Fang, K.; Dou, J.; Zhou, N.; Ma, X.; Gu, N. The effects of macroporosity and stiffness of poly[(methyl vinyl ether)-alt-(maleic acid)] cross-linked egg white simulations of an aged extracellular matrix on the proliferation of ovarian cancer cells. RSC Adv. 2016, 6, 43892–43900. [Google Scholar] [CrossRef]
- Loessner, D.; Flegg, J.A.; Byrne, H.M.; Clements, J.A.; Hutmacher, D.W. Growth of confined cancer spheroids: A combined experimental and mathematical modelling approach. Integr. Biol. 2013, 5, 597–605. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; Chen, J.; Zhang, Q.; Dou, J.; Gu, N. Poly(ethylene glycol)-cross linked poly(methyl vinyl ether-co-maleic acid)hydrogels for three-dimensional human ovarian cancer cell culture. Colloids Surf. A Physicochem. Eng. Asp. 2013, 422, 81–89. [Google Scholar] [CrossRef]
- Kaemmerer, E.; Melchels, F.P.; Holzapfel, B.M.; Meckel, T.; Hutmacher, D.W.; Loessner, D. Gelatine methacrylamide-based hydrogels: An alternative three-dimensional cancer cell culture system. Acta Biomater. 2014, 10, 2551–2562. [Google Scholar] [CrossRef]
- Lee, J.M.; Park, D.Y.; Yang, L.; Kim, E.-J.; Ahrberg, C.D.; Lee, K.-B.; Chung, B.G. Generation of uniform-sized multicellular tumor spheroids using hydrogel microwells for advanced drug screening. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Loessner, D.; Kobel, S.; Clements, J.A.; Lutolf, M.P.; Hutmacher, D.W. Hydrogel Microwell Arrays Allow the Assessment of Protease-Associated Enhancement of Cancer Cell Aggregation and Survival. Microarrays 2013, 2, 208–227. [Google Scholar] [CrossRef] [Green Version]
- Loessner, D.; Rizzi, S.C.; Stok, K.S.; Führmann, T.; Hollier, B.; Magdolen, V.; Hutmacher, D.W.; Clements, J. A bioengineered 3D ovarian cancer model for the assessment of peptidase–mediated enhancement of spheroid growth and intraperitoneal spread. Biomaterials 2013, 34, 7389–7400. [Google Scholar] [CrossRef]
- Loessner, D.; Rockstroh, A.; Shokoohmand, A.; Holzapfel, B.M.; Wagner, F.; Baldwin, J.; Boxberg, M.; Schmalfeldt, B.; Lengyel, E.; Clements, J.A.; et al. A 3D tumor microenvironment regulates cell proliferation, peritoneal growth and expression patterns. Biomaterials 2019, 190–191, 63–75. [Google Scholar] [CrossRef]
- Brooks, E.A.; Gencoglu, M.F.; Corbett, D.C.; Stevens, K.R.; Peyton, S.R. An omentum-inspired 3D PEG hydrogel for identifying ECM-drivers of drug resistant ovarian cancer. APL Bioeng. 2019, 3, 026106. [Google Scholar] [CrossRef] [Green Version]
- Abu-Yousif, A.O.; Rizvi, I.; Evans, C.L.; Celli, J.P.; Hasan, T. PuraMatrix Encapsulation of Cancer Cells. J. Vis. Exp. 2009, e1692. [Google Scholar] [CrossRef]
- Yang, Z.; Zhao, X. A 3D model of ovarian cancer cell lines on peptide nanofiber scaffold to explore the cell–scaffold interaction and chemotherapeutic resistance of anticancer drugs. Int. J. Nanomed. 2011, 6, 303–310. [Google Scholar] [CrossRef] [Green Version]
- Song, H.; Cai, G.-H.; Liang, J.; Ao, D.-S.; Wang, H.; Yang, Z.-H. Three-dimensional culture and clinical drug responses of a highly metastatic human ovarian cancer HO-8910PM cells in nanofibrous microenvironments of three hydrogel biomaterials. J. Nanobiotechnol. 2020, 18, 1–19. [Google Scholar] [CrossRef]
- Yang, Z.; Zhuang, H.; Song, H.; Liu, J.; Zhao, X.; Lin, W. A Miniature Cell Pattern Formation of Ovarian Cancer Cell Lines on Self-Assembling Peptide Nanofiber-Coated Coverslip and In Vitro Chemosensitivity Assay. J. Nanosci. Nanotechnol. 2018, 18, 2370–2378. [Google Scholar] [CrossRef]
- Hedegaard, C.L.; Redondo-Gómez, C.; Tan, B.Y.; Ng, K.W.; Loessner, D.; Mata, A. Peptide-protein coassembling matrices as a biomimetic 3D model of ovarian cancer. Sci. Adv. 2020, 6, eabb3298. [Google Scholar] [CrossRef]
- Hirschhaeuser, F.; Menne, H.; Dittfeld, C.; West, J.; Mueller-Klieser, W.; Kunz-Schughart, L.A. Multicellular tumor spheroids: An underestimated tool is catching up again. J. Biotechnol. 2010, 148, 3–15. [Google Scholar] [CrossRef]
- Huang, B.-W.; Gao, J.-Q. Application of 3D cultured multicellular spheroid tumor models in tumor-targeted drug delivery system research. J. Control. Release 2018, 270, 246–259. [Google Scholar] [CrossRef]
- Pinto, B.; Henriques, A.C.; Silva, P.M.A.; Bousbaa, H. Three-Dimensional Spheroids as In Vitro Preclinical Models for Cancer Research. Pharmaceutics 2020, 12, 1186. [Google Scholar] [CrossRef]
- Mehta, G.; Hsiao, A.Y.; Ingram, M.; Luker, G.D.; Takayama, S. Opportunities and challenges for use of tumor spheroids as models to test drug delivery and efficacy. J. Control. Release 2012, 164, 192–204. [Google Scholar] [CrossRef] [Green Version]
- Zietarska, M.; Maugard, C.M.; Filali-Mouhim, A.; Alam-Fahmy, M.; Tonin, P.N.; Provencher, D.M.; Mes-Masson, A.-M. Molecular description of a 3D in vitro model for the study of epithelial ovarian cancer (EOC). Mol. Carcinog. 2007, 46, 872–885. [Google Scholar] [CrossRef]
- Lee, J.M.; Mhawech-Fauceglia, P.; Lee, N.; Parsanian, L.C.; Lin, Y.G.; Gayther, S.A.; Lawrenson, K. A three-dimensional microenvironment alters protein expression and chemosensitivity of epithelial ovarian cancer cells in vitro. Lab. Investig. 2013, 93, 528–542. [Google Scholar] [CrossRef] [Green Version]
- Shishido, A.; Mori, S.; Yokoyama, Y.; Hamada, Y.; Minami, K.; Qian, Y.; Wang, J.; Hirose, H.; Wu, X.; Kawaguchi, N.; et al. Mesothelial cells facilitate cancer stem-like properties in spheroids of ovarian cancer cells. Oncol. Rep. 2018, 40, 2105–2114. [Google Scholar] [CrossRef] [Green Version]
- Costa, E.C.; Moreira, A.F.; Diogo, D.M.D.M.; Gaspar, V.; Carvalho, M.P.; Correia, I.J. 3D tumor spheroids: An overview on the tools and techniques used for their analysis. Biotechnol. Adv. 2016, 34, 1427–1441. [Google Scholar] [CrossRef]
- Chau, W.K.; Ip, C.K.; Mak, A.S.C.; Lai, H.-C.; Wong, A.S.T. c-Kit mediates chemoresistance and tumor-initiating capacity of ovarian cancer cells through activation of Wnt/β-catenin–ATP-binding cassette G2 signaling. Oncogene 2012, 32, 2767–2781. [Google Scholar] [CrossRef] [Green Version]
- Liao, J.; Qian, F.; Tchabo, N.; Mhawech-Fauceglia, P.; Beck, A.; Qian, Z.; Wang, X.; Huss, W.J.; Lele, S.B.; Morrison, C.D.; et al. Ovarian Cancer Spheroid Cells with Stem Cell-Like Properties Contribute to Tumor Generation, Metastasis and Chemotherapy Resistance through Hypoxia-Resistant Metabolism. PLoS ONE 2014, 9, e84941. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, Y.; Kurokawa, T.; Nishikawa, Y.; Orisa, M.; Kleinman, H.K.; Kotsuji, F. Laminin-1-derived scrambled peptide AG73T disaggregates laminin-1-induced ovarian cancer cell spheroids and improves the efficacy of cisplatin. Int. J. Oncol. 2008, 32, 673–681. [Google Scholar] [CrossRef] [Green Version]
- Minchinton, A.I.; Tannock, I.F. Drug penetration in solid tumours. Nat. Rev. Cancer 2006, 6, 583–592. [Google Scholar] [CrossRef]
- Lazzari, G.; Couvreur, P.; Mura, S. Multicellular tumor spheroids: A relevant 3D model for the in vitro preclinical investigation of polymer nanomedicines. Polym. Chem. 2017, 8, 4947–4969. [Google Scholar] [CrossRef] [Green Version]
- Raghavan, S.; Mehta, P.; Horst, E.N.; Ward, M.R.; Rowley, K.R.; Mehta, G. Comparative analysis of tumor spheroid generation techniques for differential in vitro drug toxicity. Oncotarget 2016, 7, 16948–16961. [Google Scholar] [CrossRef]
- Shoval, H.; Karsch-Bluman, A.; Brill-Karniely, Y.; Stern, T.; Zamir, G.; Hubert, A.; Benny, O. Tumor cells and their crosstalk with endothelial cells in 3D spheroids. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Tang, P.; Cai, S.; Peng, J.; Hua, G. Organoid based personalized medicine: From bench to bedside. Cell Regen. 2020, 9, 1–33. [Google Scholar] [CrossRef]
- Xu, H.; Lyu, X.; Yi, M.; Zhao, W.; Song, Y.; Wu, K. Organoid technology and applications in cancer research. J. Hematol. Oncol. 2018, 11, 1–15. [Google Scholar] [CrossRef]
- Liu, H.-D.; Xia, B.-R.; Jin, M.-Z.; Lou, G. Organoid of ovarian cancer: Genomic analysis and drug screening. Clin. Transl. Oncol. 2020, 22, 1240–1251. [Google Scholar] [CrossRef] [Green Version]
- Pauli, C.; Hopkins, B.D.; Prandi, D.; Shaw, R.; Fedrizzi, T.; Sboner, A.; Sailer, V.; Augello, M.; Puca, L.; Rosati, R.; et al. Personalized In Vitro and In Vivo Cancer Models to Guide Precision Medicine. Cancer Discov. 2017, 7, 462–477. [Google Scholar] [CrossRef] [Green Version]
- Maru, Y.; Hippo, Y. Current Status of Patient-Derived Ovarian Cancer Models. Cells 2019, 8, 505. [Google Scholar] [CrossRef] [Green Version]
- Verduin, M.; Hoeben, A.; De Ruysscher, D.; Vooijs, M. Patient-Derived Cancer Organoids as Predictors of Treatment Response. Front. Oncol. 2021, 11, 820. [Google Scholar] [CrossRef]
- Drost, J.; Clevers, H. Organoids in cancer research. Nat. Rev. Cancer 2018, 18, 407–418. [Google Scholar] [CrossRef]
- Gunti, S.; Hoke, A.; Vu, K.; London, N. Organoid and Spheroid Tumor Models: Techniques and Applications. Cancers 2021, 13, 874. [Google Scholar] [CrossRef]
- Es, H.A.; Montazeri, L.; Aref, A.R.; Vosough, M.; Baharvand, H. Personalized Cancer Medicine: An Organoid Approach. Trends Biotechnol. 2018, 36, 358–371. [Google Scholar] [CrossRef]
- Corro’, C.; Novellasdemunt, L.; Li, V.S. A brief history of organoids. Am. J. Physiol. Physiol. 2020, 319, C151–C165. [Google Scholar] [CrossRef]
- Ong, L.J.Y.; Chong, L.H.; Jin, L.; Singh, P.K.; Lee, P.; Yu, H.; Ananthanarayanan, A.; Leo, H.L.; Toh, Y.-C. A pump-free microfluidic 3D perfusion platform for the efficient differentiation of human hepatocyte-like cells. Biotechnol. Bioeng. 2017, 114, 2360–2370. [Google Scholar] [CrossRef]
- Tsai, H.-F.; Trubelja, A.; Shen, A.Q.; Bao, G. Tumour-on-a-chip: Microfluidic models of tumour morphology, growth and microenvironment. J. R. Soc. Interface 2017, 14, 20170137. [Google Scholar] [CrossRef] [Green Version]
- Sato, K.; Sato, M.; Yokoyama, M.; Hirai, M.; Furuta, A. Influence of Culture Conditions on Cell Proliferation in a Microfluidic Channel. Anal. Sci. 2019, 35, 49–56. [Google Scholar] [CrossRef] [Green Version]
- Six, K.R.; Sicot, G.; Devloo, R.; Feys, H.B.; Baruch, D.; Compernolle, V. A comparison of haematopoietic stem cells from umbilical cord blood and peripheral blood for platelet production in a microfluidic device. Vox Sang. 2019, 114, 330–339. [Google Scholar] [CrossRef]
- Sun, J.; Warden, A.R.; Ding, X. Recent advances in microfluidics for drug screening. Biomicrofluidics 2019, 13, 061503. [Google Scholar] [CrossRef]
- Jeon, J.; Zervantonakis, I.; Chung, S.; Kamm, R.D.; Charest, J.L. In Vitro Model of Tumor Cell Extravasation. PLoS ONE 2013, 8, e56910. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Yang, X.; Zou, J.; Jia, C.; Hu, Y.; Du, H.; Wang, H. Evaluation of photodynamic therapy efficiency using an in vitro three-dimensional microfluidic breast cancer tissue model. Lab Chip 2015, 15, 735–744. [Google Scholar] [CrossRef]
- Gori, M.; Simonelli, M.C.; Giannitelli, S.M.; Businaro, L.; Trombetta, M.; Rainer, A. Investigating Nonalcoholic Fatty Liver Disease in a Liver-on-a-Chip Microfluidic Device. PLoS ONE 2016, 11, e0159729. [Google Scholar] [CrossRef]
- Kasendra, M.; Tovaglieri, A.; Sontheimer-Phelps, A.; Jalili-Firoozinezhad, S.; Bein, A.; Chalkiadaki, A.; Scholl, W.; Zhang, C.; Rickner, H.; Richmond, C.A.; et al. Development of a primary human Small Intestine-on-a-Chip using biopsy-derived organoids. Sci. Rep. 2018, 8, 1–14. [Google Scholar] [CrossRef]
- Pocock, K.; Delon, L.; Bala, V.; Rao, S.; Priest, C.; Prestidge, C.; Thierry, B. Intestine-on-a-Chip Microfluidic Model for Efficient in Vitro Screening of Oral Chemotherapeutic Uptake. ACS Biomater. Sci. Eng. 2017, 3, 951–959. [Google Scholar] [CrossRef]
- Agarwal, A.; Goss, J.A.; Cho, A.; McCain, M.L.; Parker, K.K. Microfluidic heart on a chip for higher throughput pharmacological studies. Lab Chip 2013, 13, 3599–3608. [Google Scholar] [CrossRef] [Green Version]
- Huh, D.; Matthews, B.D.; Mammoto, A.; Montoya-Zavala, M.; Hsin, H.Y.; Ingber, D.E. Reconstituting Organ-Level Lung Functions on a Chip. Science 2010, 328, 1662–1668. [Google Scholar] [CrossRef] [Green Version]
- Stucki, A.O.; Stucki, J.D.; Hall, S.R.R.; Felder, M.; Mermoud, Y.; Schmid, R.A.; Geiser, T.; Guenat, O.T. A lung-on-a-chip array with an integrated bio-inspired respiration mechanism. Lab Chip 2015, 15, 1302–1310. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Chen, H.; Wu, D.; Chen, Q.; Zhou, Z.; Zhang, R.; Peng, X.; Su, Y.-C.; Sun, D. 3D printed microfluidic chip for multiple anticancer drug combinations. Sens. Actuators B Chem. 2018, 276, 507–516. [Google Scholar] [CrossRef]
- Wufuer, M.; Lee, G.H.; Hur, W.; Jeon, B.; Kim, B.J.; Choi, T.H.; Lee, S.H. Skin-on-a-chip model simulating inflammation, edema and drug-based treatment. Sci. Rep. 2016, 6, 37471. [Google Scholar] [CrossRef] [Green Version]
- Trujillo-de Santiago, G.; Flores-Garza, B.G.; Tavares-Negrete, J.A.; Lara-Mayorga, I.M.; González-Gamboa, I.; Zhang, Y.S.; Rojas-Martínez, A.; Ortiz-López, R.; Álvarez, M.M. The Tumor-on-Chip: Recent Advances in the Development of Microfluidic Systems to Recapitulate the Physiology of Solid Tumors. Materials 2019, 12, 2945. [Google Scholar] [CrossRef] [Green Version]
- Komeya, M.; Kimura, H.; Nakamura, H.; Yokonishi, T.; Sato, T.; Kojima, K.; Hayashi, K.; Katagiri, K.; Yamanaka, H.; Sanjo, H.; et al. Long-term ex vivo maintenance of testis tissues producing fertile sperm in a microfluidic device. Sci. Rep. 2016, 6, 21472. [Google Scholar] [CrossRef]
- Abaci, H.E.; Gledhill, K.; Guo, Z.; Christiano, A.M.; Shuler, M.L. Pumpless microfluidic platform for drug testing on human skin equivalents. Lab Chip 2015, 15, 882–888. [Google Scholar] [CrossRef] [Green Version]
- Shang, M.; Soon, R.H.; Lim, C.T.; Khoo, B.L.; Han, J. Microfluidic modelling of the tumor microenvironment for anti-cancer drug development. Lab Chip 2019, 19, 369–386. [Google Scholar] [CrossRef]
- Onal, S.; Alkaisi, M.M.; Nock, V. A Flexible Microdevice for Mechanical Cell Stimulation and Compression in Microfluidic Settings. Front. Phys. 2021, 9, 1–19. [Google Scholar] [CrossRef]
- Novak, C.M.; Horst, E.N.; Lin, E.; Mehta, G. Compressive Stimulation Enhances Ovarian Cancer Proliferation, Invasion, Chemoresistance, and Mechanotransduction via CDC42 in a 3D Bioreactor. Cancers 2020, 12, 1521. [Google Scholar] [CrossRef]
- Ma, Y.-H.V.; Middleton, K.; You, L.; Sun, Y. A review of microfluidic approaches for investigating cancer extravasation during metastasis. Microsystems Nanoeng. 2018, 4, 17104. [Google Scholar] [CrossRef] [Green Version]
- McGrail, D.J.; Kieu, Q.M.N.; Dawson, M.R. Metastatic ovarian cancer cell malignancy is increased on soft matrices through a mechanosensitive Rho/ROCK pathway. J. Cell Sci. 2014, 127, 2621–2626. [Google Scholar] [CrossRef] [Green Version]
- Anguiano, M.; Castilla, C.; Maška, M.; Ederra, C.; Peláez, R.; Morales, X.; Muñoz-Arrieta, G.; Mujika, M.; Kozubek, M.; Muñoz-Barrutia, A.; et al. Characterization of three-dimensional cancer cell migration in mixed collagen-Matrigel scaffolds using microfluidics and image analysis. PLoS ONE 2017, 12, e0171417. [Google Scholar] [CrossRef] [Green Version]
- Pathak, A.; Kumar, S. Independent regulation of tumor cell migration by matrix stiffness and confinement. Proc. Natl. Acad. Sci. USA 2012, 109, 10334–10339. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Ho, W.; Zhang, X.; Bertrand, N.; Farokhzad, O. Cancer nanomedicine: From targeted delivery to combination therapy. Trends Mol. Med. 2015, 21, 223–232. [Google Scholar] [CrossRef] [Green Version]
- Katt, M.E.; Placone, A.L.; Wong, A.D.; Xu, Z.S.; Searson, P.C. In Vitro Tumor Models: Advantages, Disadvantages, Variables, and Selecting the Right Platform. Front. Bioeng. Biotechnol. 2016, 4, 12. [Google Scholar] [CrossRef]
- Huh, D.; Leslie, D.C.; Matthews, B.D.; Fraser, J.P.; Jurek, S.; Hamilton, G.A.; Thorneloe, K.S.; McAlexander, M.A.; Ingber, D.E. A Human Disease Model of Drug Toxicity–Induced Pulmonary Edema in a Lung-on-a-Chip Microdevice. Sci. Transl. Med. 2012, 4, 159ra147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duzagac, F.; Saorin, G.; Memeo, L.; Canzonieri, V.; Rizzolio, F. Microfluidic Organoids-on-a-Chip: Quantum Leap in Cancer Research. Cancers 2021, 13, 737. [Google Scholar] [CrossRef]
- Kim, M.M.; Huang, Y.; Choi, K.; Hidrovo, C.H. The improved resistance of PDMS to pressure-induced deformation and chemical solvent swelling for microfluidic devices. Microelectron. Eng. 2014, 124, 66–75. [Google Scholar] [CrossRef]
- Sontheimer-Phelps, A.; Hassell, B.A.; Ingber, D.E. Modelling cancer in microfluidic human organs-on-chips. Nat. Rev. Cancer 2019, 19, 65–81. [Google Scholar] [CrossRef]
- Sun, Z.; Song, C.; Wang, C.; Hu, Y.; Wu, J. Hydrogel-Based Controlled Drug Delivery for Cancer Treatment: A Review. Mol. Pharm. 2019, 17, 373–391. [Google Scholar] [CrossRef]
- Wang, C.; Tang, Z.; Zhao, Y.; Yao, R.; Li, L.; Sun, W. Three-dimensional in vitro cancer models: A short review. Biofabrication 2014, 6, 022001. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.; Mooney, D.J. Biomaterials and emerging anticancer therapeutics: Engineering the microenvironment. Nat. Rev. Cancer 2016, 16, 56–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Kumacheva, E. Hydrogel microenvironments for cancer spheroid growth and drug screening. Sci. Adv. 2018, 4, eaas8998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stock, K.; Estrada, M.; Vidic, S.; Gjerde, K.; Rudisch, A.; Santo, V.E.; Barbier, M.; Blom, S.; Arundkar, S.C.; Selvam, I.; et al. Capturing tumor complexity in vitro: Comparative analysis of 2D and 3D tumor models for drug discovery. Sci. Rep. 2016, 6, 28951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grosskopf, A.K.; Correa, S.; Baillet, J.; Maikawa, C.L.; Gale, E.C.; Brown, R.A.; Appel, E.A. Consistent tumorigenesis with self-assembled hydrogels enables high-powered murine cancer studies. Commun. Biol. 2021, 4, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Lewin Mejia, D.; Chiang, B.; Luker, K.E.; Luker, G.D. Hybrid collagen alginate hydrogel as a platform for 3D tumor spheroid invasion. Acta Biomater. 2018, 75, 213–225. [Google Scholar] [CrossRef]
- Kamatar, A.; Gunay, G.; Acar, H. Natural and Synthetic Biomaterials for Engineering Multicellular Tumor Spheroids. Polymers 2020, 12, 2506. [Google Scholar] [CrossRef]
- Alemany-Ribes, M.; Semino, C.E. Bioengineering 3D environments for cancer models. Adv. Drug Deliv. Rev. 2014, 79–80, 40–49. [Google Scholar] [CrossRef]
- Li, X.; Sun, Q.; Li, Q.; Kawazoe, N.; Chen, G. Functional Hydrogels With Tunable Structures and Properties for Tissue Engineering Applications. Front. Chem. 2018, 6, 499. [Google Scholar] [CrossRef] [Green Version]
- Bray, L.J.; Binner, M.; Holzheu, A.; Friedrichs, J.; Freudenberg, U.; Hutmacher, D.W.; Werner, C. Multi-parametric hydrogels support 3D in vitro bioengineered microenvironment models of tumour angiogenesis. Biomaterials 2015, 53, 609–620. [Google Scholar] [CrossRef] [Green Version]
- Pradhan, S.; Clary, J.M.; Seliktar, D.; Lipke, E.A. A three-dimensional spheroidal cancer model based on PEG-fibrinogen hydrogel microspheres. Biomaterials 2017, 115, 141–154. [Google Scholar] [CrossRef]
- Taubenberger, A.V.; Bray, L.J.; Haller, B.; Shaposhnykov, A.; Binner, M.; Freudenberg, U.; Guck, J.; Werner, C. 3D extracellular matrix interactions modulate tumour cell growth, invasion and angiogenesis in engineered tumour microenvironments. Acta Biomater. 2016, 36, 73–85. [Google Scholar] [CrossRef] [Green Version]
- Mendes, A.C.L.; Baran, E.T.; Reis, R.L.; Azevedo, H.S. Self-assembly in nature: Using the principles of nature to create complex nanobiomaterials. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2013, 5, 582–612. [Google Scholar] [CrossRef]
- Loo, Y.; Zhang, S.; Hauser, C.A. From short peptides to nanofibers to macromolecular assemblies in biomedicine. Biotechnol. Adv. 2012, 30, 593–603. [Google Scholar] [CrossRef]
- Yu, Z.; Xu, Q.; Dong, C.; Lee, S.S.; Gao, L.; Li, Y.; D’Ortenzio, M.; Wu, J. Self-Assembling Peptide Nanofibrous Hydrogel as a Versatile Drug Delivery Platform. Curr. Pharm. Des. 2015, 21, 4342–4354. [Google Scholar] [CrossRef]
- Pashuck, E.T.; Cui, H.; Stupp, S.I. Tuning Supramolecular Rigidity of Peptide Fibers through Molecular Structure. J. Am. Chem. Soc. 2010, 132, 6041–6046. [Google Scholar] [CrossRef] [Green Version]
- Hurley, S.K.; Cutrone, N.M.; Fath, K.; Pajovich, H.T.; Garcia, J.; Smith, A.M.; Banerjee, I.A. Self-assembled phenylisoxazole-peptide hybrid assemblies and their interactions with breast and ovarian tumor cells. Int. J. Polym. Mater. 2018, 68, 978–992. [Google Scholar] [CrossRef]
- Lee, S.; Trinh, T.H.; Yoo, M.; Shin, J.; Lee, H.; Kim, J.; Hwang, E.; Lim, Y.-B.; Ryou, C. Self-Assembling Peptides and Their Application in the Treatment of Diseases. Int. J. Mol. Sci. 2019, 20, 5850. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, J.; Radvar, E.; Azevedo, H. Self-assembling peptides and their application in tissue engineering and regenerative medicine. In Peptides and Proteins as Biomaterials for Tissue Regeneration and Repair; Woodhead Publishing: Sawston, UK, 2018; pp. 245–281. [Google Scholar] [CrossRef]
- Yadav, S.; Sharma, A.K.; Kumar, P. Nanoscale Self-Assembly for Therapeutic Delivery. Front. Bioeng. Biotechnol. 2020, 8, 127. [Google Scholar] [CrossRef]
- Azevedo, H.S.; da Silva, R.M.P. Self-Assembling Biomaterials: Molecular Design, Characterization and Application in Biology and Medicine, 1st ed.; Woodhead Publishing: Sawston, UK; Elsevier: Amsterdam, The Netherlands, 2018; pp. 1–612. [Google Scholar] [CrossRef]
- Zhang, S.; Holmes, T.; Lockshin, C.; Rich, A. Spontaneous assembly of a self-complementary oligopeptide to form a stable macroscopic membrane. Proc. Natl. Acad. Sci. USA 1993, 90, 3334–3338. [Google Scholar] [CrossRef] [Green Version]
- Lachowski, D.; Matellan, C.; Cortes, E.; Saiani, A.; Miller, A.; Hernández, A.D.R. Self-Assembling Polypeptide Hydrogels as a Platform to Recapitulate the Tumor Microenvironment. Cancers 2021, 13, 3286. [Google Scholar] [CrossRef]
- Clough, H.C.; O’Brien, M.; Zhu, X.; Miller, A.F.; Saiani, A.; Tsigkou, O. Neutrally charged self-assembling peptide hydrogel recapitulates in vitro mechanisms of breast cancer progression. Mater. Sci. Eng. C 2021, 127, 112200. [Google Scholar] [CrossRef]
- Edwards-Gayle, C.J.C.; Hamley, I.W. Self-assembly of bioactive peptides, peptide conjugates, and peptide mimetic materials. Org. Biomol. Chem. 2017, 15, 5867–5876. [Google Scholar] [CrossRef] [Green Version]
- Derkus, B.; Okesola, B.O.; Barrett, D.W.; D’Este, M.; Chowdhury, T.T.; Eglin, D.; Mata, A. Multicomponent hydrogels for the formation of vascularized bone-like constructs in vitro. Acta Biomater. 2020, 109, 82–94. [Google Scholar] [CrossRef]
- Barrett, D.W.; Okesola, B.O.; Costa, E.; Thrasivoulou, C.; Becker, D.L.; Mata, A.; Deprest, J.A.; David, A.L.; Chowdhury, T.T. Potential sealing and repair of human FM defects after trauma with peptide amphiphiles and Cx43 antisense. Prenat. Diagn. 2021, 41, 89–99. [Google Scholar] [CrossRef]
- Cui, H.; Webber, M.J.; Stupp, S.I. Self-assembly of peptide amphiphiles: From molecules to nanostructures to biomaterials. Biopolymers 2010, 94, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, D.S.; Marques, A.P.; Reis, R.L.; Azevedo, H.S. Hyaluronan and self-assembling peptides as building blocks to reconstruct the extracellular environment in skin tissue. Biomater. Sci. 2013, 1, 952–964. [Google Scholar] [CrossRef]
- Alvero, A.B.; Kim, N.; Lima, E.; Sumi, N.J.; Lee, J.S.; Cardenas, C.; Pitruzzello, M.; Silasi, D.-A.; Buza, N.; Fahmy, T.; et al. Novel approach for the detection of intraperitoneal micrometastasis using an ovarian cancer mouse model. Sci. Rep. 2017, 7, 40989. [Google Scholar] [CrossRef] [Green Version]
- Shah, R.N.; Shah, N.A.; Lim, M.M.D.R.; Hsieh, C.; Nuber, G.; Stupp, S.I. Supramolecular design of self-assembling nanofibers for cartilage regeneration. Proc. Natl. Acad. Sci. USA 2010, 107, 3293–3298. [Google Scholar] [CrossRef] [Green Version]
- Kopesky, P.W.; Vanderploeg, E.J.; Sandy, J.S.; Kurz, B.; Grodzinsky, A.J. Self-Assembling Peptide Hydrogels Modulate In Vitro Chondrogenesis of Bovine Bone Marrow Stromal Cells. Tissue Eng. Part A 2010, 16, 465–477. [Google Scholar] [CrossRef] [Green Version]
- Galler, K.M.; Hartgerink, J.D.; Cavender, A.C.; Schmalz, G.; D’Souza, R.N. A Customized Self-Assembling Peptide Hydrogel for Dental Pulp Tissue Engineering. Tissue Eng. Part A 2012, 18, 176–184. [Google Scholar] [CrossRef]
- Okesola, B.O.; Ni, S.; Derkus, B.; Galeano, C.C.; Hasan, A.; Wu, Y.; Ramis, J.; Buttery, L.; Dawson, J.I.; D’Este, M.; et al. Growth-Factor Free Multicomponent Nanocomposite Hydrogels That Stimulate Bone Formation. Adv. Funct. Mater. 2020, 30, 1906205. [Google Scholar] [CrossRef]
- Miotto, M.; Gouveia, R.M.; Connon, C.J. Peptide Amphiphiles in Corneal Tissue Engineering. J. Funct. Biomater. 2015, 6, 687–707. [Google Scholar] [CrossRef] [Green Version]
- Hendricks, M.P.; Sato, K.; Palmer, L.C.; Stupp, S.I. Supramolecular Assembly of Peptide Amphiphiles. Acc. Chem. Res. 2017, 50, 2440–2448. [Google Scholar] [CrossRef]
- Hedegaard, C.L.; Mata, A. Integrating self-assembly and biofabrication for the development of structures with enhanced complexity and hierarchical control. Biofabrication 2020, 12, 032002. [Google Scholar] [CrossRef]
- Hedegaard, C.; Collin, E.C.; Redondo-Gómez, C.; Nguyen, L.; Ng, K.W.; Castrejón-Pita, A.A.; Castrejón-Pita, J.R.; Mata, A. Hydrodynamically Guided Hierarchical Self-Assembly of Peptide-Protein Bioinks. Adv. Funct. Mater. 2018, 28, 1703716. [Google Scholar] [CrossRef]
- Harper, M.M.; Connolly, M.L.; Goldie, L.; Irvine, E.J.; Shaw, J.E.; Jayawarna, V.; Richardson, S.M.; Dalby, M.J.; Lightbody, D.; Ulijn, R.V. Biogelx: Cell Culture on Self-Assembling Peptide Gels. Methods Mol. Biol. 2018, 1777, 283–303. [Google Scholar]
- Biogelx. Peptide-Based Bioinks for 3D Bioprinting: BiogelxTM-INK. Available online: https://www.biogelx.com/bioink-product-range/ (accessed on 15 August 2021).
- de la Peña, D.O.; Trabulo, S.; Collin, E.; Loessner, D.; Mata, Á.; Heeschen, C. Self-assembling biomimetic hydrogels as a novel 3D in vitro platform for pancreatic cancer research. Pancreatology 2020, 20, e7. [Google Scholar] [CrossRef]
- Goktas, M.; Cinar, G.; Orujalipoor, I.; Ide, S.; Tekinay, A.B.; Guler, M.O. Self-Assembled Peptide Amphiphile Nanofibers and PEG Composite Hydrogels as Tunable ECM Mimetic Microenvironment. Biomacromolecules 2015, 16, 1247–1258. [Google Scholar] [CrossRef]
- Bregenzer, M.E.; Horst, E.N.; Mehta, P.; Novak, C.M.; Raghavan, S.; Snyder, C.S.; Mehta, G. Integrated cancer tissue engineering models for precision medicine. PLoS ONE 2019, 14, e0216564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paradiso, F.; Serpelloni, S.; Francis, L.W.; Taraballi, F. Mechanical Studies of the Third Dimension in Cancer: From 2D to 3D Model. Int. J. Mol. Sci. 2021, 22, 10098. [Google Scholar] [CrossRef] [PubMed]
- Guimarães, C.F.; Gasperini, L.; Marques, A.P.; Reis, R.L. The stiffness of living tissues and its implications for tissue engineering. Nat. Rev. Mater. 2020, 5, 351–370. [Google Scholar] [CrossRef]
- Chaudhuri, O.; Cooper-White, J.; Janmey, P.A.; Mooney, D.J.; Shenoy, V.B. Effects of extracellular matrix viscoelasticity on cellular behaviour. Nat. Cell Biol. 2020, 584, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Dolega, M.E.; Delarue, M.; Ingremeau, F.; Prost, J.; Delon, A.; Cappello, G. Cell-like pressure sensors reveal increase of mechanical stress towards the core of multicellular spheroids under compression. Nat. Commun. 2017, 8, 14056. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.; Kalashnikov, N.; Mok, S.; Halaoui, R.; Kuzmin, E.; Putnam, A.J.; Takayama, S.; Park, M.; McCaffrey, L.; Zhao, R.; et al. Dispersible hydrogel force sensors reveal patterns of solid mechanical stress in multicellular spheroid cultures. Nat. Commun. 2019, 10, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, I.L.; Ahmad, M.R. Trends in characterizing single cell’s stiffness properties. Micro Nano Syst. Lett. 2014, 2, 8. [Google Scholar] [CrossRef] [Green Version]
- Taubenberger, A.V.; Girardo, S.; Träber, N.; Fischer-Friedrich, E.; Kräter, M.; Wagner, K.; Kurth, T.; Richter, I.; Haller, B.; Binner, M.; et al. 3D Microenvironment Stiffness Regulates Tumor Spheroid Growth and Mechanics via p21 and ROCK. Adv. Biosyst. 2019, 3, e1900128. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Mezencev, R.; Kim, B.; Wang, L.; McDonald, J.F.; Sulchek, T. Cell Stiffness Is a Biomarker of the Metastatic Potential of Ovarian Cancer Cells. PLoS ONE 2012, 7, e46609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, T.; Anvari, B. Characterization of the Viscoelastic Properties of Ovarian Cancer Cells Membranes by Optical Tweezers and Quantitative Phase Imaging. Front. Phys. 2020, 8, 1–10. [Google Scholar] [CrossRef]
- Conrad, C.; Gray, K.M.; Stroka, K.M.; Rizvi, I.; Scarcelli, G. Mechanical Characterization of 3D Ovarian Cancer Nodules Using Brillouin Confocal Microscopy. Cell. Mol. Bioeng. 2019, 12, 215–226. [Google Scholar] [CrossRef]
- Mahajan, V.; Beck, T.; Gregorczyk, P.; Ruland, A.; Alberti, S.; Guck, J.; Werner, C.; Schlüßler, R.; Taubenberger, A.V. Mapping Tumor Spheroid Mechanics in Dependence of 3D Microenvironment Stiffness and Degradability by Brillouin Microscopy. Cancers 2021, 13, 5549. [Google Scholar] [CrossRef] [PubMed]
- Martikainen, L.; Bertula, K.; Turunen, M.; Ikkala, O. Strain Stiffening and Negative Normal Force of Agarose Hydrogels. Macromolecules 2020, 53, 9983–9992. [Google Scholar] [CrossRef]
- Kalli, M.; Stylianopoulos, T. Defining the Role of Solid Stress and Matrix Stiffness in Cancer Cell Proliferation and Metastasis. Front. Oncol. 2018, 8, 55. [Google Scholar] [CrossRef]
- Raphael, B.; Khalil, T.; Workman, V.; Smith, A.; Brown, C.; Streuli, C.; Saiani, A.; Domingos, M. 3D cell bioprinting of self-assembling peptide-based hydrogels. Mater. Lett. 2017, 190, 103–106. [Google Scholar] [CrossRef] [Green Version]





| Model | Characteristics and Advantages | Disadvantages and Limitations | Applications in Cancer Research | Cell Types | Refs. |
|---|---|---|---|---|---|
| Mouse models: | Captures in vivo complexity | Ethical concerns | - | - | - |
| Costly | |||||
| Time-consuming | |||||
| Special facilities required for housing | |||||
| Requires licenses | |||||
| Murine biology and stroma different from human TME | |||||
| Xenografts | Cell lines or patient-derived | Low success rate | Analysis of cancer development and heterogeneity of tumors | HO-8910PM, from patient-derived tissue and ascites | [57,59,61,62,63,64] |
| Resemble tumor histology, formation of ascites, gene expression, vasculature, metastatic potential and response to chemotherapy | Possibility of leakage of cancer cells after injection | ||||
| Establishment of tumor biobanks. | Possible downregulation of certain genes and replacement of human stroma by murine stroma | Evaluation of tumor responses to drugs. | |||
| Resemble patient heterogeneity | Immunodeficient host | Used in parallel with 3D in vitro studies | |||
| Syngeneic | Immunocompetent model | Lack of heterogeneity. Few host strains | Evaluate tumor growth. Model metastasis in peritoneal cavity Study anoikis resistance | ID8 | [68,69] |
| Rapid growth | |||||
| Easily manipulated | |||||
| Induce metastasis with ascites formation | |||||
| Recapitulate anoikis resistance | |||||
| Genetically engineered | Display genetic heterogeneity | Longer time for tumor development. Lack of promoters to develop these models | Model metastasis and cancer progression Study mutation combinations | - | [72,73,74,75] |
| Resemble tumor histology | |||||
| Genetically manipulated. | |||||
| Laying hen | Display pathological and genetical features similar to patient tumors | Ethical concerns. | Study cancer origin | - | [78,79] |
| Lack of native TME | |||||
| Similar developmental pattern to human tumors | Lack of technology-specific for host (e.g., antibodies) | ||||
| High incidence of disease | Lack of protocols | ||||
| Spheroids | Resemble cell aggregates found in ascites | Require inclusion of vasculature, immune system components, mechanical signals and fluid dynamics | Study spheroid formation mechanisms. | Ascites-derived cells, SKOV-3, OV-90, OVCAR-3, OVCAR-8, TOV-112, TOV-21, TOV-155 | [81,82,83,84,85,86,87,88,89,90,91,92] |
| Support different ratios of cancer and stromal cells | |||||
| Mimic nutrient transport, growth kinetics and cell–cell interactions found in solid tumors | Difficulty to image them | Evaluate tumor invasion. | |||
| Diverse spheroid production techniques | Not all cell lines are capable of forming spheroids | ||||
| Resemble chemoresistance | Different morphology depending on protocol used | Testing of drug delivery systems, drug efficacy and penetration, receptor targeting, cell recruitment abilities and tumor biology. | |||
| Low cost, ease of use, reproducible, and high-throughput | Lack of native ECM | ||||
| Organoids | Maintain histological features | Lack of immune system elements, stromal cells and vasculature. | Study carcinogenesis High-throughput drug screening Genomic analysis | Patient-derived tissue fragments, ascites-derived cells | [93,94,95,96,97,98,99,100,101] |
| Mimic genetic features including intra-tumoral | |||||
| High-throughput screening | Costly. | ||||
| Derived from small pieces of tissue | Require supplemental growth factors | ||||
| Can be genetically modified | Intra-tumoral heterogeneity can be lost during passages | ||||
| Creation of biobanks | Mutations are subsequently acquired | ||||
| Maintain cell viability over long periods of time | Need of culture protocols and drug screening strategies | ||||
| Microfluidic devices | Commercially available or custom-made devices | Costly | Study tumor development | A2780, TOV112D, OV90, OVCAR5, SKOV-3, ascites-derived cells | [83,102,103,104,105,106,107] |
| Include multiple chambers and cell populations | |||||
| Enable fluid perfusion | Special facilities required for manufacture | Resemble cancer dissemination and metastasis | |||
| Enable formation of spheroids | Predesigned devices cannot be customized | ||||
| Some platforms enable testing pharmcokinetics/dynamics of drugs | Limited recollection of spheroids | Drug screening | |||
| Variable shear stress | Complex design and use | ||||
| Include nutrient supply and waste removal | Limited material choice | Genomic analysis | |||
| Maintain cell viability over long periods of time | Lack of cell–cell and cell–matrix interactions | ||||
| Natural hydrogels: Matrigel | Contains collagen, laminin, enactin, other ECM molecules and growth factors | Chemically not well-defined | Study tumor biology | SKOV-3, OVCAR-10 | [108] |
| Cyto-compatible | High batch-to-batch variation | ||||
| Minimally processed | Undefined impurities | ||||
| Mimics in vivo conditions | Limited flexibility to tune the mechanical properties | ||||
| Enables cell–matrix interactions | Quick gelation time | ||||
| Promotes cell growth | Contains growth factors that can cause activation of signaling cascades | ||||
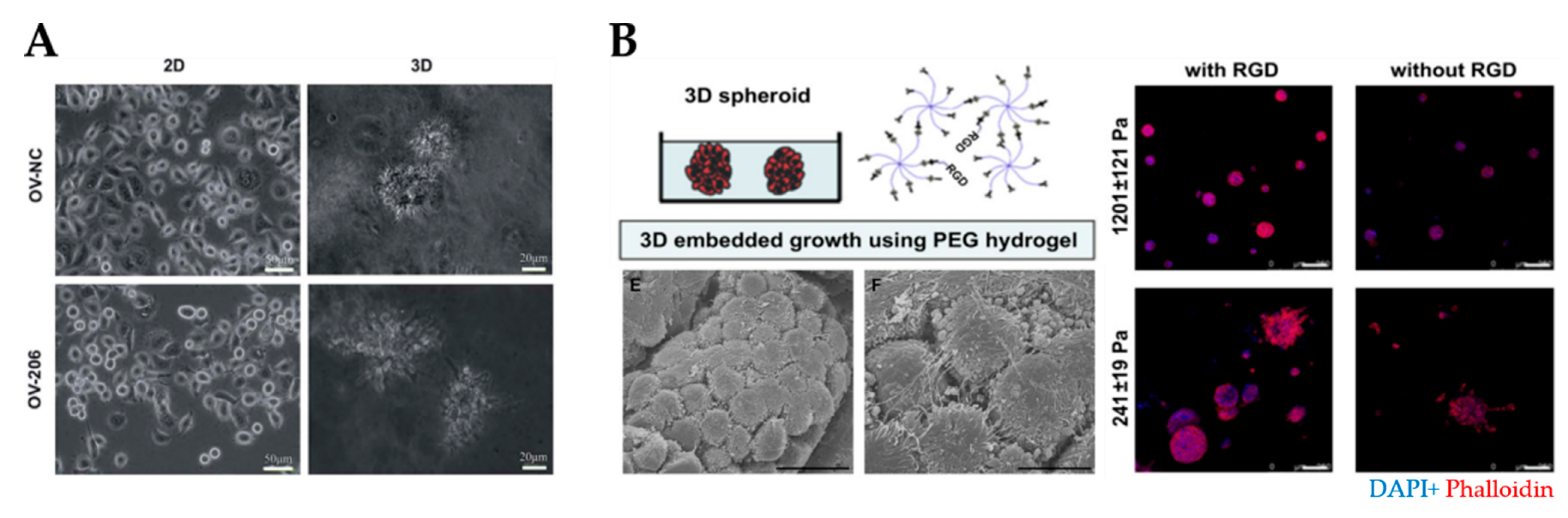
| Collagen | Primary constituent of ECM | Batch-to-batch variation | Study tumor biology Evaluate tumor invasion | A2780, OV-NC, OV-206, SKOV-3, OVCAR-3, OvCa433, DOV13, OVSAHO | [38,109,110,111,112] |
| Intrinsic cues for cell recognition | |||||
| Similar stiffness to tissues | Limited control over physical and mechanical properties | ||||
| Maintains cell viability over long periods of time | Inability to tailor its composition | ||||
| Enhances cell spheroid and invasion | TME contains different types of collagen and other ECM molecules, not only collagen of a single type | ||||
| Stimulates EMT phenotype | Low mechanical strength | ||||
| Synthetic polymer hydrogels (e.g., PEG, GelMA) | Biocompatible | Require cell-binding moieties due to inert nature | Study influence of matrix stiffness on spheroid formation and disease progression | OV-MZ-6, SKOV-3, HO8910, ascites-derived cells. | [113,114,115,116,117,118,119,120,121,122] |
| Tunable architecture and stiffness | |||||
| Tailorable with functional ligands | Limited cell recovery | ||||
| Functionalized with ECM proteins or proteolytic degradation sites | Drug screening | ||||
| Enable spheroid formation | Lack of nanofibrous network | Genomic analysis | |||
| Maintain cell viability over long periods of time | Spheroid formation technique | ||||
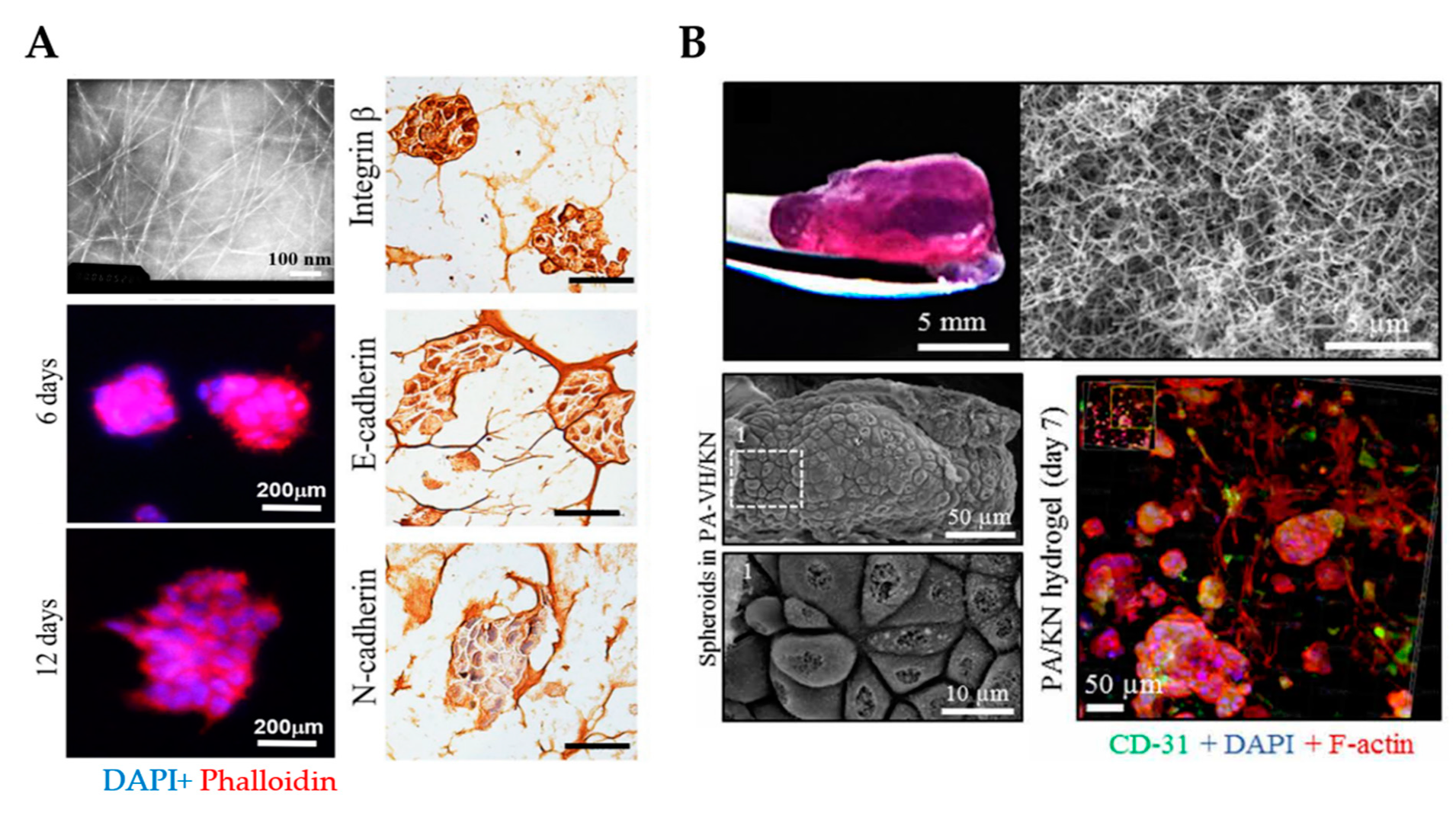
| Self-assembling peptide hydrogels | Chemically synthesized to enable tunability of properties | Costly | - | - | - |
| High design flexibility | |||||
| Reproducible | |||||
| Stable nanofiber network that resembles the ECM | |||||
| Supportive of cell proliferation, invasion and spheroid formation | |||||
| PuraMatrix™ | Commercially available Low immunogenicity | Poor mechanical strength | Model tumorigenesis and metastasis. | SKOV-3, A2780, A2780/DDP, OVCAR-5. | [123,124,125,126]. |
| Study influence of matrix stiffness on spheroid formation and disease progression. | |||||
| Drug screening. | |||||
| Peptide amphiphiles | Available through custom peptide synthesis. | Low scalability | Study tumor biology | NIH:OVCAR-4. | [127] |
| Tailorable with specific signaling motifs | |||||
| Incorporation of ECM proteins | Evaluate influence of matrix stiffness on spheroid formation and disease progression | ||||
| Maintains cell viability over long periods of time | Peptide sequences not normally found in the ECM | ||||
| Supports co-cultures | Drug screening | ||||
| Minimal batch-to batch-variation |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mendoza-Martinez, A.K.; Loessner, D.; Mata, A.; Azevedo, H.S. Modeling the Tumor Microenvironment of Ovarian Cancer: The Application of Self-Assembling Biomaterials. Cancers 2021, 13, 5745. https://doi.org/10.3390/cancers13225745
Mendoza-Martinez AK, Loessner D, Mata A, Azevedo HS. Modeling the Tumor Microenvironment of Ovarian Cancer: The Application of Self-Assembling Biomaterials. Cancers. 2021; 13(22):5745. https://doi.org/10.3390/cancers13225745
Chicago/Turabian StyleMendoza-Martinez, Ana Karen, Daniela Loessner, Alvaro Mata, and Helena S. Azevedo. 2021. "Modeling the Tumor Microenvironment of Ovarian Cancer: The Application of Self-Assembling Biomaterials" Cancers 13, no. 22: 5745. https://doi.org/10.3390/cancers13225745
APA StyleMendoza-Martinez, A. K., Loessner, D., Mata, A., & Azevedo, H. S. (2021). Modeling the Tumor Microenvironment of Ovarian Cancer: The Application of Self-Assembling Biomaterials. Cancers, 13(22), 5745. https://doi.org/10.3390/cancers13225745








