An Immunohistochemical Evaluation of Tumor-Associated Glycans and Mucins as Targets for Molecular Imaging of Pancreatic Ductal Adenocarcinoma
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
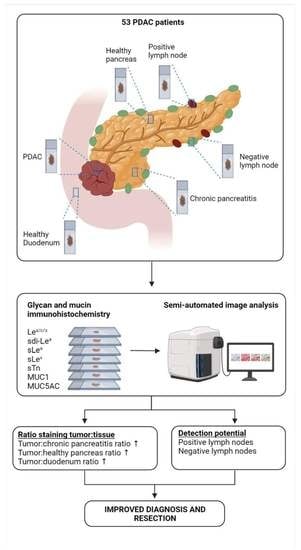
2.1. Patient and Tissue Selection
2.2. Monoclonal Antibodies and Reagents
2.3. Immunohistochemistry (IHC)
2.4. Semi-Automated Imaging Analysis
2.5. Statistical Analysis
3. Results
3.1. Patient Characteristics
3.2. Object Classifier Training and Validation
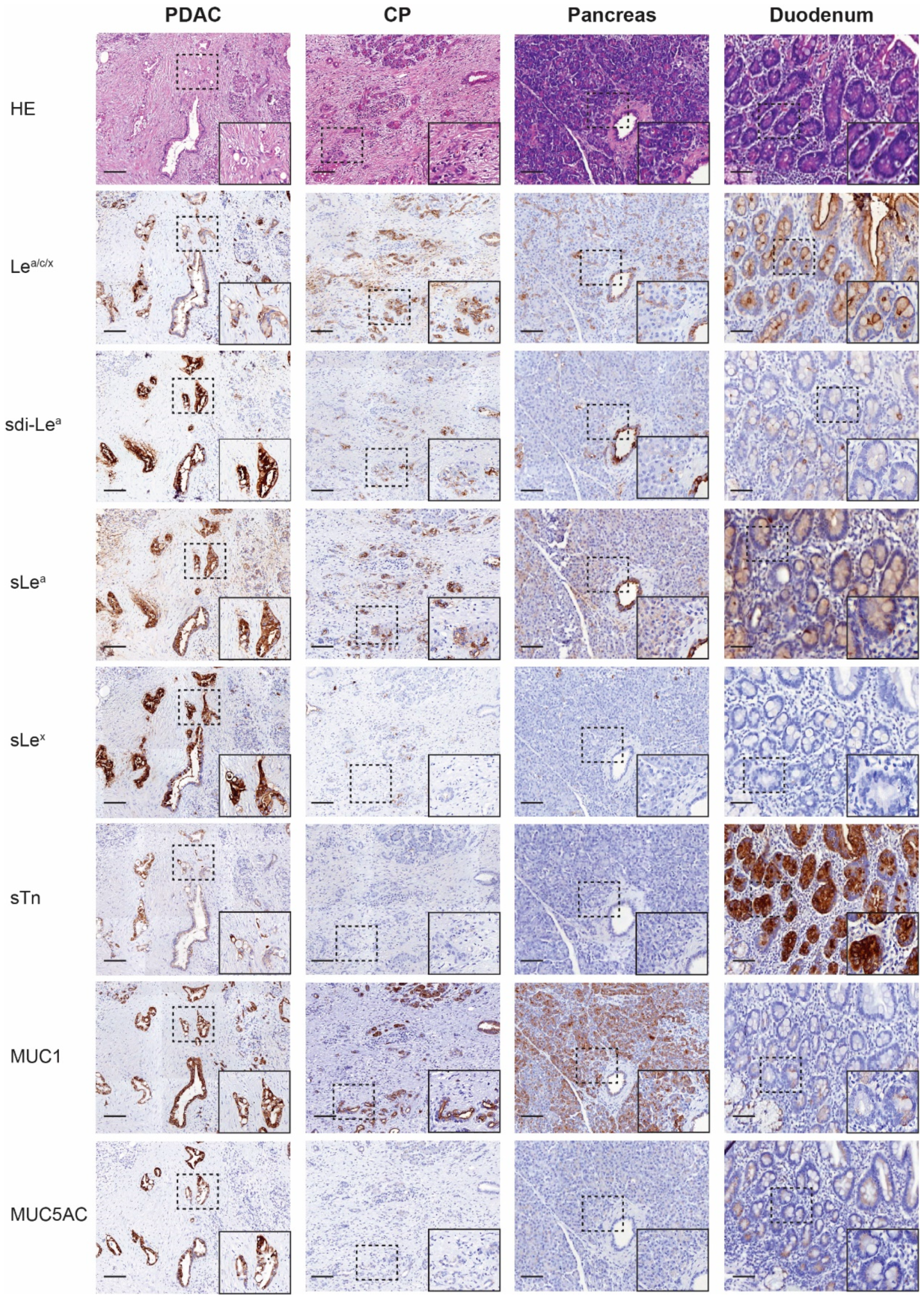
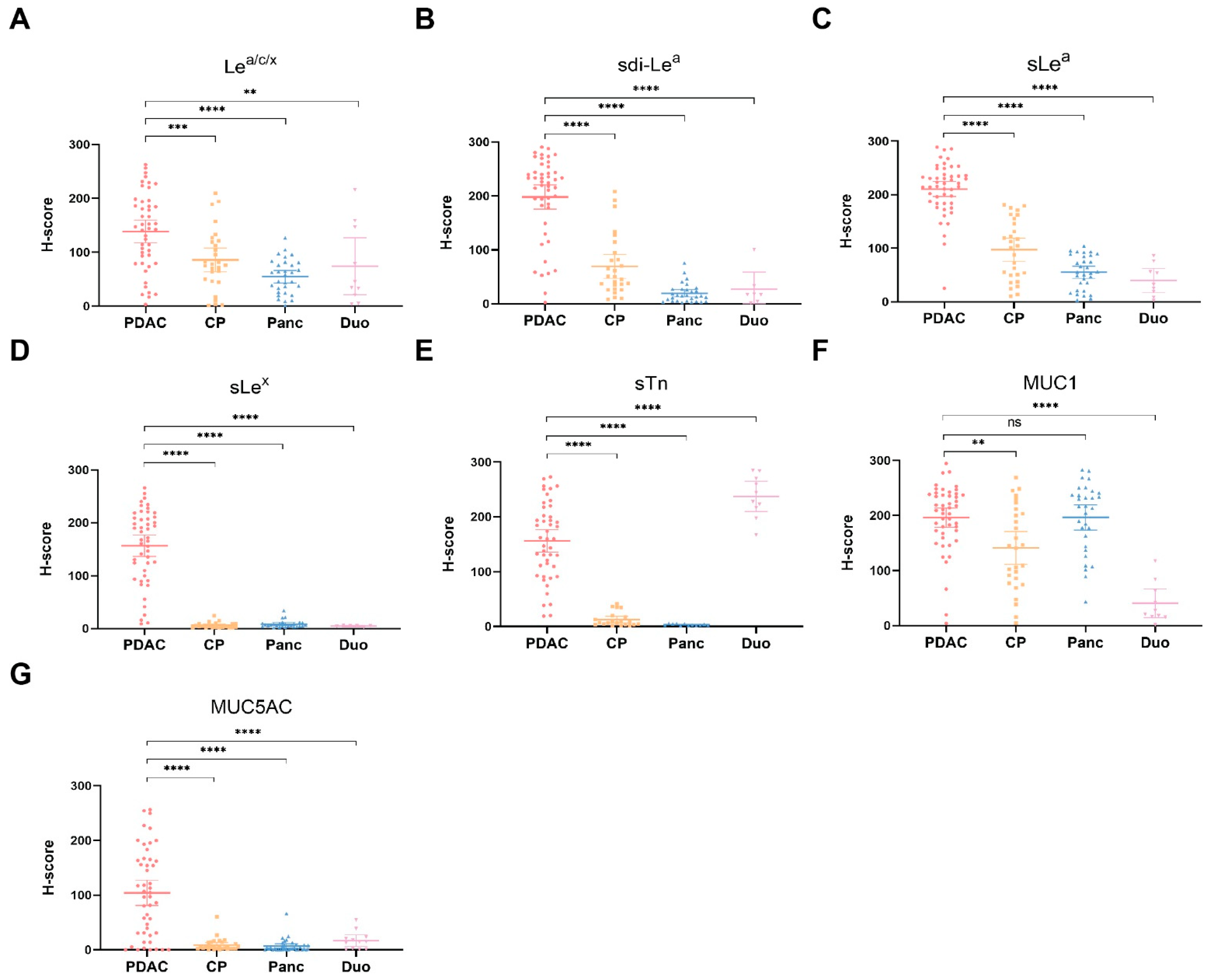
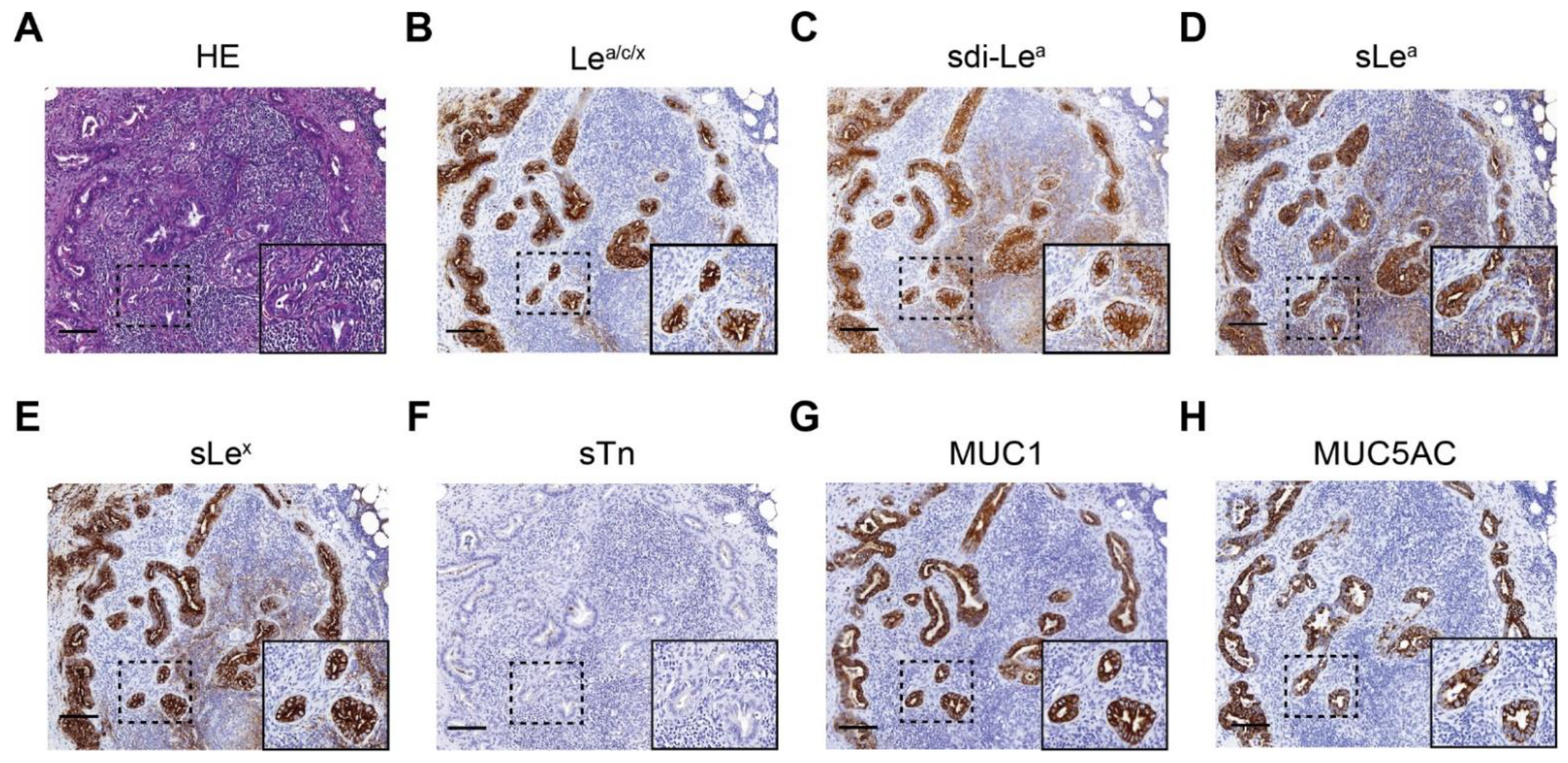
3.3. Biomarker Expression on PDAC, CP, Healthy Pancreatic and Duodenal Tissues
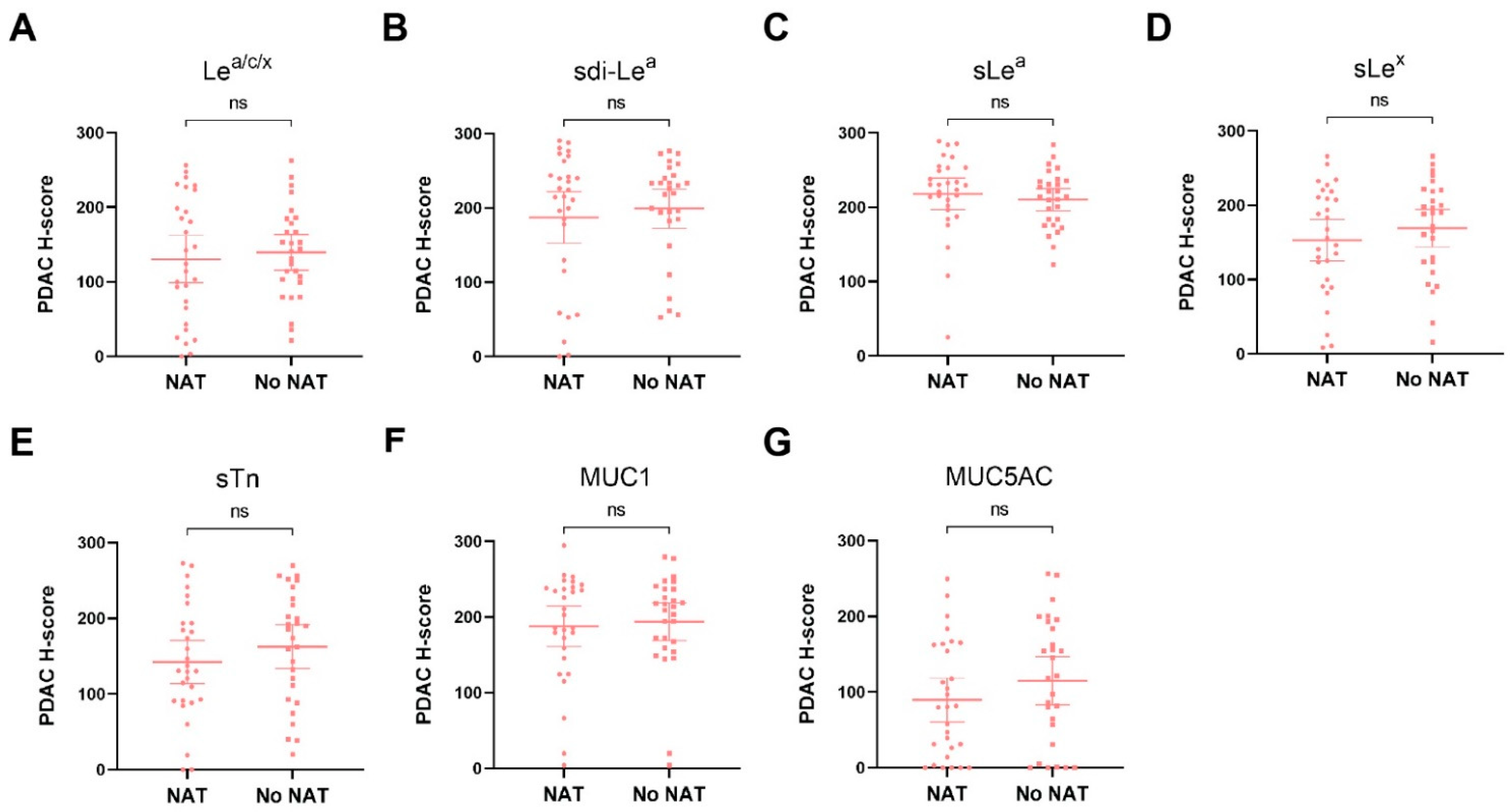
3.4. Biomarker Expression on PDAC Tissues after NAT
3.5. Biomarker Co-Expression on PDAC Tissues
3.6. Detection of Lymph Node Metastases
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Rawla, P.; Sunkara, T.; Gaduputi, V. Epidemiology of pancreatic cancer: Global trends, etiology and risk factors. World J. Oncol. 2019, 10, 10–27. [Google Scholar] [CrossRef] [PubMed]
- Versteijne, E.; Vogel, J.A.; Besselink, M.G.; Busch, O.R.C.; Wilmink, J.W.; Daams, J.G.; van Eijck, C.H.J.; Groot Koerkamp, B.; Rasch, C.R.N.; van Tienhoven, G.; et al. Meta-analysis comparing upfront surgery with neoadjuvant treatment in patients with resectable or borderline resectable pancreatic cancer. Br. J. Surg. 2018, 105, 946–958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papalezova, K.T.; Tyler, D.S.; Blazer, D.G., 3rd; Clary, B.M.; Czito, B.G.; Hurwitz, H.I.; Uronis, H.E.; Pappas, T.N.; Willett, C.G.; White, R.R. Does preoperative therapy optimize outcomes in patients with resectable pancreatic cancer? J. Surg. Oncol. 2012, 106, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Pietryga, J.A.; Morgan, D.E. Imaging preoperatively for pancreatic adenocarcinoma. J. Gastrointest. Oncol. 2015, 6, 343–357. [Google Scholar] [CrossRef]
- Tummers, W.S.; Groen, J.V.; Sibinga Mulder, B.G.; Farina-Sarasqueta, A.; Morreau, J.; Putter, H.; van de Velde, C.J.; Vahrmeijer, A.L.; Bonsing, B.A.; Mieog, J.S.; et al. Impact of resection margin status on recurrence and survival in pancreatic cancer surgery. Br. J. Surg. 2019, 106, 1055–1065. [Google Scholar] [CrossRef] [Green Version]
- Strobel, O.; Hank, T.; Hinz, U.; Bergmann, F.; Schneider, L.; Springfeld, C.; Jäger, D.; Schirmacher, P.; Hackert, T.; Büchler, M.W. Pancreatic cancer surgery: The new R-status counts. Ann. Surg. 2017, 265, 565–573. [Google Scholar] [CrossRef]
- Gerritsen, A.; Molenaar, I.Q.; Bollen, T.L.; Nio, C.Y.; Dijkgraaf, M.G.; van Santvoort, H.C.; Offerhaus, G.J.; Brosens, L.A.; Biermann, K.; Sieders, E.; et al. Preoperative characteristics of patients with presumed pancreatic cancer but ultimately benign disease: A multicenter series of 344 pancreatoduodenectomies. Ann. Surg. Oncol. 2014, 21, 3999–4006. [Google Scholar] [CrossRef]
- Pandol, S.; Edderkaoui, M.; Gukovsky, I.; Lugea, A.; Gukovskaya, A. Desmoplasia of pancreatic ductal adenocarcinoma. Clin. Gastroenterol. Hepatol. 2009, 7, S44–S47. [Google Scholar] [CrossRef] [Green Version]
- Chatterjee, D.; Katz, M.H.; Rashid, A.; Estrella, J.S.; Wang, H.; Varadhachary, G.R.; Wolff, R.A.; Lee, J.E.; Pisters, P.W.; Abbruzzese, J.L.; et al. Pancreatic intraepithelial neoplasia and histological changes in non-neoplastic pancreas associated with neoadjuvant therapy in patients with pancreatic ductal adenocarcinoma. Histopathology 2013, 63, 841–851. [Google Scholar] [CrossRef] [Green Version]
- Vahrmeijer, A.L.; Hutteman, M.; van der Vorst, J.R.; van de Velde, C.J.H.; Frangioni, J.V. Image-guided cancer surgery using near-infrared fluorescence. Nat. Rev. Clin. Oncol. 2013, 10, 507–518. [Google Scholar] [CrossRef] [Green Version]
- Houvast, R.D.; Vankemmelbeke, M.; Durrant, L.G.; Wuhrer, M.; Baart, V.M.; Kuppen, P.J.K.; de Geus-Oei, L.F.; Vahrmeijer, A.L.; Sier, C.F.M. Targeting glycans and heavily glycosylated proteins for tumor imaging. Cancers 2020, 12, 3870. [Google Scholar] [CrossRef]
- Fuster, M.M.; Esko, J.D. The sweet and sour of cancer: Glycans as novel therapeutic targets. Nat. Rev. Cancer 2005, 5, 526–542. [Google Scholar] [CrossRef]
- Chua, J.X.; Vankemmelbeke, M.; McIntosh, R.S.; Clarke, P.A.; Moss, R.; Parsons, T.; Spendlove, I.; Zaitoun, A.M.; Madhusudan, S.; Durrant, L.G. Monoclonal antibodies targeting LecLex-related Glycans with potent antitumor activity. Clin. Cancer Res. 2015, 21, 2963–2974. [Google Scholar] [CrossRef] [Green Version]
- Dube, D.H.; Bertozzi, C.R. Glycans in cancer and inflammation—Potential for therapeutics and diagnostics. Nat. Rev. Drug Discov. 2005, 4, 477–488. [Google Scholar] [CrossRef]
- Tivadar, S.T.; McIntosh, R.S.; Chua, J.X.; Moss, R.; Parsons, T.; Zaitoun, A.M.; Madhusudan, S.; Durrant, L.G.; Vankemmelbeke, M. Monoclonal antibody targeting Sialyl-di-Lewis(a)-containing internalizing and noninternalizing glycoproteins with cancer immunotherapy development potential. Mol. Cancer Ther. 2020, 19, 790–801. [Google Scholar] [CrossRef] [Green Version]
- Trinchera, M.; Aronica, A.; Dall’Olio, F. Selectin ligands Sialyl-Lewis a and Sialyl-Lewis x in gastrointestinal cancers. Biology 2017, 6, 16. [Google Scholar] [CrossRef]
- Contessa, J.N.; Bhojani, M.S.; Freeze, H.H.; Rehemtulla, A.; Lawrence, T.S. Inhibition of N-linked glycosylation disrupts receptor tyrosine kinase signaling in tumor cells. Cancer Res. 2008, 68, 3803–3809. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; You, L.; Dai, M.; Zhao, Y. Mucins in pancreatic cancer: A well-established but promising family for diagnosis, prognosis and therapy. J. Cell. Mol. Med. 2020, 24, 10279–10289. [Google Scholar] [CrossRef]
- Kaur, S.; Kumar, S.; Momi, N.; Sasson, A.R.; Batra, S.K. Mucins in pancreatic cancer and its microenvironment. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 607–620. [Google Scholar] [CrossRef] [Green Version]
- Lebrilla, C.B.; An, H.J. The prospects of glycan biomarkers for the diagnosis of diseases. Mol. BioSyst. 2009, 5, 17–20. [Google Scholar] [CrossRef]
- Bankhead, P.; Loughrey, M.B.; Fernández, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.; et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017, 7, 16878. [Google Scholar] [CrossRef] [Green Version]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Thike, A.A.; Chng, M.J.; Fook-Chong, S.; Tan, P.H. Immunohistochemical expression of hormone receptors in invasive breast carcinoma: Correlation of results of H-score with pathological parameters. Pathology 2001, 33, 21–25. [Google Scholar] [CrossRef]
- Janssen, Q.P.; van Dam, J.L.; Bonsing, B.A.; Bos, H.; Bosscha, K.P.; Coene, P.; van Eijck, C.H.J.; de Hingh, I.; Karsten, T.M.; van der Kolk, M.B.; et al. Total neoadjuvant FOLFIRINOX versus neoadjuvant gemcitabine-based chemoradiotherapy and adjuvant gemcitabine for resectable and borderline resectable pancreatic cancer (PREOPANC-2 trial): Study protocol for a nationwide multicenter randomized controlled trial. BMC Cancer 2021, 21, 300. [Google Scholar] [CrossRef]
- Park, H.S.; Lee, J.M.; Choi, H.K.; Hong, S.H.; Han, J.K.; Choi, B.I. Preoperative evaluation of pancreatic cancer: Comparison of gadolinium-enhanced dynamic MRI with MR cholangiopancreatography versus MDCT. J. Magn. Reson. Imaging 2009, 30, 586–595. [Google Scholar] [CrossRef]
- Arvold, N.D.; Niemierko, A.; Mamon, H.J.; Fernandez-del Castillo, C.; Hong, T.S. Pancreatic cancer tumor size on CT scan versus pathologic specimen: Implications for radiation treatment planning. Int. J. Radiat. Oncol. Biol. Phys. 2011, 80, 1383–1390. [Google Scholar] [CrossRef] [Green Version]
- Tummers, W.S.; Miller, S.E.; Teraphongphom, N.T.; Gomez, A.; Steinberg, I.; Huland, D.M.; Hong, S.; Kothapalli, S.R.; Hasan, A.; Ertsey, R.; et al. Intraoperative pancreatic cancer detection using tumor-specific multimodality molecular imaging. Ann. Surg. Oncol. 2018, 25, 1880–1888. [Google Scholar] [CrossRef]
- Vuijk, F.A.; Hilling, D.E.; Mieog, J.S.D.; Vahrmeijer, A.L. Fluorescent-guided surgery for sentinel lymph node detection in gastric cancer and carcinoembryonic antigen targeted fluorescent-guided surgery in colorectal and pancreatic cancer. J. Surg. Oncol. 2018, 118, 315–323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tummers, W.S.; Farina-Sarasqueta, A.; Boonstra, M.C.; Prevoo, H.A.; Sier, C.F.; Mieog, J.S.; Morreau, J.; van Eijck, C.H.; Kuppen, P.J.; van de Velde, C.J.; et al. Selection of optimal molecular targets for tumor-specific imaging in pancreatic ductal adenocarcinoma. Oncotarget 2017, 8, 56816–56828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lohrmann, C.; O’Reilly, E.M.; O’Donoghue, J.A.; Pandit-Taskar, N.; Carrasquillo, J.A.; Lyashchenko, S.K.; Ruan, S.; Teng, R.; Scholz, W.; Maffuid, P.W.; et al. Retooling a blood-based biomarker: Phase I assessment of the high-affinity CA19-9 antibody HuMab-5B1 for immuno-PET imaging of pancreatic cancer. Clin. Cancer Res. 2019, 25, 7014–7023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ballehaninna, U.K.; Chamberlain, R.S. Serum CA 19-9 as a biomarker for pancreatic cancer—A comprehensive review. Indian J. Surg. Oncol. 2011, 2, 88–100. [Google Scholar] [CrossRef] [Green Version]
- Houvast, R.D.; Baart, V.M.; Bhairosingh, S.S.; Cordfunke, R.A.; Chua, J.X.; Vankemmelbeke, M.; Parsons, T.; Kuppen, P.J.K.; Durrant, L.G.; Vahrmeijer, A.L.; et al. Glycan-based near-infrared fluorescent (NIRF) imaging of gastrointestinal tumors: A preclinical proof-of-concept in vivo study. Mol. Imaging Biol. 2020, 22, 1511–1522. [Google Scholar] [CrossRef]
- Gold, D.V.; Lew, K.; Maliniak, R.; Hernandez, M.; Cardillo, T. Characterization of monoclonal antibody PAM4 reactive with a pancreatic cancer mucin. Int. J. Cancer 1994, 57, 204–210. [Google Scholar] [CrossRef]
- Fan, X.N.; Karsten, U.; Goletz, S.; Cao, Y. Reactivity of a humanized antibody (hPankoMab) towards a tumor-related MUC1 epitope (TA-MUC1) with various human carcinomas. Pathol. Res. Pract. 2010, 206, 585–589. [Google Scholar] [CrossRef]
- Gold, D.V.; Karanjawala, Z.; Modrak, D.E.; Goldenberg, D.M.; Hruban, R.H. PAM4-reactive MUC1 is a biomarker for early pancreatic adenocarcinoma. Clin. Cancer Res. 2007, 13, 7380–7387. [Google Scholar] [CrossRef] [Green Version]
- Shi, C.; Merchant, N.; Newsome, G.; Goldenberg, D.M.; Gold, D.V. Differentiation of pancreatic ductal adenocarcinoma from chronic pancreatitis by PAM4 immunohistochemistry. Arch. Pathol. Lab. Med. 2014, 138, 220–228. [Google Scholar] [CrossRef] [Green Version]
- Loughrey, M.B.; Bankhead, P.; Coleman, H.G.; Hagan, R.S.; Craig, S.; McCorry, A.M.B.; Gray, R.T.; McQuaid, S.; Dunne, P.D.; Hamilton, P.W.; et al. Validation of the systematic scoring of immunohistochemically stained tumour tissue microarrays using QuPath digital image analysis. Histopathology 2018, 73, 327–338. [Google Scholar] [CrossRef] [Green Version]
- Adams, E.J.; Green, J.A.; Clark, A.H.; Youngson, J.H. Comparison of different scoring systems for immunohistochemical staining. J. Clin. Pathol. 1999, 52, 75–77. [Google Scholar] [CrossRef] [Green Version]
- Feuchtinger, A.; Stiehler, T.; Jütting, U.; Marjanovic, G.; Luber, B.; Langer, R.; Walch, A. Image analysis of immunohistochemistry is superior to visual scoring as shown for patient outcome of esophageal adenocarcinoma. Histochem. Cell Biol. 2015, 143, 1–9. [Google Scholar] [CrossRef]
- Bankhead, P.; Fernández, J.A.; McArt, D.G.; Boyle, D.P.; Li, G.; Loughrey, M.B.; Irwin, G.W.; Harkin, D.P.; James, J.A.; McQuaid, S.; et al. Integrated tumor identification and automated scoring minimizes pathologist involvement and provides new insights to key biomarkers in breast cancer. Lab. Investig. 2018, 98, 15–26. [Google Scholar] [CrossRef] [Green Version]

| Characteristic | Total PDAC (n = 53) | NAT (n = 22) | No NAT (n = 31) | p-Value | CP (n = 9) |
|---|---|---|---|---|---|
| Age, years, mean (SD) | 64.7 (9.8) | 61.3 (9.1) | 67.1 (9.7) | 0.033 | 53.5 (10.9) |
| Gender, n (%) | |||||
| Male | 26 (49) | 9 (41) | 17 (55) | 0.406 | 8 (89) |
| Female | 27 (51) | 13 (59) | 14 (45) | 1 (11) | |
| Surgery type, n (%) | |||||
| Pancreaticoduodenectomy | 41 (77) | 16 (73) | 25 (81) | 0.632 | 4 (44) |
| Pancreatic corpus/tail resection | 9 (17) | 4 (18) | 5 (16) | 5 (56) | |
| Total pancreatectomy | 3 (6) | 2 (9) | 1 (3) | 0 (0) | |
| Tumor differentiation, n (%) | |||||
| Good | 6 (11) | 1 (5) | 5 (16) | 0.607 | - |
| Moderate | 12 (23) | 1 (5) | 11 (36) | - | |
| Poor | 18 (34) | 4 (18) | 14 (45) | - | |
| Missing | 17 (32) | 16 (73) | 1 (3) | - | |
| Primary tumor, n (%) | |||||
| pT1 | 18 (34) | 10 (46) | 8 (26) | 0.275 | - |
| pT2 | 27 (51) | 10 (46) | 17 (55) | - | |
| pT3 | 8 (15) | 2 (9) | 6 (19) | - | |
| Regional lymph nodes, n (%) | |||||
| pN0 | 18 (34) | 13 (59) | 5 (16) | <0.001 | - |
| pN1 | 21 (40) | 9 (41) | 12 (39) | - | |
| pN2 | 14 (26) | 0 (0) | 14 (45) | - | |
| Surgical margin status, n (%) | |||||
| R0 | 29 (55) | 15 (68) | 14 (45) | 0.161 | - |
| R1 | 24 (45) | 7 (32) | 17 (55) | - | |
| NAT, n (%) | |||||
| No | 31 (59) | 0 (0) | 31 (100) | - | 8 (89) |
| Yes, chemoradiotherapy | 15 (28) | 15 (68) | 0 (0) | - | 0 (0) |
| Yes, chemotherapy | 7 (13) | 7 (32) | 0 (0) | - | 1 (11) |
| Tumor size, mm, mean (SD) | 26 (13) | 22 (11) | 30 (13) | 0.024 | - |
| Serum CEA, µg/L, median (IQR) | 3.2 (5.9) | 3.2 (6.5) | 3.5 (5.2) | 0.349 | - |
| Serum CA19-9, kU/L, median (IQR) | 74.5 (377.5) | 48.4 (69.7) | 322.8 (371.6) | 0.007 | - |
| Biomarker | PDAC Positive n (%) | Tumor: CP | p-Value | Tumor: Pancreas | p-Value | Tumor: Duodenum | p-Value |
|---|---|---|---|---|---|---|---|
| Lea/c/x | 40 (83) | 1.7 | 0.0010 | 2.5 | <0.0001 | 1.9 | 0.0073 |
| sdi-Lea | 45 (94) | 2.9 | <0.0001 | 10.3 | <0.0001 | 10.0 | <0.0001 |
| sLea | 47 (98) | 2.2 | <0.0001 | 3.8 | <0.0001 | 5.9 | <0.0001 |
| sLex | 43 (90) | 33.2 | <0.0001 | 20.9 | <0.0001 | 53.0 | <0.0001 |
| sTn | 42 (88) | 15.6 | <0.0001 | 100.9 | <0.0001 | 0.6 | <0.0001 |
| MUC1 | 46 (96) | 1.4 | 0.0012 | 1.0 | >0.9999 | 4.8 | <0.0001 |
| MUC5AC | 32 (67) | 11.5 | <0.0001 | 13.6 | <0.0001 | 5.6 | <0.0001 |
| PDAC Expression | ||||
|---|---|---|---|---|
| Biomarker | Negative n (%) | Low n (%) | Moderate n (%) | High n (%) |
| Lea/c/x | 8 (17) | 8 (17) | 23 (48) | 9 (19) |
| sdi-Lea | 3 (6) | 5 (10) | 11 (23) | 29 (60) |
| sLea | 1 (2) | 0 (0) | 15 (31) | 32 (67) |
| sLex | 5 (10) | 7 (15) | 20 (42) | 16 (33) |
| sTn | 6 (13) | 7 (15) | 23 (48) | 12 (25) |
| MUC1 | 2 (4) | 1 (2) | 18 (38) | 27 (56) |
| MUC5AC | 16 (33) | 9 (19) | 17 (35) | 6 (13) |
| Biomarker | Panel | Lea/c/x (%) | sdi-Lea (%) | sLea (%) | sLex (%) | sTn (%) | MUC1 (%) | MUC5AC (%) |
|---|---|---|---|---|---|---|---|---|
| Lea/c/x | ≥1 Both | - | - | - | - | - | - | - |
| sdi-Lea | ≥1 Both | 94 83 | - | - | - | - | - | - |
| sLea | ≥1 Both | 100 81 | 100 92 | - | - | - | - | - |
| sLex | ≥1 Both | 98 75 | 100 83 | 100 88 | - | - | - | - |
| sTn | ≥1 Both | 100 71 | 100 81 | 100 85 | 98 79 | - | - | - |
| MUC1 | ≥1 Both | 100 79 | 100 90 | 100 94 | 100 85 | 100 83 | - | - |
| MUC5AC | ≥1 Both | 96 54 | 96 65 | 98 67 | 96 60 | 90 65 | 96 67 | - |
| Biomarker | Sens. (%) | Spec. (%) | PPV (%) | NPV (%) | Accuracy (%) | AUC (95% CI) | p-Value |
|---|---|---|---|---|---|---|---|
| Lea/c/x | 78 | 98 | 96 | 87 | 90 | 0.929 (0.846–1.000) | <0.0001 |
| sdi-Lea | 70 | 88 | 79 | 82 | 81 | 0.955 (0.896–1.000) | <0.0001 |
| sLea | 78 | 83 | 75 | 85 | 81 | 0.927 (0.858–0.995) | <0.0001 |
| sLex | 59 | 100 | 100 | 79 | 84 | 0.960 (0.913–1.000) | <0.0001 |
| sTn | 52 | 100 | 100 | 76 | 81 | 0.954 (0.894–1.000) | <0.0001 |
| MUC1 | 93 | 100 | 100 | 95 | 97 | 1.000 (1.000–1.000) | <0.0001 |
| MUC5AC | 78 | 100 | 100 | 87 | 91 | 0.972 (0.912–1.000) | <0.0001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Houvast, R.D.; Thijse, K.; Groen, J.V.; Chua, J.; Vankemmelbeke, M.; Durrant, L.G.; Mieog, J.S.D.; Bonsing, B.A.; Vahrmeijer, A.L.; Kuppen, P.J.K.; et al. An Immunohistochemical Evaluation of Tumor-Associated Glycans and Mucins as Targets for Molecular Imaging of Pancreatic Ductal Adenocarcinoma. Cancers 2021, 13, 5777. https://doi.org/10.3390/cancers13225777
Houvast RD, Thijse K, Groen JV, Chua J, Vankemmelbeke M, Durrant LG, Mieog JSD, Bonsing BA, Vahrmeijer AL, Kuppen PJK, et al. An Immunohistochemical Evaluation of Tumor-Associated Glycans and Mucins as Targets for Molecular Imaging of Pancreatic Ductal Adenocarcinoma. Cancers. 2021; 13(22):5777. https://doi.org/10.3390/cancers13225777
Chicago/Turabian StyleHouvast, Ruben D., Kira Thijse, Jesse V. Groen, JiaXin Chua, Mireille Vankemmelbeke, Lindy G. Durrant, J. Sven D. Mieog, Bert A. Bonsing, Alexander L. Vahrmeijer, Peter J. K. Kuppen, and et al. 2021. "An Immunohistochemical Evaluation of Tumor-Associated Glycans and Mucins as Targets for Molecular Imaging of Pancreatic Ductal Adenocarcinoma" Cancers 13, no. 22: 5777. https://doi.org/10.3390/cancers13225777
APA StyleHouvast, R. D., Thijse, K., Groen, J. V., Chua, J., Vankemmelbeke, M., Durrant, L. G., Mieog, J. S. D., Bonsing, B. A., Vahrmeijer, A. L., Kuppen, P. J. K., Crobach, A. S. L. P., & Sier, C. F. M. (2021). An Immunohistochemical Evaluation of Tumor-Associated Glycans and Mucins as Targets for Molecular Imaging of Pancreatic Ductal Adenocarcinoma. Cancers, 13(22), 5777. https://doi.org/10.3390/cancers13225777