Plasma Proteome Signature to Predict the Outcome of Breast Cancer Patients Receiving Neoadjuvant Chemotherapy
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Participants and Surveillance
2.2. Sample Preparation
2.3. Nano-LC-ESI-MS/MS Analysis
2.4. Protein Identification by Database Search
2.5. Differential Data Analysis by Normalization and Filling Missing Data
2.6. Statistical Clinical Model Generation Based on Feature Selection
2.7. Mining Public Microarray Data
2.8. Statistical Methods
3. Results
3.1. Baseline Characteristics
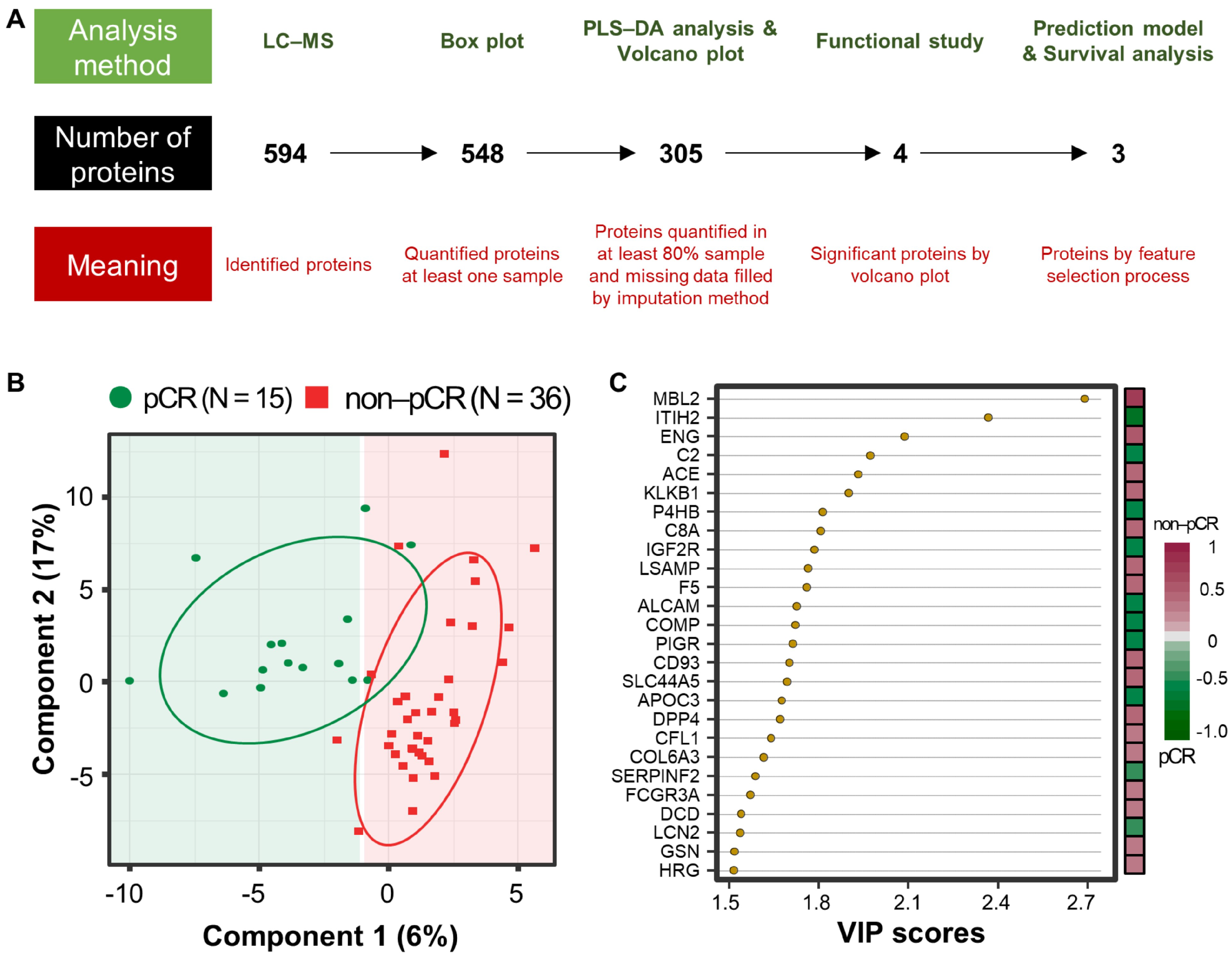
3.2. Proteome Results from Clinical Plasma Samples by LC-MS/MS
3.3. Differentially Abundant Plasma Proteins between pCR and Non-pCR BC Patients
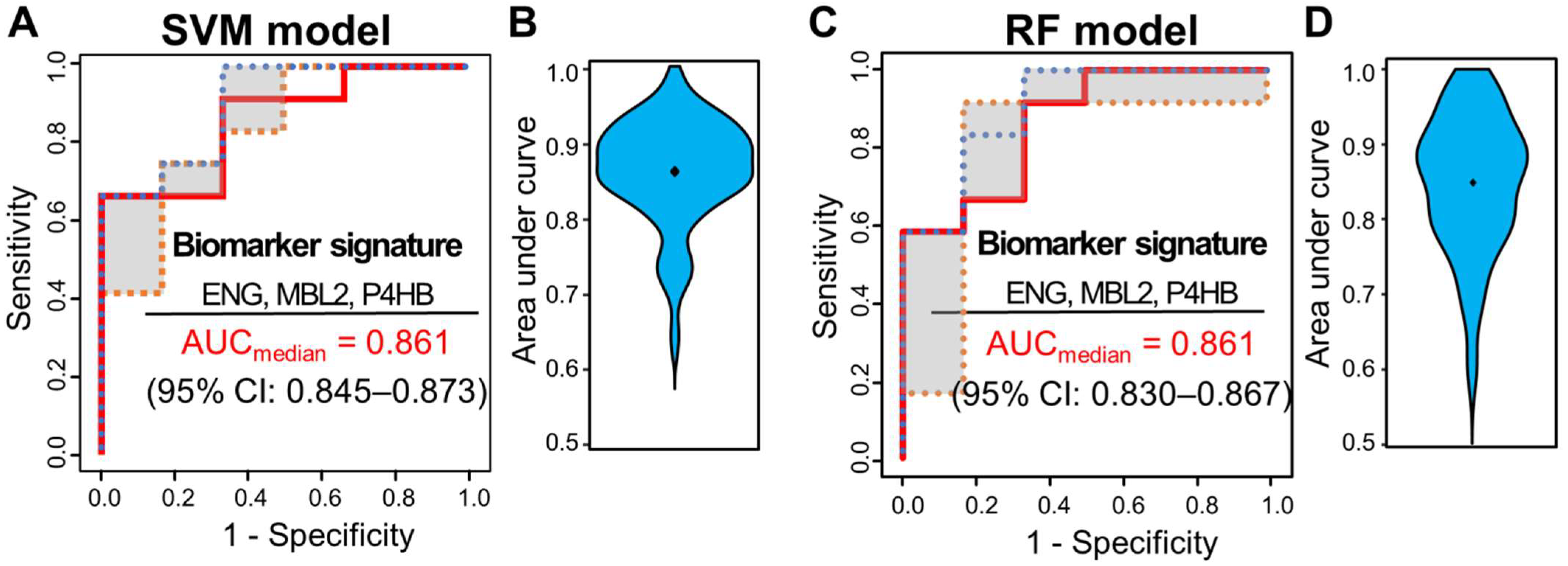
3.4. Multivariate Analysis for Predicting pCR Outcome
3.5. Survival Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fisher, B.; Brown, A.; Mamounas, E.; Wieand, S.; Robidoux, A.; Margolese, R.G.; Cruz, A.B., Jr.; Fisher, E.R.; Wickerham, D.L.; Wolmark, N.; et al. Effect of preoperative chemotherapy on local-regional disease in women with operable breast cancer: Findings from National Surgical Adjuvant Breast and Bowel Project B-18. J. Clin. Oncol. 1997, 15, 2483–2493. [Google Scholar] [CrossRef] [PubMed]
- van der Hage, J.A.; van de Velde, C.J.; Julien, J.P.; Tubiana-Hulin, M.; Vandervelden, C.; Duchateau, L. Preoperative chemotherapy in primary operable breast cancer: Results from the European Organization for Research and Treatment of Cancer trial 10902. J. Clin. Oncol. 2001, 19, 4224–4237. [Google Scholar] [CrossRef]
- Caudle, A.S.; Bedrosian, I.; Milton, D.R.; DeSnyder, S.M.; Kuerer, H.M.; Hunt, K.K.; Mittendorf, E.A. Use of Sentinel Lymph Node Dissection After Neoadjuvant Chemotherapy in Patients with Node-Positive Breast Cancer at Diagnosis: Practice Patterns of American Society of Breast Surgeons Members. Ann. Surg. Oncol. 2017, 10, 2925–2934. [Google Scholar] [CrossRef]
- Meisamy, S.; Bolan, P.J.; Baker, E.H.; Bliss, R.L.; Gulbahce, E.; Everson, L.I.; Nelson, M.T.; Emory, T.H.; Tuttle, T.M.; Yee, D. Neoadjuvant chemotherapy of locally advanced breast cancer: Predicting response with in vivo 1H MR spectroscopy—A pilot study at 4 T. Radiology 2004, 233, 424–431. [Google Scholar] [CrossRef]
- Cerussi, A.; Hsiang, D.; Shah, N.; Mehta, R.; Durkin, A.; Butler, J.; Tromberg, B.J. Predicting response to breast cancer neoadjuvant chemotherapy using diffuse optical spectroscopy. Proc. Natl. Acad. Sci. USA 2007, 104, 4014–4019. [Google Scholar] [CrossRef] [Green Version]
- Sharma, U.; Danishad, K.K.A.; Seenu, V.; Jagannathan, N.R. Longitudinal study of the assessment by MRI and diffusion-weighted imaging of tumor response in patients with locally advanced breast cancer undergoing neoadjuvant chemotherapy. NMR Biomed. Int. J. Devoted Dev. Appl. Magn. Reson. In Vivo 2009, 22, 104–113. [Google Scholar] [CrossRef] [PubMed]
- Manton, D.; Chaturvedi, A.; Hubbard, A.; Lind, M.; Lowry, M.; Maraveyas, A.; Pickles, M.; Tozer, D.; Turnbull, L. Neoadjuvant chemotherapy in breast cancer: Early response prediction with quantitative MR imaging and spectroscopy. Br. J. Cancer 2006, 94, 427. [Google Scholar] [CrossRef] [PubMed]
- Rajan, R.; Poniecka, A.; Smith, T.L.; Yang, Y.; Frye, D.; Pusztai, L.; Fiterman, D.J.; Gal-Gombos, E.; Whitman, G.; Rouzier, R. Change in tumor cellularity of breast carcinoma after neoadjuvant chemotherapy as a variable in the pathologic assessment of response. Cancer Interdiscip. Int. J. Am. Cancer Soc. 2004, 100, 1365–1373. [Google Scholar] [CrossRef]
- Baek, H.-M.; Chen, J.-H.; Nie, K.; Yu, H.J.; Bahri, S.; Mehta, R.S.; Nalcioglu, O.; Su, M.-Y. Predicting pathologic response to neoadjuvant chemotherapy in breast cancer by using MR imaging and quantitative 1H MR spectroscopy. Radiology 2009, 251, 653–662. [Google Scholar] [CrossRef] [Green Version]
- Liedtke, C.; Mazouni, C.; Hess, K.R.; André, F.; Tordai, A.; Mejia, J.A.; Symmans, W.F.; Gonzalez-Angulo, A.M.; Hennessy, B.; Green, M. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J. Clin. Oncol. 2008, 26, 1275–1281. [Google Scholar] [CrossRef] [PubMed]
- Houssami, N.; Macaskill, P.; von Minckwitz, G.; Marinovich, M.L.; Mamounas, E. Meta-analysis of the association of breast cancer subtype and pathologic complete response to neoadjuvant chemotherapy. Eur. J. Cancer 2012, 48, 3342–3354. [Google Scholar] [CrossRef]
- Carey, L.A.; Dees, E.C.; Sawyer, L.; Gatti, L.; Moore, D.T.; Collichio, F.; Ollila, D.W.; Sartor, C.I.; Graham, M.L.; Perou, C.M. The triple negative paradox: Primary tumor chemosensitivity of breast cancer subtypes. Clin. Cancer Res. 2007, 13, 2329–2334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schott, A.F.; Roubidoux, M.A.; Helvie, M.A.; Hayes, D.F.; Kleer, C.G.; Newman, L.A.; Pierce, L.J.; Griffith, K.A.; Murray, S.; Hunt, K.A.; et al. Clinical and radiologic assessments to predict breast cancer pathologic complete response to neoadjuvant chemotherapy. Breast Cancer Res. Treat. 2005, 92, 231–238. [Google Scholar] [CrossRef]
- Baehner, F.L. The analytical validation of the Oncotype DX Recurrence Score assay. Ecancermedicalscience 2016, 10, 675. [Google Scholar] [CrossRef]
- Safi, F.; Kohler, I.; Beger, H.G.; Röttinger, E. The value of the tumor marker CA 15-3 in diagnosing and monitoring breast cancer. A comparative study with carcinoembryonic antigen. Cancer 1991, 68, 574–582. [Google Scholar] [CrossRef]
- Lumachi, F.; Basso, S.M.M. Serum tumor markers in patients with breast cancer. Expert Rev. Anticancer Ther. 2004, 4, 921–931. [Google Scholar] [CrossRef]
- Bayo, J.; Castaño, M.; Rivera, F.; Navarro, F. Analysis of blood markers for early breast cancer diagnosis. Clin. Transl. Oncol. 2017, 20, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Trufero, J.; de Lobera, A.R.; Lao, J.; Puértolas, T.; Artal-Cortés, A.; Zorrilla, M.; Alonso, V.; Pazo, R.; Valero, M.I.; Ríos-Mitchell, M.J. Serum markers and prognosis in locally advanced breast cancer. Tumori J. 2005, 91, 522–530. [Google Scholar] [CrossRef]
- Alix-Panabieres, C.; Pantel, K. Liquid Biopsy: From Discovery to Clinical Application. Cancer Discov. 2021, 11, 858–873. [Google Scholar] [CrossRef]
- Alix-Panabieres, C.; Pantel, K. Circulating tumor cells: Liquid biopsy of cancer. Clin. Chem. 2013, 59, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Matera, J.; Miller, M.C.; Reuben, J.M.; Doyle, G.V.; Allard, W.J.; Terstappen, L.W.; et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 2004, 351, 781–791. [Google Scholar] [CrossRef] [Green Version]
- Hayes, D.F.; Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Miller, M.C.; Matera, J.; Allard, W.J.; Doyle, G.V.; Terstappen, L.W. Circulating tumor cells at each follow-up time point during therapy of metastatic breast cancer patients predict progression-free and overall survival. Clin. Cancer Res. 2006, 12, 4218–4224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Dalum, G.; Van der Stam, G.J.; Tibbe, A.G.; Franken, B.; Mastboom, W.J.; Vermes, I.; De Groot, M.R.; Terstappen, L.W. Circulating tumor cells before and during follow-up after breast cancer surgery. Int. J. Oncol. 2015, 46, 407–413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rack, B.; Schindlbeck, C.; Jückstock, J.; Andergassen, U.; Hepp, P.; Zwingers, T.; Friedl, T.W.P.; Lorenz, R.; Tesch, H.; Fasching, P.A.; et al. Circulating Tumor Cells Predict Survival in Early Average-to-High Risk Breast Cancer Patients. JNCI J. Natl. Cancer Inst. 2014, 106, dju066. [Google Scholar] [CrossRef] [PubMed]
- Franken, B.; de Groot, M.R.; Mastboom, W.J.; Vermes, I.; van der Palen, J.; Tibbe, A.G.; Terstappen, L.W. Circulating tumor cells, disease recurrence and survival in newly diagnosed breast cancer. Breast Cancer Res. 2012, 14, R133. [Google Scholar] [CrossRef] [Green Version]
- Bidard, F.C.; Mathiot, C.; Delaloge, S.; Brain, E.; Giachetti, S.; de Cremoux, P.; Marty, M.; Pierga, J.Y. Single circulating tumor cell detection and overall survival in nonmetastatic breast cancer. Ann. Oncol. 2010, 21, 729–733. [Google Scholar] [CrossRef]
- Lucci, A.; Hall, C.S.; Lodhi, A.K.; Bhattacharyya, A.; Anderson, A.E.; Xiao, L.; Bedrosian, I.; Kuerer, H.M.; Krishnamurthy, S. Circulating tumour cells in non-metastatic breast cancer: A prospective study. Lancet Oncol. 2012, 13, 688–695. [Google Scholar] [CrossRef]
- Hall, C.; Karhade, M.; Laubacher, B.; Anderson, A.; Kuerer, H.; DeSynder, S.; Lucci, A. Circulating tumor cells after neoadjuvant chemotherapy in stage I–III triple-negative breast cancer. Ann. Surg. Oncol. 2015, 22, 552–558. [Google Scholar] [CrossRef]
- Fei, F.; Du, Y.; Di, G.; Wu, J.; Shao, Z. Are changes in circulating tumor cell (CTC) count associated with the response to neoadjuvant chemotherapy in local advanced breast cancer? A meta-analysis. Oncol. Res. Treat. 2014, 37, 250–254. [Google Scholar] [CrossRef]
- Kasimir-Bauer, S.; Bittner, A.-K.; König, L.; Reiter, K.; Keller, T.; Kimmig, R.; Hoffmann, O. Does primary neoadjuvant systemic therapy eradicate minimal residual disease? Analysis of disseminated and circulating tumor cells before and after therapy. Breast Cancer Res. 2016, 18, 20. [Google Scholar] [CrossRef]
- Gwark, S.; Kim, J.; Kwon, N.-J.; Kim, K.-Y.; Kim, Y.; Lee, C.H.; Kim, Y.H.; Kim, M.S.; Hong, S.W.; Choi, M.Y. Analysis of the serial circulating tumor cell count during neoadjuvant chemotherapy in breast cancer patients. Sci. Rep. 2020, 10, 17466. [Google Scholar] [CrossRef] [PubMed]
- Bertucci, F.; Birnbaum, D.; Goncalves, A. Proteomics of breast cancer—Principles and potential clinical applications. Mol. Cell. Proteom. 2006, 5, 1772–1786. [Google Scholar] [CrossRef] [Green Version]
- Galvao, E.R.; Martins, L.M.; Ibiapina, J.O.; Andrade, H.M.; Monte, S.J. Breast cancer proteomics: A review for clinicians. J. Cancer Res. Clin. Oncol. 2011, 137, 915–925. [Google Scholar] [CrossRef]
- Gast, M.C.; Schellens, J.H.; Beijnen, J.H. Clinical proteomics in breast cancer: A review. Breast Cancer Res. Treat. 2009, 116, 17–29. [Google Scholar] [CrossRef] [Green Version]
- Goncalves, A.; Bertucci, F. Clinical application of proteomics in breast cancer: State of the art and perspectives. Med. Princ. Pract. 2011, 20, 4–18. [Google Scholar] [CrossRef]
- Lumachi, F.; Chiara, G.B.; Foltran, L.; Basso, S.M. Proteomics as a Guide for Personalized Adjuvant Chemotherapy in Patients with Early Breast Cancer. Cancer Genom. Proteom. 2015, 12, 385–390. [Google Scholar]
- Tyanova, S.; Albrechtsen, R.; Kronqvist, P.; Cox, J.; Mann, M.; Geiger, T. Proteomic maps of breast cancer subtypes. Nat. Commun. 2016, 7, 10259. [Google Scholar] [CrossRef] [Green Version]
- Shenoy, A.; Belugali Nataraj, N.; Perry, G.; Loayza Puch, F.; Nagel, R.; Marin, I.; Balint, N.; Bossel, N.; Pavlovsky, A.; Barshack, I.; et al. Proteomic patterns associated with response to breast cancer neoadjuvant treatment. Mol. Syst. Biol. 2020, 16, e9443. [Google Scholar] [CrossRef]
- Gonzalez-Angulo, A.M.; Hennessy, B.T.; Meric-Bernstam, F.; Sahin, A.; Liu, W.; Ju, Z.; Carey, M.S.; Myhre, S.; Speers, C.; Deng, L.; et al. Functional proteomics can define prognosis and predict pathologic complete response in patients with breast cancer. Clin. Proteom. 2011, 8, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McNamara, K.L.; Caswell-Jin, J.L.; Joshi, R.; Ma, Z.; Kotler, E.; Bean, G.R.; Kriner, M.; Zhou, Z.; Hoang, M.; Beechem, J.; et al. Spatial proteomic characterization of HER2-positive breast tumors through neoadjuvant therapy predicts response. Nat. Cancer 2021, 2, 400–413. [Google Scholar] [CrossRef]
- Haugen, M.H.; Lingjaerde, O.C.; Hedenfalk, I.; Garred, O.; Borgen, E.; Loman, N.; Hatschek, T.; Borresen-Dale, A.L.; Naume, B.; Mills, G.B.; et al. Protein Signature Predicts Response to Neoadjuvant Treatment With Chemotherapy and Bevacizumab in HER2-Negative Breast Cancers. JCO Precis. Oncol. 2021, 5, 286–306. [Google Scholar] [CrossRef]
- Bonneterre, J.; Revillion, F.; Desauw, C.; Blot, E.; Kramar, A.; Fournier, C.; Hornez, L.; Peyrat, J.P. Plasma and tissue proteomic prognostic factors of response in primary breast cancer patients receiving neoadjuvant chemotherapy. Oncol. Rep. 2013, 29, 355–361. [Google Scholar] [CrossRef]
- He, J.; Whelan, S.A.; Lu, M.; Shen, D.; Chung, D.U.; Saxton, R.E.; Faull, K.F.; Whitelegge, J.P.; Chang, H.R. Proteomic-based biosignatures in breast cancer classification and prediction of therapeutic response. Int. J. Proteom. 2011, 2011, 896476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazouni, C.; Baggerly, K.; Hawke, D.; Tsavachidis, S.; Andre, F.; Buzdar, A.U.; Martin, P.M.; Kobayashi, R.; Pusztai, L. Evaluation of changes in serum protein profiles during neoadjuvant chemotherapy in HER2-positive breast cancer using an LC-MALDI-TOF/MS procedure. Proteomics 2010, 10, 3525–3532. [Google Scholar] [CrossRef]
- Michlmayr, A.; Bachleitner-Hofmann, T.; Baumann, S.; Marchetti-Deschmann, M.; Rech-Weichselbraun, I.; Burghuber, C.; Pluschnig, U.; Bartsch, R.; Graf, A.; Greil, R.; et al. Modulation of plasma complement by the initial dose of epirubicin/docetaxel therapy in breast cancer and its predictive value. Br. J. Cancer 2010, 103, 1201–1208. [Google Scholar] [CrossRef] [Green Version]
- Yang, T.; Fu, Z.; Zhang, Y.; Wang, M.; Mao, C.; Ge, W. Serum proteomics analysis of candidate predictive biomarker panel for the diagnosis of trastuzumab-based therapy resistant breast cancer. Biomed. Pharm. 2020, 129, 110465. [Google Scholar] [CrossRef] [PubMed]
- Koh, Y.Q.; Ng, D.Q.; Ng, C.C.; Boey, A.; Wei, M.; Sze, S.K.; Ho, H.K.; Acharya, M.; Limoli, C.L.; Chan, A. Extracellular Vesicle Proteome of Breast Cancer Patients with and Without Cognitive Impairment Following Anthracycline-based Chemotherapy: An Exploratory Study. Biomark Insights 2021, 16, 11772719211018204. [Google Scholar] [CrossRef]
- Johansson, H.J.; Socciarelli, F.; Vacanti, N.M.; Haugen, M.H.; Zhu, Y.; Siavelis, I.; Fernandez-Woodbridge, A.; Aure, M.R.; Sennblad, B.; Vesterlund, M.; et al. Breast cancer quantitative proteome and proteogenomic landscape. Nat. Commun. 2019, 10, 1600. [Google Scholar] [CrossRef] [PubMed]
- De Marchi, T.; Pyl, P.T.; Sjostrom, M.; Klasson, S.; Sartor, H.; Tran, L.; Pekar, G.; Malmstrom, J.; Malmstrom, L.; Nimeus, E. Proteogenomic Workflow Reveals Molecular Phenotypes Related to Breast Cancer Mammographic Appearance. J. Proteome Res. 2021, 20, 2983–3001. [Google Scholar] [CrossRef] [PubMed]
- Krug, K.; Jaehnig, E.J.; Satpathy, S.; Blumenberg, L.; Karpova, A.; Anurag, M.; Miles, G.; Mertins, P.; Geffen, Y.; Tang, L.C.; et al. Proteogenomic Landscape of Breast Cancer Tumorigenesis and Targeted Therapy. Cell 2020, 183, 1436–1456.e31. [Google Scholar] [CrossRef]
- Mertins, P.; Mani, D.R.; Ruggles, K.V.; Gillette, M.A.; Clauser, K.R.; Wang, P.; Wang, X.; Qiao, J.W.; Cao, S.; Petralia, F.; et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature 2016, 534, 55–62. [Google Scholar] [CrossRef] [Green Version]
- Moo, T.A.; Sanford, R.; Dang, C.; Morrow, M. Overview of Breast Cancer Therapy. PET Clin. 2018, 13, 339–354. [Google Scholar] [CrossRef]
- McShane, L.M.; Altman, D.G.; Sauerbrei, W.; Taube, S.E.; Gion, M.; Clark, G.M. Reporting recommendations for tumor marker prognostic studies (REMARK). J. Natl. Cancer Inst. 2005, 97, 1180–1184. [Google Scholar] [CrossRef] [Green Version]
- Allred, D.C.; Harvey, J.M.; Berardo, M.; Clark, G.M. Prognostic and predictive factors in breast cancer by immunohistochemical analysis. Mod. Pathol. 1998, 11, 155–168. [Google Scholar]
- Wolff, A.C.; Hammond, M.E.H.; Hicks, D.G.; Dowsett, M.; McShane, L.M.; Allison, K.H.; Allred, D.C.; Bartlett, J.M.S.; Bilous, M.; Fitzgibbons, P.; et al. Recommendations for Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Update. Arch. Pathol. Lab. Med. 2014, 138, 241–256. [Google Scholar] [CrossRef] [Green Version]
- Edge, S.B.; Compton, C.C. The American Joint Committee on Cancer: The 7th edition of the AJCC cancer staging manual and the future of TNM. Ann. Surg. Oncol. 2010, 17, 1471–1474. [Google Scholar] [CrossRef]
- Eisenhauer, E.A.; Therasse, P.; Bogaerts, J.; Schwartz, L.H.; Sargent, D.; Ford, R.; Dancey, J.; Arbuck, S.; Gwyther, S.; Mooney, M. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur. J. Cancer 2009, 45, 228–247. [Google Scholar] [CrossRef]
- Therasse, P.; Arbuck, S.G.; Eisenhauer, E.A.; Wanders, J.; Kaplan, R.S.; Rubinstein, L.; Verweij, J.; Van Glabbeke, M.; van Oosterom, A.T.; Christian, M.C. New guidelines to evaluate the response to treatment in solid tumors. J. Natl. Cancer Inst. 2000, 92, 205–216. [Google Scholar] [CrossRef] [Green Version]
- HaileMariam, M.; Eguez, R.V.; Singh, H.; Bekele, S.; Ameni, G.; Pieper, R.; Yu, Y. S-Trap, an Ultrafast Sample-Preparation Approach for Shotgun Proteomics. J. Proteome Res. 2018, 17, 2917–2924. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef]
- Bateman, A. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Kall, L.; Canterbury, J.D.; Weston, J.; Noble, W.S.; MacCoss, M.J. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods 2007, 4, 923–925. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Aguilar, J.; Molloy, M.P. Label-free selected reaction monitoring enables multiplexed quantitation of S100 protein isoforms in cancer cells. J. Proteome Res. 2013, 12, 3679–3688. [Google Scholar] [CrossRef] [PubMed]
- Wisniewski, J.R.; Hein, M.Y.; Cox, J.; Mann, M. A “proteomic ruler” for protein copy number and concentration estimation without spike-in standards. Mol. Cell. Proteom. 2014, 13, 3497–3506. [Google Scholar] [CrossRef] [Green Version]
- Andersen, C.L.; Jensen, J.L.; Orntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [Green Version]
- Ahn, H.S.; Ho, J.Y.; Yu, J.; Yeom, J.; Lee, S.; Hur, S.Y.; Jung, Y.; Kim, K.; Choi, Y.J. Plasma Protein Biomarkers Associated with Higher Ovarian Cancer Risk in BRCA1/2 Carriers. Cancers 2021, 13, 2300. [Google Scholar] [CrossRef]
- Ahn, H.S.; Kim, J.H.; Jeong, H.; Yu, J.; Yeom, J.; Song, S.H.; Kim, S.S.; Kim, I.J.; Kim, K. Differential Urinary Proteome Analysis for Predicting Prognosis in Type 2 Diabetes Patients with and without Renal Dysfunction. Int. J. Mol. Sci. 2020, 21, 4236. [Google Scholar] [CrossRef]
- Ahn, H.S.; Sohn, T.S.; Kim, M.J.; Cho, B.K.; Kim, S.M.; Kim, S.T.; Yi, E.C.; Lee, C. SEPROGADIC—Serum protein-based gastric cancer prediction model for prognosis and selection of proper adjuvant therapy. Sci. Rep. 2018, 8, 16892. [Google Scholar] [CrossRef]
- Kim, H.; Golub, G.H.; Park, H. Missing value estimation for DNA microarray gene expression data: Local least squares imputation. Bioinformatics 2005, 21, 187–198. [Google Scholar] [CrossRef]
- Calle, M.L.; Urrea, V.; Boulesteix, A.L.; Malats, N. AUC-RF: A new strategy for genomic profiling with random forest. Hum. Hered. 2011, 72, 121–132. [Google Scholar] [CrossRef] [Green Version]
- Cortes, C.; Vapnik, V. Support-Vector Networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Adams, S.; Chakravarthy, A.B.; Donach, M.; Spicer, D.; Lymberis, S.; Singh, B.; Bauer, J.A.; Hochman, T.; Goldberg, J.D.; Muggia, F.; et al. Preoperative concurrent paclitaxel-radiation in locally advanced breast cancer: Pathologic response correlates with five-year overall survival. Breast Cancer Res. Treat. 2010, 124, 723–732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bauer, J.A.; Chakravarthy, A.B.; Rosenbluth, J.M.; Mi, D.; Seeley, E.H.; De Matos Granja-Ingram, N.; Olivares, M.G.; Kelley, M.C.; Mayer, I.A.; Meszoely, I.M.; et al. Identification of markers of taxane sensitivity using proteomic and genomic analyses of breast tumors from patients receiving neoadjuvant paclitaxel and radiation. Clin. Cancer Res. 2010, 16, 681–690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lehmann, B.D.; Bauer, J.A.; Chen, X.; Sanders, M.E.; Chakravarthy, A.B.; Shyr, Y.; Pietenpol, J.A. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Investig. 2011, 121, 2750–2767. [Google Scholar] [CrossRef] [Green Version]
- Iwamoto, T.; Bianchini, G.; Booser, D.; Qi, Y.; Coutant, C.; Shiang, C.Y.; Santarpia, L.; Matsuoka, J.; Hortobagyi, G.N.; Symmans, W.F.; et al. Gene pathways associated with prognosis and chemotherapy sensitivity in molecular subtypes of breast cancer. J. Natl. Cancer Inst. 2011, 103, 264–272. [Google Scholar] [CrossRef] [Green Version]
- Shen, K.; Song, N.; Kim, Y.; Tian, C.; Rice, S.D.; Gabrin, M.J.; Symmans, W.F.; Pusztai, L.; Lee, J.K. A systematic evaluation of multi-gene predictors for the pathological response of breast cancer patients to chemotherapy. PLoS ONE 2012, 7, e49529. [Google Scholar] [CrossRef]
- Barrett, T.; Edgar, R. Gene expression omnibus: Microarray data storage, submission, retrieval, and analysis. Methods Enzym. 2006, 411, 352–369. [Google Scholar] [CrossRef] [Green Version]
- Rohart, F.; Gautier, B.; Singh, A.; Le Cao, K.A. mixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef] [Green Version]
- Rouzier, R.; Perou, C.M.; Symmans, W.F.; Ibrahim, N.; Cristofanilli, M.; Anderson, K.; Hess, K.R.; Stec, J.; Ayers, M.; Wagner, P. Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clin. Cancer Res. 2005, 11, 5678–5685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dziura, J.D.; Post, L.A.; Zhao, Q.; Fu, Z.; Peduzzi, P. Strategies for dealing with missing data in clinical trials: From design to analysis. Yale J. Biol. Med. 2013, 86, 343–358. [Google Scholar]
- Karpievitch, Y.V.; Dabney, A.R.; Smith, R.D. Normalization and missing value imputation for label-free LC-MS analysis. BMC Bioinform. 2012, 13 (Suppl. 16), S5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nanjappa, V.; Thomas, J.K.; Marimuthu, A.; Muthusamy, B.; Radhakrishnan, A.; Sharma, R.; Ahmad Khan, A.; Balakrishnan, L.; Sahasrabuddhe, N.A.; Kumar, S.; et al. Plasma Proteome Database as a resource for proteomics research: 2014 update. Nucleic Acids Res. 2014, 42, D959–D965. [Google Scholar] [CrossRef] [Green Version]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cortazar, P.; Zhang, L.; Untch, M.; Mehta, K.; Costantino, J.P.; Wolmark, N.; Bonnefoi, H.; Cameron, D.; Gianni, L.; Valagussa, P.; et al. Pathological complete response and long-term clinical benefit in breast cancer: The CTNeoBC pooled analysis. Lancet 2014, 384, 164–172. [Google Scholar] [CrossRef] [Green Version]
- Koboldt, D.; Fulton, R.; McLellan, M.; Schmidt, H.; Kalicki-Veizer, J.; McMichael, J.; Fulton, L.; Dooling, D.; Ding, L.; Mardis, E. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [Google Scholar]
- Heer, E.; Harper, A.; Escandor, N.; Sung, H.; McCormack, V.; Fidler-Benaoudia, M.M. Global burden and trends in premenopausal and postmenopausal breast cancer: A population-based study. Lancet Glob. Health 2020, 8, e1027–e1037. [Google Scholar] [CrossRef]
- Redig, A.J.; McAllister, S.S. Breast cancer as a systemic disease: A view of metastasis. J. Intern. Med. 2013, 274, 113–126. [Google Scholar] [CrossRef] [Green Version]
- Fehm, T.; Hoffmann, O.; Aktas, B.; Becker, S.; Solomayer, E.F.; Wallwiener, D.; Kimmig, R.; Kasimir-Bauer, S. Detection and characterization of circulating tumor cells in blood of primary breast cancer patients by RT-PCR and comparison to status of bone marrow disseminated cells. Breast Cancer Res. 2009, 11, R59. [Google Scholar] [CrossRef] [Green Version]
- Von Minckwitz, G.; Untch, M.; Blohmer, J.-U.; Costa, S.D.; Eidtmann, H.; Fasching, P.A.; Gerber, B.; Eiermann, W.; Hilfrich, J.; Huober, J. Definition and impact of pathologic complete response on prognosis after neoadjuvant chemotherapy in various intrinsic breast cancer subtypes. J. Clin. Oncol. 2012, 30, 1796–1804. [Google Scholar] [CrossRef] [Green Version]
- Broglio, K.R.; Quintana, M.; Foster, M.; Olinger, M.; McGlothlin, A.; Berry, S.M.; Boileau, J.-F.; Brezden-Masley, C.; Chia, S.; Dent, S. Association of pathologic complete response to neoadjuvant therapy in HER2-positive breast cancer with long-term outcomes: A meta-analysis. JAMA Oncol. 2016, 2, 751–760. [Google Scholar] [CrossRef] [Green Version]
- Pierga, J.-Y.; Bidard, F.-C.; Mathiot, C.; Brain, E.; Delaloge, S.; Giachetti, S.; de Cremoux, P.; Salmon, R.; Vincent-Salomon, A.; Marty, M. Circulating tumor cell detection predicts early metastatic relapse after neoadjuvant chemotherapy in large operable and locally advanced breast cancer in a phase II randomized trial. Clin. Cancer Res. 2008, 14, 7004–7010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wülfing, P.; Borchard, J.; Buerger, H.; Heidl, S.; Zänker, K.S.; Kiesel, L.; Brandt, B. HER2-positive circulating tumor cells indicate poor clinical outcome in stage I to III breast cancer patients. Clin. Cancer Res. 2006, 12, 1715–1720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Budd, G.T.; Cristofanilli, M.; Ellis, M.J.; Stopeck, A.; Borden, E.; Miller, M.C.; Matera, J.; Repollet, M.; Doyle, G.V.; Terstappen, L.W. Circulating tumor cells versus imaging—predicting overall survival in metastatic breast cancer. Clin. Cancer Res. 2006, 12, 6403–6409. [Google Scholar] [CrossRef] [Green Version]
- Bidard, F.-C.; Vincent-Salomon, A.; Sigal-Zafrani, B.; Dieras, V.; Mathiot, C.; Mignot, L.; Thiery, J.-P.; Sastre-Garau, X.; Pierga, J.-Y. Prognosis of women with stage IV breast cancer depends on detection of circulating tumor cells rather than disseminated tumor cells. Ann. Oncol. 2008, 19, 496–500. [Google Scholar] [CrossRef] [PubMed]
- Camara, O.; Rengsberger, M.; Egbe, A.; Koch, A.; Gajda, M.; Hammer, U.; Jörke, C.; Rabenstein, C.; Untch, M.; Pachmann, K. The relevance of circulating epithelial tumor cells (CETC) for therapy monitoring during neoadjuvant (primary systemic) chemotherapy in breast cancer. Ann. Oncol. 2007, 18, 1484–1492. [Google Scholar] [CrossRef] [PubMed]
- Riethdorf, S.; Müller, V.; Loibl, S.; Nekljudova, V.; Weber, K.; Huober, J.; Fehm, T.; Schrader, I.; Hilfrich, J.; Holms, F. Prognostic impact of circulating tumor cells for breast cancer patients treated in the neoadjuvant “Geparquattro” trial. Clin. Cancer Res. 2017, 23, 5384–5393. [Google Scholar] [CrossRef] [Green Version]
- Wulfkuhle, J.D.; McLean, K.C.; Paweletz, C.P.; Sgroi, D.C.; Trock, B.J.; Steeg, P.S.; Petricoin, E.F., III. New approaches to proteomic analysis of breast cancer. Proteom. Int. Ed. 2001, 1, 1205–1215. [Google Scholar] [CrossRef]
- Sawyers, C.L. The cancer biomarker problem. Nature 2008, 452, 548–552. [Google Scholar] [CrossRef]
- Vogelstein, B.; Kinzler, K.W. Cancer genes and the pathways they control. Nat. Med. 2004, 10, 789–799. [Google Scholar] [CrossRef]
- Zhang, J.; Guo, S.; Wu, Y.; Zheng, Z.-c.; Wang, Y.; Zhao, Y. P4HB, a novel hypoxia target gene related to gastric cancer invasion and metastasis. BioMed Res. Int. 2019, 2019, 9749751. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Yang, J.; Zhang, Q.; Xu, Q.; Lu, L.; Wang, J.; Xia, W. P4HB knockdown induces human HT29 colon cancer cell apoptosis through the generation of reactive oxygen species and inactivation of STAT3 signaling. Mol. Med. Rep. 2019, 19, 231–237. [Google Scholar] [CrossRef]
- Ma, X.; Wang, J.; Zhuang, J.; Ma, X.; Zheng, N.; Song, Y.; Xia, W. P4HB modulates epithelial-mesenchymal transition and the β-catenin/Snail pathway influencing chemoresistance in liver cancer cells. Oncol. Lett. 2020, 20, 257–265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, L.; Li, H.; Zhang, L.; Ma, X.; Dang, Y.; Guo, J.; Liu, J.; Ge, L.; Nan, F.; Dong, H. Autophagy-related gene P4HB: A novel diagnosis and prognosis marker for kidney renal clear cell carcinoma. Aging 2020, 12, 1828. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Wu, X.; Zhou, F. Collagen Type X Alpha 1 (COL10A1) Contributes to Cell Proliferation, Migration, and Invasion by Targeting Prolyl 4-Hydroxylase Beta Polypeptide (P4HB) in Breast Cancer. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2021, 27, e928919-1–e928919-17. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Hampson, I.N.; HAMPSON, L.; KUMAR, P.; Bernabeu, C.; Kumar, S. CD105 antagonizes the inhibitory signaling of transforming growth factor βl on human vascular endothelial cells. FASEB J. 2000, 14, 55–64. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, F.; Otake, Y.; Yanagihara, K.; Kawano, Y.; Miyahara, R.; Li, M.; Yamada, T.; Hanaoka, N.; Inui, K.; Wada, H. Evaluation of angiogenesis in non-small cell lung cancer: Comparison between anti-CD34 antibody and anti-CD105 antibody. Clin. Cancer Res. 2001, 7, 3410–3415. [Google Scholar]
- Miller, D.W.; Graulich, W.; Karges, B.; Stahl, S.; Ernst, M.; Ramaswamy, A.; Sedlacek, H.H.; Müller, R.; Adamkiewicz, J. Elevated expression of endoglin, a component of the TGF-β-receptor complex, correlates with proliferation of tumor endothelial cells. Int. J. Cancer 1999, 81, 568–572. [Google Scholar] [CrossRef]
- Nassiri, F.; Cusimano, M.D.; Scheithauer, B.W.; Rotondo, F.; Fazio, A.; Yousef, G.M.; Syro, L.V.; Kovacs, K.; Lloyd, R.V. Endoglin (CD105): A review of its role in angiogenesis and tumor diagnosis, progression and therapy. Anticancer Res. 2011, 31, 2283–2290. [Google Scholar]
- Li, C.; Guo, B.; Wilson, P.B.; Stewart, A.; Byrne, G.; Bundred, N.; Kumar, S. Plasma levels of soluble CD105 correlate with metastasis in patients with breast cancer. Int. J. Cancer 2000, 89, 122–126. [Google Scholar] [CrossRef]
- Kumar, S.; Ghellal, A.; Li, C.; Byrne, G.; Haboubi, N.; Wang, J.M.; Bundred, N. Breast carcinoma: Vascular density determined using CD105 antibody correlates with tumor prognosis. Cancer Res. 1999, 59, 856–861. [Google Scholar]
- Coussens, L.M.; Werb, Z. Inflammation and cancer. Nature 2002, 420, 860–867. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.P.; Hofseth, L.J.; Harris, C.C. Radical causes of cancer. Nat. Rev. Cancer 2003, 3, 276–285. [Google Scholar] [CrossRef] [PubMed]
- Ohta, M.; Kawasaki, T. Complement-dependent cytotoxic activity of serum mannan-binding protein towards mammalian cells with surface-exposed high-mannose type glycans. Glycoconj. J. 1994, 11, 304–308. [Google Scholar] [CrossRef]
- Nakagawa, T.; Ma, B.Y.; Uemura, K.; Oka, S.; Kawasaki, N.; Kawasaki, T. Role of mannan-binding protein, MBP, in innate immunity. Anticancer Res. 2003, 23, 4467–4471. [Google Scholar] [PubMed]
- Nakagawa, T.; Kawasaki, N.; Ma, Y.; Uemura, K.; Kawasaki, T. Antitumor activity of mannan-binding protein. Methods Enzymol. 2003, 363, 26–33. [Google Scholar]
- Holm, M.; Joenväärä, S.; Saraswat, M.; Mustonen, H.; Tohmola, T.; Ristimäki, A.; Renkonen, R.; Haglund, C. Identification of several plasma proteins whose levels in colorectal cancer patients differ depending on outcome. FASEB BioAdvances 2019, 1, 723–730. [Google Scholar] [CrossRef] [Green Version]
- Ytting, H.; Jensenius, J.; Christensen, I.; Thiel, S.; Nielsen, H. Increased activity of the mannan-binding lectin complement activation pathway in patients with colorectal cancer. Scand. J. Gastroenterol. 2004, 39, 674–679. [Google Scholar] [CrossRef]
- Swierzko, A.S.; Szala, A.; Sawicki, S.; Szemraj, J.; Sniadecki, M.; Sokolowska, A.; Kaluzynski, A.; Wydra, D.; Cedzynski, M. Mannose-Binding Lectin (MBL) and MBL-associated serine protease-2 (MASP-2) in women with malignant and benign ovarian tumours. Cancer Immunol. Immunother. 2014, 63, 1129–1140. [Google Scholar] [CrossRef] [Green Version]


| Variables | HR+/HER2- (n = 20) | HR+/HER2+ (n = 5) | HER2+ (n = 5) | Triple-Negative (n = 21) | p | |||
|---|---|---|---|---|---|---|---|---|
| Age at diagnosis (range) | 32–58 | 41–66 | 45–59 | 35–53 | 0.463 | |||
| ≤40 | 18 (90.0%) | 3 (60.0%) | 4 (80.0%) | 17 (81.0%) | ||||
| >40 | 2 (10.0%) | 2 (40.0%) | 1 (20.0%) | 4 (19.0%) | ||||
| Clinical T stage | 0.206 | |||||||
| T1 | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | ||||
| T2 | 11 (55.0%) | 2 (40.0%) | 5 (100%) | 15 (71.4%) | ||||
| T3 | 8 (40.0%) | 2 (40.0%) | 0 (0%) | 6 (28.6%) | ||||
| T4 | 1 (5.0%) | 1 (20.0%) | 0 (0%) | 0 (0%) | ||||
| Lymph node status | 0.473 | |||||||
| Negative | 6 (30.0%) | 0 (0%) | 2 (40.0%) | 5 (23.8%) | ||||
| Positive | 14 (70.0%) | 5 (100%) | 3 (60.0%) | 16 (76.2%) | ||||
| Nuclear grade | 0.001 | |||||||
| G1 and G2 | 19 (95.0%) | 5 (100%) | 4 (80.0%) | 9 (42.9%) | ||||
| G3 | 1 (5.0%) | 0 (0%) | 1 (20.0%) | 12 (57.1%) | ||||
| Tumor response (RECIST) | 0.295 | |||||||
| CR | 3 (15.0%) | 1 (20.0%) | 0 (0%) | 7 (33.3%) | ||||
| PR | 17 (85.0%) | 4 (80.0%) | 5 (100%) | 12 (57.2%) | ||||
| SD | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | ||||
| PD | 0 (0%) | 0 (0%) | 0 (0%) | 2 (9.5%) | ||||
| Type of Surgery (adjuvant RT) | 0.419 | |||||||
| BCS (26/26) | 8 (40.0%) | 4 (80.0%) | 3 (60.0%)) | 11 (52.4%) | ||||
| Mastectomy (20/25) | 12 (60.0%) | 1 (20.0%) | 2 (40.0%) | 10 (47.6%) | ||||
| Pathological response | 0.047 | |||||||
| pCR | 2 (10.0%) | 3 (60.0%) | 2 (40.0%) | 8 (38.1%) | ||||
| non-pCR | 18 (90.0%) | 2 (40.0%) | 3 (60.0%) | 13 (61.9%) | ||||
| NCT regimen | ||||||||
| Anthracycline based (AC#4, AC#4 > D#4, FEC#4 > D#4) | 47 (92.2%) | |||||||
| NCT02032277 * (Veliparib/Placebo + Carboplatin/Placebo + Paclitaxel) | 4 (7.8%) | |||||||
| Uniprot Accession No. | Gene Name | Importance | Prob. Select * | Selection | Univariate AUC |
|---|---|---|---|---|---|
| P11226 | MBL2 | 6.105 | 0.96 | Y | 0.807 |
| P17813 | ENG | 5.556 | 0.85 | Y | 0.739 |
| P07237 | P4HB | 3.522 | 0.58 | Y | 0.722 |
| P02656 | APOC3 | NA | NA | N | 0.654 |
| DMFS | ||
|---|---|---|
| Multivariate HR (95% CI) | p | |
| Patient age (>40 vs. ≤40) | 1.30 (0.33–5.06) | 0.709 |
| Tumor size (≤5 cm vs. >5 cm) | 2.55 (0.56–11.65) | 0.226 |
| Node negative vs. positive | * 2.1 × 105 | 0.963 |
| HR positive vs. negative | 2.95 (0.63–13.88) | 0.172 |
| HER2 negative vs. positive | 0.61 (0.07–5.44) | 0.660 |
| MBL2 abundance (low vs. high) | 9.65 (2.10–44.31) | 0.004 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gwark, S.; Ahn, H.-S.; Yeom, J.; Yu, J.; Oh, Y.; Jeong, J.H.; Ahn, J.-H.; Jung, K.H.; Kim, S.-B.; Lee, H.J.; et al. Plasma Proteome Signature to Predict the Outcome of Breast Cancer Patients Receiving Neoadjuvant Chemotherapy. Cancers 2021, 13, 6267. https://doi.org/10.3390/cancers13246267
Gwark S, Ahn H-S, Yeom J, Yu J, Oh Y, Jeong JH, Ahn J-H, Jung KH, Kim S-B, Lee HJ, et al. Plasma Proteome Signature to Predict the Outcome of Breast Cancer Patients Receiving Neoadjuvant Chemotherapy. Cancers. 2021; 13(24):6267. https://doi.org/10.3390/cancers13246267
Chicago/Turabian StyleGwark, Sungchan, Hee-Sung Ahn, Jeonghun Yeom, Jiyoung Yu, Yumi Oh, Jae Ho Jeong, Jin-Hee Ahn, Kyung Hae Jung, Sung-Bae Kim, Hee Jin Lee, and et al. 2021. "Plasma Proteome Signature to Predict the Outcome of Breast Cancer Patients Receiving Neoadjuvant Chemotherapy" Cancers 13, no. 24: 6267. https://doi.org/10.3390/cancers13246267
APA StyleGwark, S., Ahn, H.-S., Yeom, J., Yu, J., Oh, Y., Jeong, J. H., Ahn, J.-H., Jung, K. H., Kim, S.-B., Lee, H. J., Gong, G., Lee, S. B., Chung, I. Y., Kim, H. J., Ko, B. S., Lee, J. W., Son, B. H., Ahn, S. H., Kim, K., & Kim, J. (2021). Plasma Proteome Signature to Predict the Outcome of Breast Cancer Patients Receiving Neoadjuvant Chemotherapy. Cancers, 13(24), 6267. https://doi.org/10.3390/cancers13246267






