HLA Class II Histocompatibility Antigen γ Chain (CD74) Expression Is Associated with Immune Cell Infiltration and Favorable Outcome in Breast Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culturing and Reagents
2.2. CD74 Modulation
2.3. Protein Extraction and Western Blot Analysis
2.4. Clinical Samples and Tissue Microarray (TMA)
2.5. Immunohistochemistry (IHC)
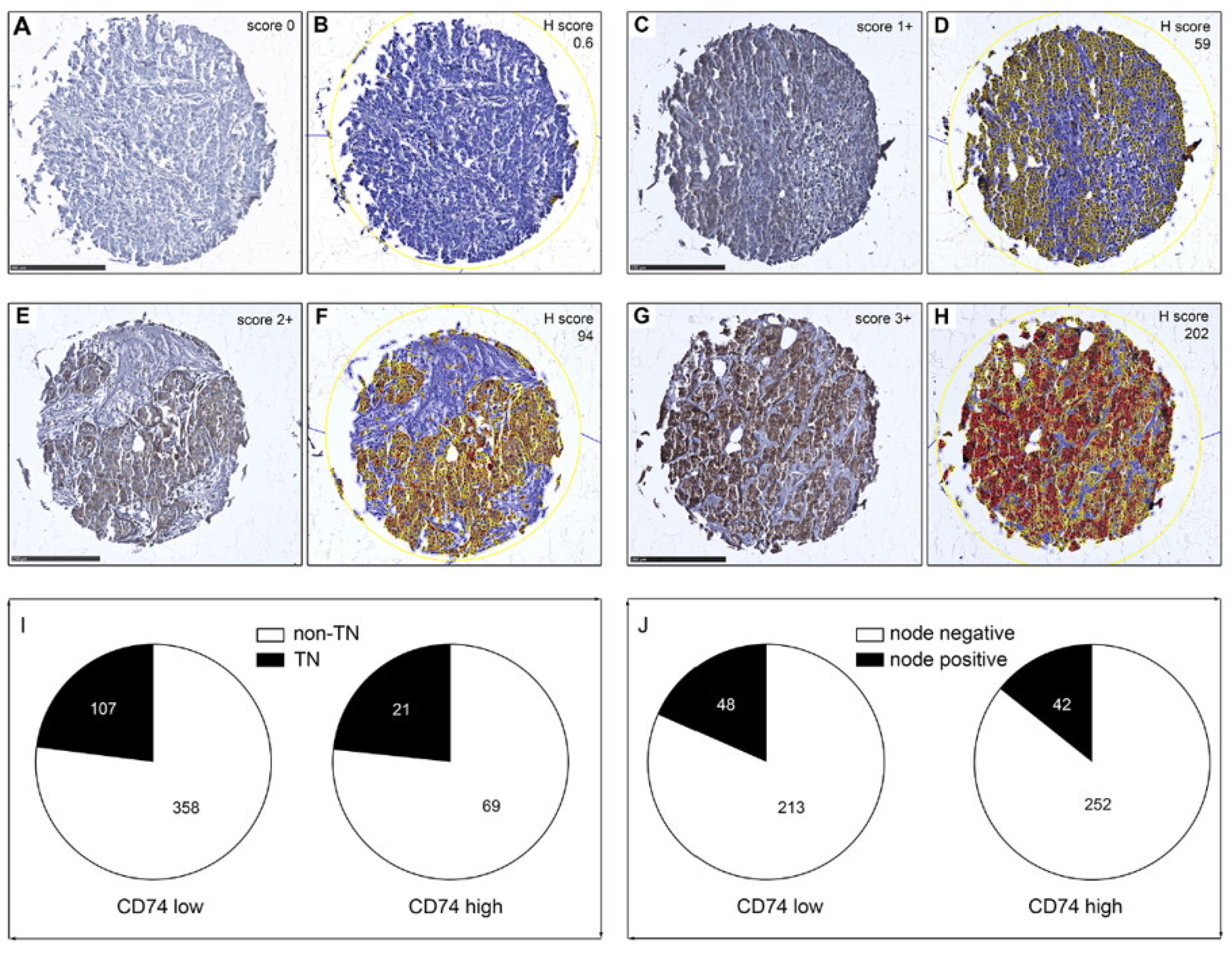
2.6. Quantitative Assessment of IHC Staining
2.7. Data Analysis
2.8. Statistical Analysis
3. Results
3.1. Validation and Use of UMAb231, a New Anti-CD74 Antibody
3.2. Tissue Microarray (TMA) Analysis of CD74 Expression in BC
3.3. Immune Cell Infiltration and CD74 Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Harbeck, N.; Gnant, M. Breast cancer. Lancet 2017, 389, 1134–1150. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brenton, J.D.; Carey, L.A.; Ahmed, A.A.; Caldas, C. Molecular classification and molecular forecasting of breast cancer: Ready for clinical application? J. Clin. Oncol. 2005, 23, 7350–7360. [Google Scholar] [CrossRef] [Green Version]
- Wirapati, P.; Sotiriou, C.; Kunkel, S.; Farmer, P.; Pradervand, S.; Haibe-Kains, B.; Desmedt, C.; Ignatiadis, M.; Sengstag, T.; Schutz, F.; et al. Meta-analysis of gene expression profiles in breast cancer: Toward a unified understanding of breast cancer subtyping and prognosis signatures. Breast Cancer Res. 2008, 10, R65. [Google Scholar] [CrossRef]
- Goldhirsch, A.; Winer, E.P.; Coates, A.S.; Gelber, R.D.; Piccart-Gebhart, M.; Thurlimann, B.; Senn, H.J.; Panel, m. Personalizing the treatment of women with early breast cancer: Highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann. Oncol. 2013, 24, 2206–2223. [Google Scholar] [CrossRef] [PubMed]
- Perou, C.M.; Sorlie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular portraits of human breast tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Sorlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef] [Green Version]
- Curtis, C.; Shah, S.P.; Chin, S.F.; Turashvili, G.; Rueda, O.M.; Dunning, M.J.; Speed, D.; Lynch, A.G.; Samarajiwa, S.; Yuan, Y.; et al. The genomic and transcriptomic architecture of 2000 breast tumours reveals novel subgroups. Nature 2012, 486, 346–352. [Google Scholar] [CrossRef]
- Anders, C.K.; Carey, L.A. Biology, metastatic patterns, and treatment of patients with triple-negative breast cancer. Clin. Breast Cancer 2009, 9 (Suppl. 2), S73–S81. [Google Scholar] [CrossRef]
- Dent, R.; Trudeau, M.; Pritchard, K.I.; Hanna, W.M.; Kahn, H.K.; Sawka, C.A.; Lickley, L.A.; Rawlinson, E.; Sun, P.; Narod, S.A. Triple-negative breast cancer: Clinical features and patterns of recurrence. Clin. Cancer Res. 2007, 13, 4429–4434. [Google Scholar] [CrossRef] [Green Version]
- Abramson, V.G.; Lehmann, B.D.; Ballinger, T.J.; Pietenpol, J.A. Subtyping of triple-negative breast cancer: Implications for therapy. Cancer 2015, 121, 8–16. [Google Scholar] [CrossRef] [Green Version]
- Robson, M.; Im, S.A.; Senkus, E.; Xu, B.; Domchek, S.M.; Masuda, N.; Delaloge, S.; Li, W.; Tung, N.; Armstrong, A.; et al. Olaparib for Metastatic Breast Cancer in Patients with a Germline BRCA Mutation. N. Engl. J. Med. 2017, 377, 523–533. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.Y.; Shanmugam, M.K.; Sethi, G.; Bishayee, A. Potential role of targeted therapies in the treatment of triple-negative breast cancer. Anticancer Drugs 2016, 27, 147–155. [Google Scholar] [CrossRef]
- Thorsson, V.; Gibbs, D.L.; Brown, S.D.; Wolf, D.; Bortone, D.S.; Ou Yang, T.H.; Porta-Pardo, E.; Gao, G.F.; Plaisier, C.L.; Eddy, J.A.; et al. The Immune Landscape of Cancer. Immunity 2018, 48, 812–830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karn, T.; Jiang, T.; Hatzis, C.; Sanger, N.; El-Balat, A.; Rody, A.; Holtrich, U.; Becker, S.; Bianchini, G.; Pusztai, L. Association Between Genomic Metrics and Immune Infiltration in Triple-Negative Breast Cancer. JAMA Oncol. 2017, 3, 1707–1711. [Google Scholar] [CrossRef]
- Schmid, P.; Adams, S.; Rugo, H.S.; Schneeweiss, A.; Barrios, C.H.; Iwata, H.; Dieras, V.; Hegg, R.; Im, S.A.; Shaw Wright, G.; et al. Atezolizumab and Nab-Paclitaxel in Advanced Triple-Negative Breast Cancer. N. Engl. J. Med. 2018, 379, 2108–2121. [Google Scholar] [CrossRef] [PubMed]
- Sabatier, R.; Finetti, P.; Mamessier, E.; Adelaide, J.; Chaffanet, M.; Ali, H.R.; Viens, P.; Caldas, C.; Birnbaum, D.; Bertucci, F. Prognostic and predictive value of PDL1 expression in breast cancer. Oncotarget 2015, 6, 5449–5464. [Google Scholar] [CrossRef] [Green Version]
- Mittendorf, E.A.; Philips, A.V.; Meric-Bernstam, F.; Qiao, N.; Wu, Y.; Harrington, S.; Su, X.; Wang, Y.; Gonzalez-Angulo, A.M.; Akcakanat, A.; et al. PD-L1 expression in triple-negative breast cancer. Cancer Immunol. Res. 2014, 2, 361–370. [Google Scholar] [CrossRef] [Green Version]
- Adams, S.; Schmid, P.; Rugo, H.S.; Winer, E.P.; Loirat, D.; Awada, A.; Cescon, D.W.; Iwata, H.; Campone, M.; Nanda, R.; et al. Pembrolizumab monotherapy for previously treated metastatic triple-negative breast cancer: Cohort A of the phase II KEYNOTE-086 study. Ann. Oncol. 2019, 30, 397–404. [Google Scholar] [CrossRef] [Green Version]
- Warmerdam, P.A.; Long, E.O.; Roche, P.A. Isoforms of the invariant chain regulate transport of MHC class II molecules to antigen processing compartments. J. Cell Biol. 1996, 133, 281–291. [Google Scholar] [CrossRef]
- Roche, P.A.; Cresswell, P. Invariant chain association with HLA-DR molecules inhibits immunogenic peptide binding. Nature 1990, 345, 615–618. [Google Scholar] [CrossRef] [PubMed]
- Roche, P.A.; Cresswell, P. Proteolysis of the class II-associated invariant chain generates a peptide binding site in intracellular HLA-DR molecules. Proc. Natl. Acad. Sci. USA 1991, 88, 3150–3154. [Google Scholar] [CrossRef] [Green Version]
- Anderson, M.S.; Miller, J. Invariant chain can function as a chaperone protein for class II major histocompatibility complex molecules. Proc. Natl. Acad. Sci. USA 1992, 89, 2282–2286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leng, L.; Metz, C.N.; Fang, Y.; Xu, J.; Donnelly, S.; Baugh, J.; Delohery, T.; Chen, Y.; Mitchell, R.A.; Bucala, R. MIF signal transduction initiated by binding to CD74. J. Exp. Med. 2003, 197, 1467–1476. [Google Scholar] [CrossRef] [Green Version]
- Schwartz, V.; Lue, H.; Kraemer, S.; Korbiel, J.; Krohn, R.; Ohl, K.; Bucala, R.; Weber, C.; Bernhagen, J. A functional heteromeric MIF receptor formed by CD74 and CXCR4. FEBS Lett. 2009, 583, 2749–2757. [Google Scholar] [CrossRef] [Green Version]
- Maharshak, N.; Cohen, S.; Lantner, F.; Hart, G.; Leng, L.; Bucala, R.; Shachar, I. CD74 is a survival receptor on colon epithelial cells. World J. Gastroenterol. 2010, 16, 3258–3266. [Google Scholar] [CrossRef]
- Yaddanapudi, K.; Rendon, B.E.; Lamont, G.; Kim, E.J.; Al Rayyan, N.; Richie, J.; Albeituni, S.; Waigel, S.; Wise, A.; Mitchell, R.A. MIF Is Necessary for Late-Stage Melanoma Patient MDSC Immune Suppression and Differentiation. Cancer Immunol. Res. 2016, 4, 101–112. [Google Scholar] [CrossRef] [Green Version]
- Yaddanapudi, K.; Putty, K.; Rendon, B.E.; Lamont, G.J.; Faughn, J.D.; Satoskar, A.; Lasnik, A.; Eaton, J.W.; Mitchell, R.A. Control of tumor-associated macrophage alternative activation by macrophage migration inhibitory factor. J. Immunol. 2013, 190, 2984–2993. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Na, N.; Zhang, X.; Zhao, Y. The biological function and significance of CD74 in immune diseases. Inflamm. Res. 2017, 66, 209–216. [Google Scholar] [CrossRef]
- Zhao, S.; Molina, A.; Yu, A.; Hanson, J.; Cheung, H.; Li, X.; Natkunam, Y. High frequency of CD74 expression in lymphomas: Implications for targeted therapy using a novel anti-CD74-drug conjugate. J. Pathol. Clin. Res. 2019, 5, 12–24. [Google Scholar] [CrossRef]
- Stein, R.; Mattes, M.J.; Cardillo, T.M.; Hansen, H.J.; Chang, C.H.; Burton, J.; Govindan, S.; Goldenberg, D.M. CD74: A new candidate target for the immunotherapy of B-cell neoplasms. Clin. Cancer Res. 2007, 13, 5556s–5563s. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richard, V.; Kindt, N.; Saussez, S. Macrophage migration inhibitory factor involvement in breast cancer (Review). Int. J. Oncol. 2015, 47, 1627–1633. [Google Scholar] [CrossRef] [Green Version]
- Moller, P.; Mattfeldt, T.; Gross, C.; Schlosshauer, P.; Koch, A.; Koretz, K.; Moldenhauer, G.; Kaufmann, M.; Otto, H.F. Expression of HLA-A, -B, -C, -DR, -DP, -DQ, and of HLA-D-associated invariant chain (Ii) in non-neoplastic mammary epithelium, fibroadenoma, adenoma, and carcinoma of the breast. Am. J. Pathol. 1989, 135, 73–83. [Google Scholar]
- Koretz, K.; Moldenhauer, G.; Majdic, O.; Moller, P. Correlation of HLA-D/Ii antigen expression in breast carcinoma with local lymphohistiocytic infiltration reveals considerable dysregulation in a subset of tumors. Int. J. Cancer 1989, 44, 816–822. [Google Scholar] [CrossRef] [PubMed]
- Greenwood, C.; Metodieva, G.; Al-Janabi, K.; Lausen, B.; Alldridge, L.; Leng, L.; Bucala, R.; Fernandez, N.; Metodiev, M.V. Stat1 and CD74 overexpression is co-dependent and linked to increased invasion and lymph node metastasis in triple-negative breast cancer. J. Proteom. 2012, 75, 3031–3040. [Google Scholar] [CrossRef]
- Tian, B.; Zhang, Y.; Li, N.; Liu, X.; Dong, J. CD74: A potential novel target for triple-negative breast cancer. Tumour Biol. 2012, 33, 2273–2277. [Google Scholar] [CrossRef] [PubMed]
- Metodieva, G.; Nogueira-de-Souza, N.C.; Greenwood, C.; Al-Janabi, K.; Leng, L.; Bucala, R.; Metodiev, M.V. CD74-dependent deregulation of the tumor suppressor scribble in human epithelial and breast cancer cells. Neoplasia 2013, 15, 660–668. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.Q.; Milne, K.; Webb, J.R.; Watson, P.H. CD74 and intratumoral immune response in breast cancer. Oncotarget 2017, 8, 12664–12674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Chu, S.; Yao, S.; Li, Y.; Fan, S.; Sun, X.; Su, L.; Liu, X. CD74 interacts with CD44 and enhances tumorigenesis and metastasis via RHOA-mediated cofilin phosphorylation in human breast cancer cells. Oncotarget 2016, 7, 68303–68313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richard, V.; Kindt, N.; Decaestecker, C.; Gabius, H.J.; Laurent, G.; Noel, J.C.; Saussez, S. Involvement of macrophage migration inhibitory factor and its receptor (CD74) in human breast cancer. Oncol. Rep. 2014, 32, 523–529. [Google Scholar] [CrossRef] [Green Version]
- Oldford, S.A.; Robb, J.D.; Codner, D.; Gadag, V.; Watson, P.H.; Drover, S. Tumor cell expression of HLA-DM associates with a Th1 profile and predicts improved survival in breast carcinoma patients. Int. Immunol. 2006, 18, 1591–1602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forero, A.; Li, Y.; Chen, D.; Grizzle, W.E.; Updike, K.L.; Merz, N.D.; Downs-Kelly, E.; Burwell, T.C.; Vaklavas, C.; Buchsbaum, D.J.; et al. Expression of the MHC Class II Pathway in Triple-Negative Breast Cancer Tumor Cells Is Associated with a Good Prognosis and Infiltrating Lymphocytes. Cancer Immunol. Res. 2016, 4, 390–399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cabezon, T.; Gromova, I.; Gromov, P.; Serizawa, R.; Timmermans Wielenga, V.; Kroman, N.; Celis, J.E.; Moreira, J.M. Proteomic profiling of triple-negative breast carcinomas in combination with a three-tier orthogonal technology approach identifies Mage-A4 as potential therapeutic target in estrogen receptor negative breast cancer. Mol. Cell Proteom. 2013, 12, 381–394. [Google Scholar] [CrossRef] [Green Version]
- Dolled-Filhart, M.; McCabe, A.; Giltnane, J.; Cregger, M.; Camp, R.L.; Rimm, D.L. Quantitative in situ analysis of beta-catenin expression in breast cancer shows decreased expression is associated with poor outcome. Cancer Res. 2006, 66, 5487–5494. [Google Scholar] [CrossRef] [Green Version]
- Bankhead, P.; Loughrey, M.B.; Fernandez, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.; et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017, 7, 16878. [Google Scholar] [CrossRef] [Green Version]
- Loughrey, M.B.; Bankhead, P.; Coleman, H.G.; Hagan, R.S.; Craig, S.; McCorry, A.M.B.; Gray, R.T.; McQuaid, S.; Dunne, P.D.; Hamilton, P.W.; et al. Validation of the systematic scoring of immunohistochemically stained tumour tissue microarrays using QuPath digital image analysis. Histopathology 2018, 73, 327–338. [Google Scholar] [CrossRef] [Green Version]
- McCarty, K.S., Jr.; Szabo, E.; Flowers, J.L.; Cox, E.B.; Leight, G.S.; Miller, L.; Konrath, J.; Soper, J.T.; Budwit, D.A.; Creasman, W.T.; et al. Use of a monoclonal anti-estrogen receptor antibody in the immunohistochemical evaluation of human tumors. Cancer Res. 1986, 46, 4244s–4248s. [Google Scholar] [PubMed]
- Gyorffy, B.; Lanczky, A.; Eklund, A.C.; Denkert, C.; Budczies, J.; Li, Q.; Szallasi, Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1809 patients. Breast Cancer Res. Treat. 2010, 123, 725–731. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Birkbak, N.J.; Gyorffy, B.; Szallasi, Z.; Eklund, A.C. Jetset: Selecting the optimal microarray probe set to represent a gene. BMC Bioinf. 2011, 12, 474. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Severson, E.; Pignon, J.C.; Zhao, H.; Li, T.; Novak, J.; Jiang, P.; Shen, H.; Aster, J.C.; Rodig, S.; et al. Comprehensive analyses of tumor immunity: Implications for cancer immunotherapy. Genome Biol. 2016, 17, 174. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Fan, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S.; Li, B.; Liu, X.S. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef] [Green Version]
- Lazova, R.; Moynes, R.; May, D.; Scott, G. LN-2 (CD74). A marker to distinguish atypical fibroxanthoma from malignant fibrous histiocytoma. Cancer 1997, 79, 2115–2124. [Google Scholar] [CrossRef]
- Epstein, A.L.; Marder, R.J.; Winter, J.N.; Fox, R.I. Two new monoclonal antibodies (LN-1, LN-2) reactive in B5 formalin-fixed, paraffin-embedded tissues with follicular center and mantle zone human B lymphocytes and derived tumors. J. Immunol. 1984, 133, 1028–1036. [Google Scholar] [PubMed]
- Freeman, G.J.; Cardoso, A.A.; Boussiotis, V.A.; Anumanthan, A.; Groves, R.W.; Kupper, T.S.; Clark, E.A.; Nadler, L.M. The BB1 monoclonal antibody recognizes both cell surface CD74 (MHC class II-associated invariant chain) as well as B7-1 (CD80), resolving the question regarding a third CD28/CTLA-4 counterreceptor. J. Immunol. 1998, 161, 2708–2715. [Google Scholar] [PubMed]
- Schroder, B. The multifaceted roles of the invariant chain CD74--More than just a chaperone. Biochim. Biophys. Acta 2016, 1863, 1269–1281. [Google Scholar] [CrossRef] [PubMed]
- Alabdulmonem, W.; Alhomaidan, H.T.; Rasheed, Z.; Madar, I.H.; Alasmael, N.; Alkhatib, S.; Al Ssadh, H. CD74 a Potential Therapeutic Target for Breast Cancer Therapy: Interferon Gamma Up-regulates its Expression in CAMA-1 and MDA-MB-231 Cancer Cells. Int. J. Cancer Res. 2018, 14, 11. [Google Scholar] [CrossRef] [Green Version]
- Pozner-Moulis, S.; Cregger, M.; Camp, R.L.; Rimm, D.L. Antibody validation by quantitative analysis of protein expression using expression of Met in breast cancer as a model. Lab. Investig. 2007, 87, 251–260. [Google Scholar] [CrossRef]
- Kampgen, E.; Koch, N.; Koch, F.; Stoger, P.; Heufler, C.; Schuler, G.; Romani, N. Class II major histocompatibility complex molecules of murine dendritic cells: Synthesis, sialylation of invariant chain, and antigen processing capacity are down-regulated upon culture. Proc. Natl. Acad. Sci. USA 1991, 88, 3014–3018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, Z.A.; Moore, B.B.; Quezada, D.; Chang, C.H.; Jones, P.P. Identification of an IFN-gamma responsive region in an intron of the invariant chain gene. Eur. J. Immunol. 2000, 30, 2604–2611. [Google Scholar] [CrossRef]
- Tanese, K.; Hashimoto, Y.; Berkova, Z.; Wang, Y.; Samaniego, F.; Lee, J.E.; Ekmekcioglu, S.; Grimm, E.A. Cell Surface CD74-MIF Interactions Drive Melanoma Survival in Response to Interferon-gamma. J. Investig. Dermatol. 2015, 135, 2775–2784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicolini, A.; Carpi, A.; Rossi, G. Cytokines in breast cancer. Cytokine Growth Factor Rev. 2006, 17, 325–337. [Google Scholar] [CrossRef]
- Schupp, J.; Krebs, F.K.; Zimmer, N.; Trzeciak, E.; Schuppan, D.; Tuettenberg, A. Targeting myeloid cells in the tumor sustaining microenvironment. Cell Immunol. 2019, 343, 103713. [Google Scholar] [CrossRef] [PubMed]
- Ascierto, M.L.; Kmieciak, M.; Idowu, M.O.; Manjili, R.; Zhao, Y.; Grimes, M.; Dumur, C.; Wang, E.; Ramakrishnan, V.; Wang, X.Y.; et al. A signature of immune function genes associated with recurrence-free survival in breast cancer patients. Breast Cancer Res. Treat. 2012, 131, 871–880. [Google Scholar] [CrossRef] [Green Version]
- Ekmekcioglu, S.; Davies, M.A.; Tanese, K.; Roszik, J.; Shin-Sim, M.; Bassett, R.L., Jr.; Milton, D.R.; Woodman, S.E.; Prieto, V.G.; Gershenwald, J.E.; et al. Inflammatory Marker Testing Identifies CD74 Expression in Melanoma Tumor Cells, and Its Expression Associates with Favorable Survival for Stage III Melanoma. Clin. Cancer Res. 2016, 22, 3016–3024. [Google Scholar] [CrossRef] [Green Version]
- Zeiner, P.S.; Zinke, J.; Kowalewski, D.J.; Bernatz, S.; Tichy, J.; Ronellenfitsch, M.W.; Thorsen, F.; Berger, A.; Forster, M.T.; Muller, A.; et al. CD74 regulates complexity of tumor cell HLA class II peptidome in brain metastasis and is a positive prognostic marker for patient survival. Acta Neuropathol. Commun. 2018, 6, 18. [Google Scholar] [CrossRef] [Green Version]
- Otterstrom, C.; Soltermann, A.; Opitz, I.; Felley-Bosco, E.; Weder, W.; Stahel, R.A.; Triponez, F.; Robert, J.H.; Serre-Beinier, V. CD74: A new prognostic factor for patients with malignant pleural mesothelioma. Br. J. Cancer 2014, 110, 2040–2046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nathan, M.R.; Schmid, P. The emerging world of breast cancer immunotherapy. Breast 2018, 37, 200–206. [Google Scholar] [CrossRef] [Green Version]
- Verjans, E.; Noetzel, E.; Bektas, N.; Schutz, A.K.; Lue, H.; Lennartz, B.; Hartmann, A.; Dahl, E.; Bernhagen, J. Dual role of macrophage migration inhibitory factor (MIF) in human breast cancer. BMC Cancer 2009, 9, 230. [Google Scholar] [CrossRef] [Green Version]
- Meyer-Siegler, K.L.; Iczkowski, K.A.; Leng, L.; Bucala, R.; Vera, P.L. Inhibition of macrophage migration inhibitory factor or its receptor (CD74) attenuates growth and invasion of DU-145 prostate cancer cells. J. Immunol. 2006, 177, 8730–8739. [Google Scholar] [CrossRef] [Green Version]



| Parameter | N | Mean/Percentage of Cases (%) |
|---|---|---|
| Age (mean) | 555 | 58.0 years |
| Lymph node status | ||
| Negative | 261 | 47% |
| Positive | 294 | 53% |
| ER status | ||
| Negative | 270 | 49% |
| Positive | 285 | 51% |
| PR status | ||
| Negative | 294 | 53% |
| Positive | 261 | 47% |
| HER2 status | ||
| Negative | 359 | 65% |
| Positive | 196 | 35% |
| TN status | ||
| Non-TN | 427 | 77% |
| TN | 128 | 23% |
| CD74 Low | CD74 High | p-Value | |
|---|---|---|---|
| Age (years) | 58.0 (56.9–59.2) | 57.7 (55.2–60.2) | 0.7601 |
| ER status | 0.8567 | ||
| Negative | 227 | 43 | |
| Positive | 238 | 47 | |
| PR status | 0.1235 | ||
| Negative | 253 | 41 | |
| Positive | 212 | 49 | |
| HER2 status | 0.9585 | ||
| Negative | 301 | 58 | |
| Positive | 164 | 32 | |
| Nodal status | 0.1904 | ||
| Negative | 213 | 48 | |
| Positive | 252 | 42 | |
| TN status | 0.947 | ||
| Not TN | 358 | 69 | |
| TN | 107 | 21 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Noer, J.B.; Talman, M.-L.M.; Moreira, J.M.A. HLA Class II Histocompatibility Antigen γ Chain (CD74) Expression Is Associated with Immune Cell Infiltration and Favorable Outcome in Breast Cancer. Cancers 2021, 13, 6179. https://doi.org/10.3390/cancers13246179
Noer JB, Talman M-LM, Moreira JMA. HLA Class II Histocompatibility Antigen γ Chain (CD74) Expression Is Associated with Immune Cell Infiltration and Favorable Outcome in Breast Cancer. Cancers. 2021; 13(24):6179. https://doi.org/10.3390/cancers13246179
Chicago/Turabian StyleNoer, Julie B., Maj-Lis M. Talman, and José M. A. Moreira. 2021. "HLA Class II Histocompatibility Antigen γ Chain (CD74) Expression Is Associated with Immune Cell Infiltration and Favorable Outcome in Breast Cancer" Cancers 13, no. 24: 6179. https://doi.org/10.3390/cancers13246179







