Expression of SnoRNA U50A Is Associated with Better Prognosis and Prolonged Mitosis in Breast Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Specimens
2.2. Total RNA Extraction
2.3. RNA-Sequencing Analysis
2.4. Reverse Transcription
2.5. SnoRNA Detection and Absolute/Relative Real-Time PCR
2.6. Plasmid and Transfection
2.7. SnoRNA Inhibition
2.8. Lentiviral Knockdown
2.9. Western Blot
2.10. Immunofluorescence
2.11. In Situ Hybridization (ISH) Detection
2.12. Statistical Analysis
2.13. Colony Formation
3. Results
3.1. U50A Expression Level Is Associated with Better Prognosis in Breast Cancer Patients
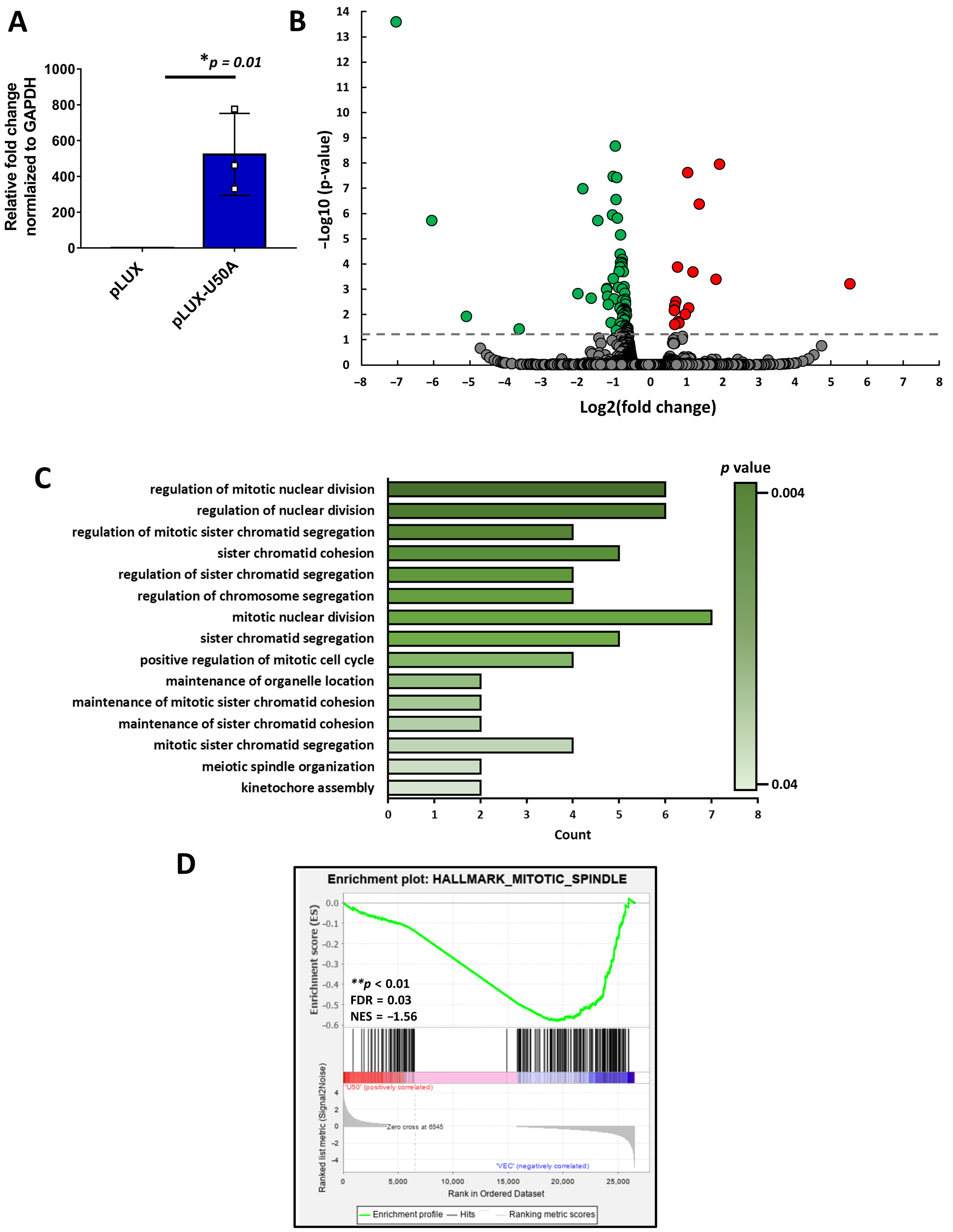
3.2. Mitosis-Related Transcriptomic Changes Regulated by U50A
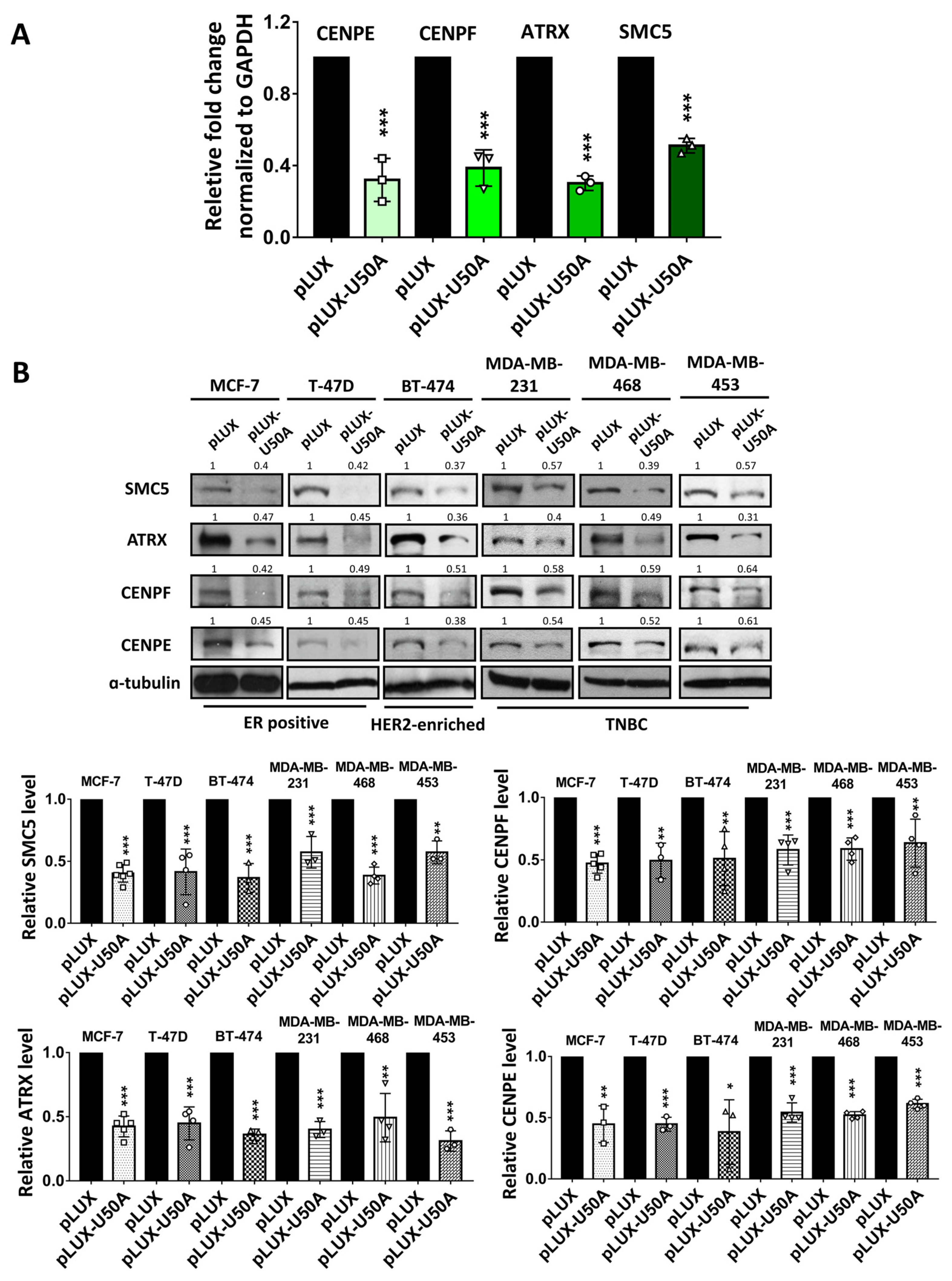
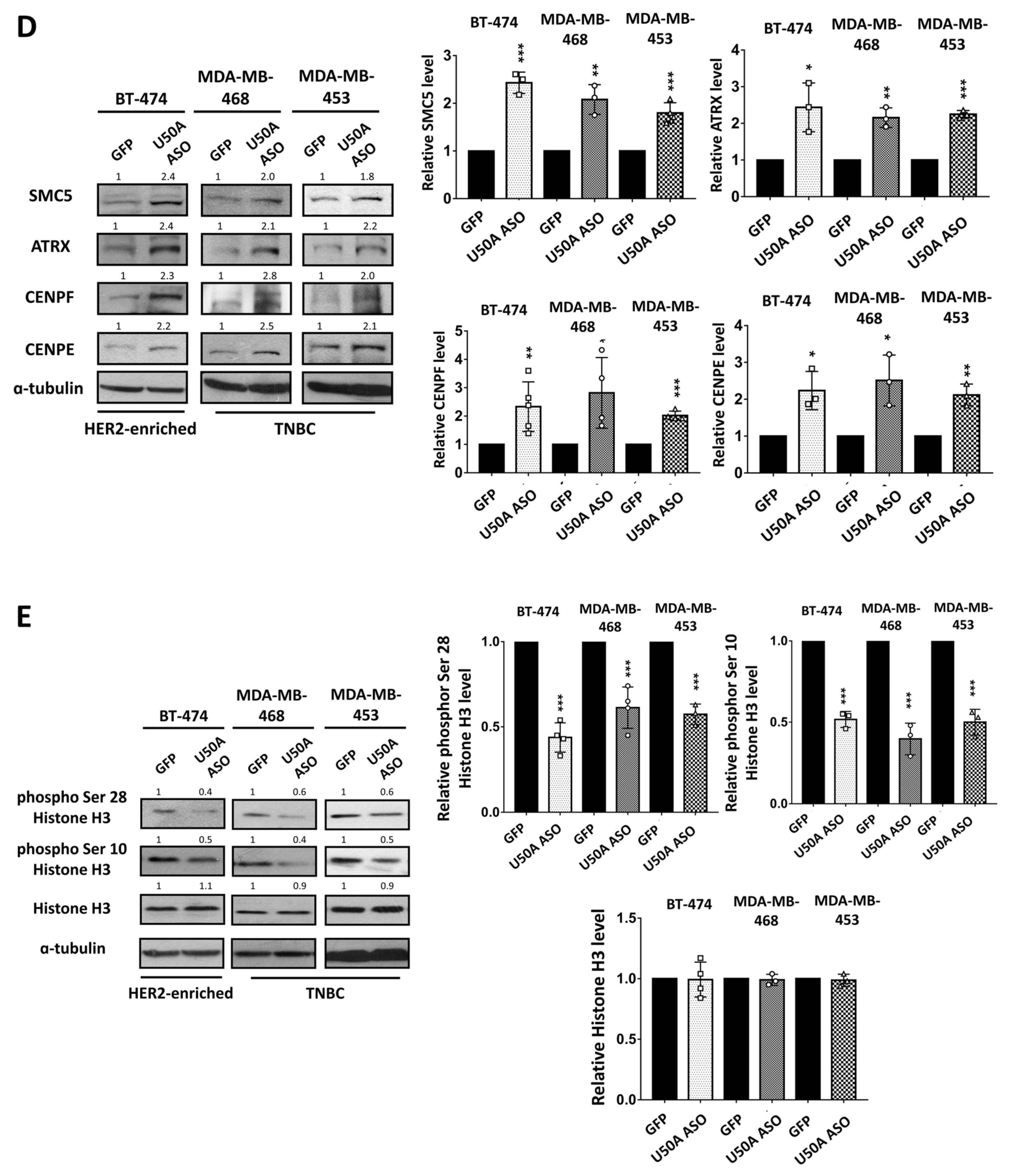
3.3. Identification of the Downstream Targets of U50A Involved in Mitosis
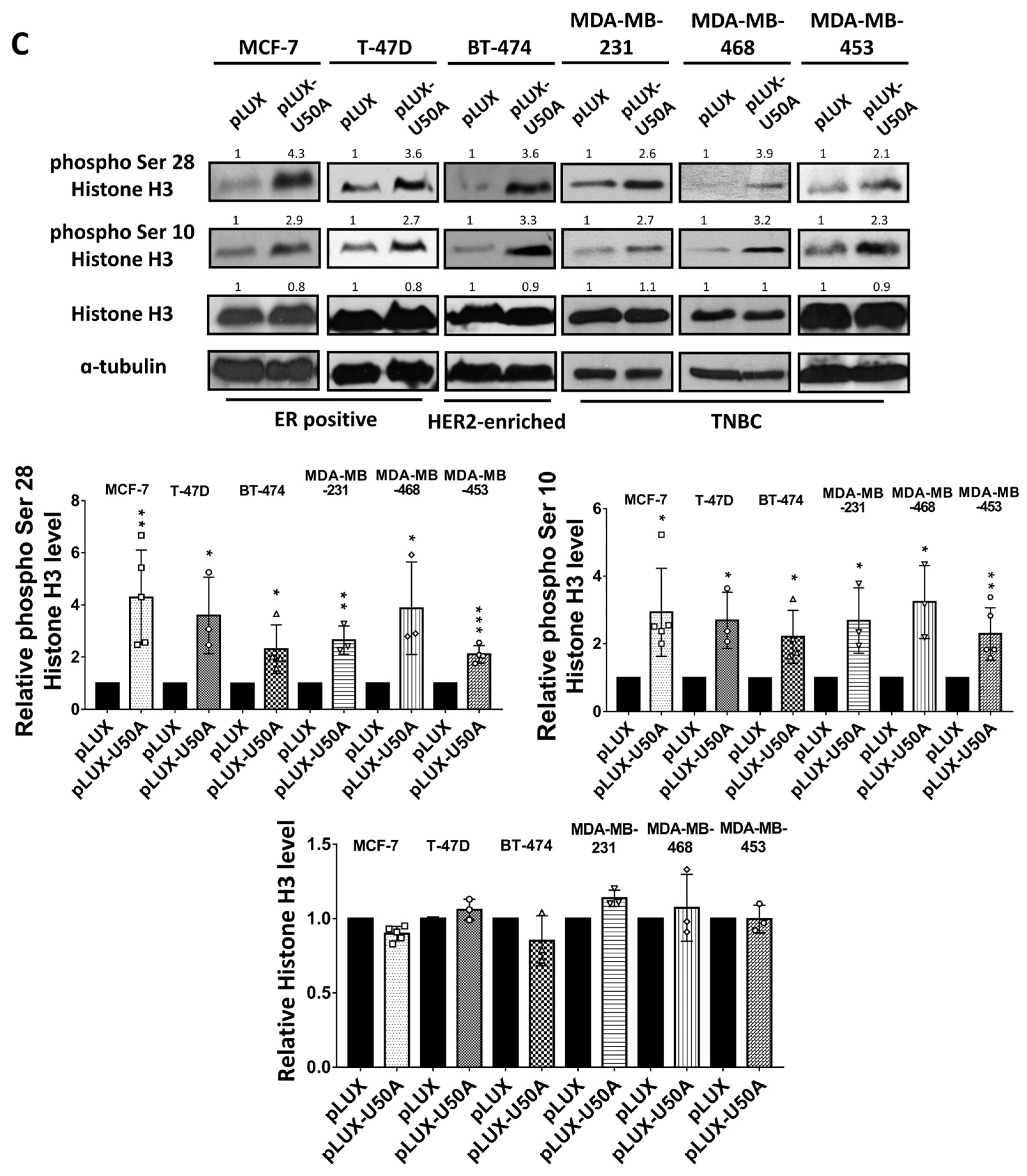
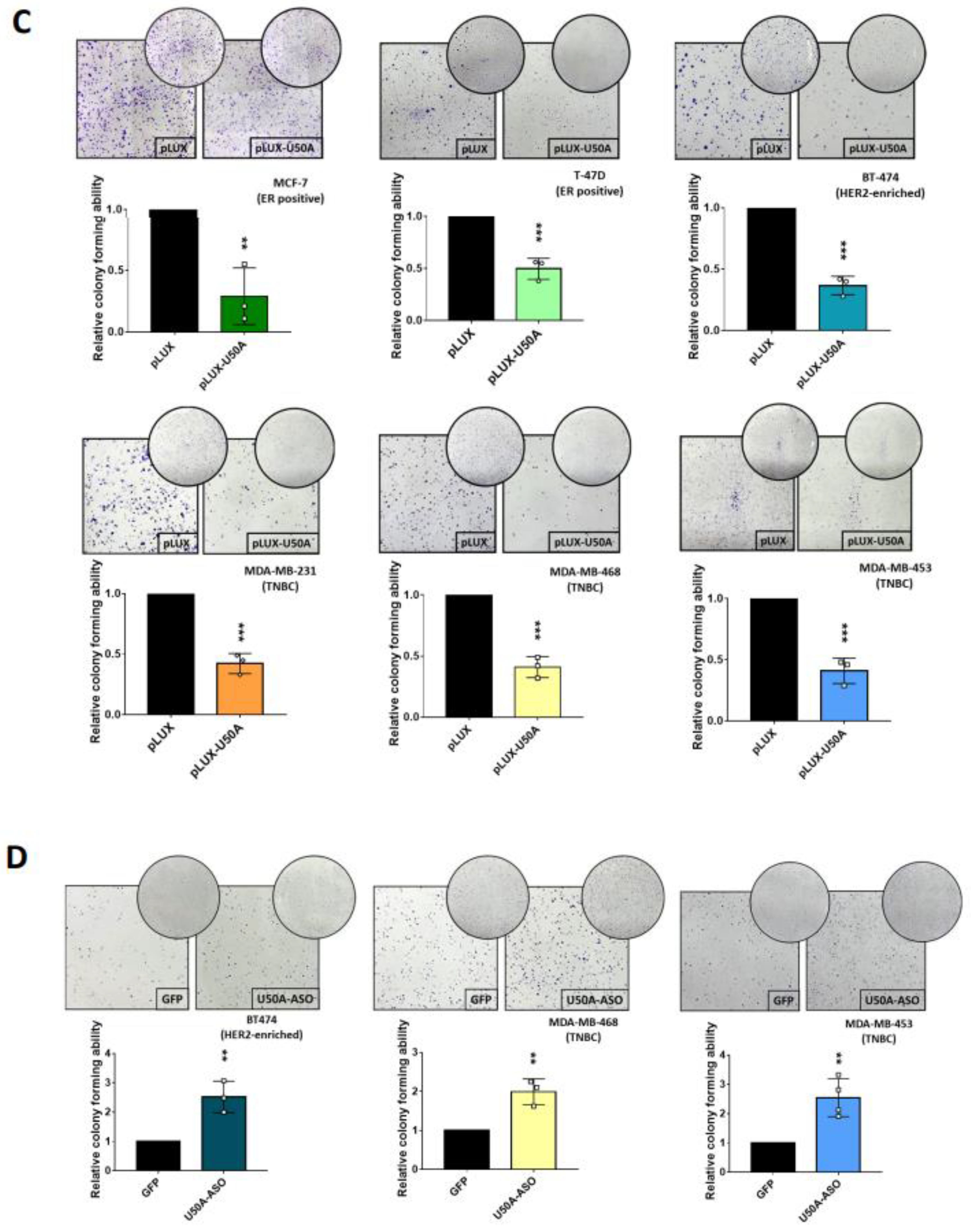
3.4. U50A Prolongs Mitosis and Suppresses Colony-Forming Ability in Breast Cancer Cells
3.5. Correlation of U50A Expression with Mitotic Cells in Human Breast Cancer Tissues
4. Discussion
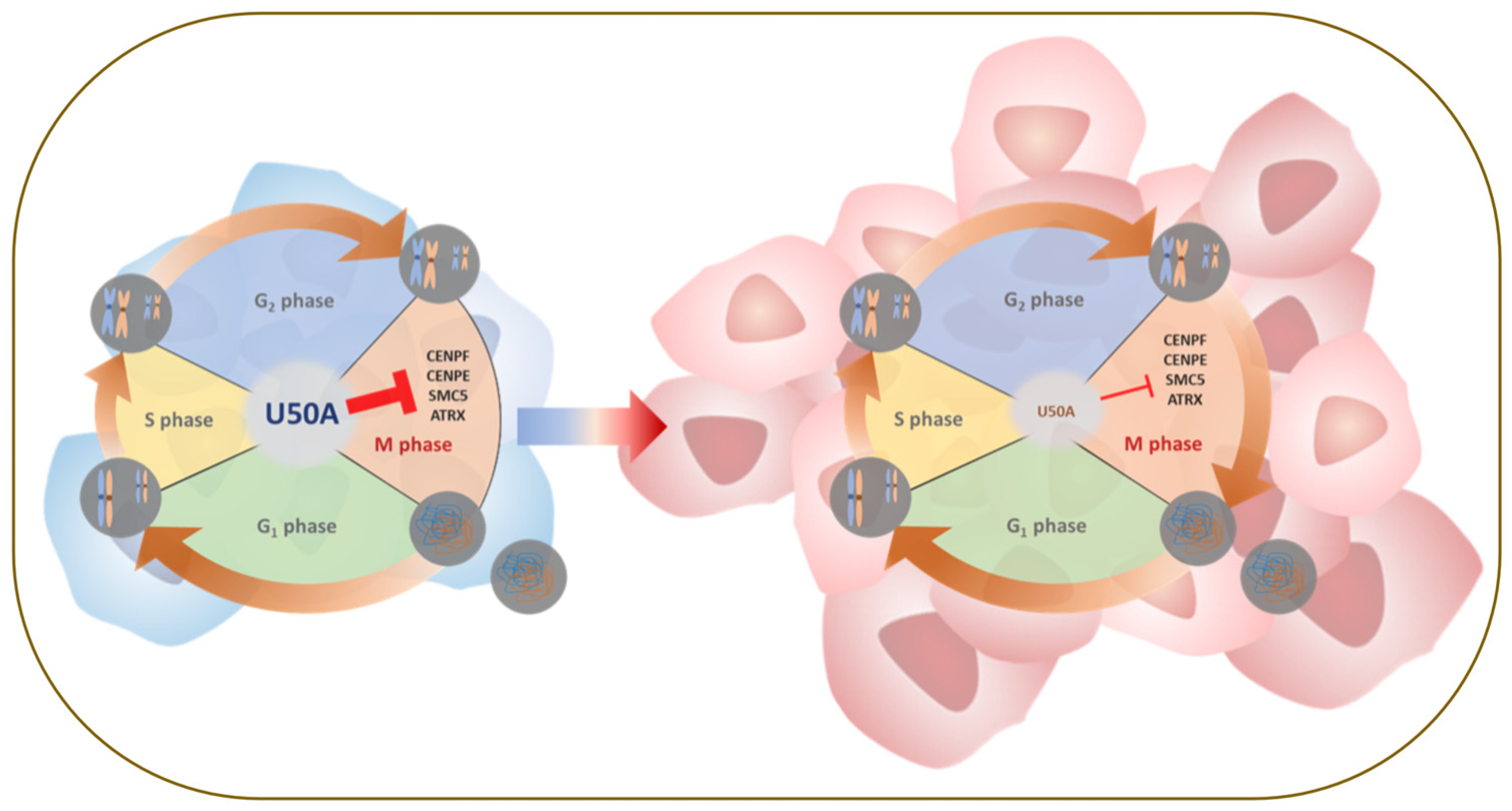
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dai, X.; Li, T.; Bai, Z.; Yang, Y.; Liu, X.; Zhan, J.; Shi, B. Breast Cancer Intrinsic Subtype Classification, Clinical Use and Future Trends. Am. J. Cancer Res. 2015, 5, 2929–2943. [Google Scholar] [PubMed]
- Domeyer, P.J.; Sergentanis, T.N. New Insights into the Screening, Prompt Diagnosis, Management, and Prognosis of Breast. Cancer J. Oncol. 2020, 2020, 8597892. [Google Scholar] [CrossRef] [Green Version]
- Lumachi, F.; Santeufemia, D.A.; Basso, S.M. Current Medical Treatment of Estrogen Receptor-Positive Breast Cancer. World J. Biol. Chem. 2015, 6, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Mutebi, M.; Anderson, B.O.; Duggan, C.; Adebamowo, C.; Agarwal, G.; Ali, Z.; Bird, P.; Bourque, J.-M.; DeBoer, R.; Gebrim, L.H.; et al. Breast Cancer Treatment: A Phased Approach to Implementation. Cancer 2020, 126, 2365–2378. [Google Scholar] [CrossRef]
- Filipowicz, W.; Pogacic, V. Biogenesis of Small Nucleolar Ribonucleoproteins. Curr. Opin. Cell Biol. 2002, 14, 319–327. [Google Scholar] [CrossRef]
- Kufel, J.; Grzechnik, P. Small Nucleolar RNAs Tell a Different Tale. Trends Genet. 2019, 35, 104–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dragon, F.O.; Lemay, V.; Trahan, C. snoRNAs: Biogenesis, Structure and Function. Encycl. Life Sci. 2006. [Google Scholar] [CrossRef]
- Lui, L.; Lowe, T. Small Nucleolar RNAs and RNA-Guided Post-Transcriptional Modification. Essays Biochem. 2013, 54, 53–77. [Google Scholar]
- Massenet, S.; Bertrand, E.; Verheggen, C. Assembly and Trafficking of box C/D and H/ACA snoRNPs. RNA Biol. 2017, 14, 680–692. [Google Scholar] [CrossRef] [Green Version]
- Janin, M.; Coll-SanMartin, L.; Esteller, M. Disruption of the RNA Modifications that Target the Ribosome Translation Machinery in Human Cancer. Mol. Cancer 2020, 19, 70. [Google Scholar] [CrossRef] [Green Version]
- Williams, G.T.; Farzaneh, F. Are snoRNAs and snoRNA Host Genes New Players in Cancer? Nat. Rev. Cancer 2012, 12, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Thorenoor, N.; Slaby, O. Small Nucleolar RNAs Functioning and Potential Roles in Cancer. Tumour Biol. 2015, 36, 41–53. [Google Scholar] [CrossRef]
- Liang, J.N.; Wen, J.Y.; Huang, Z.; Chen, X.-P.; Zhang, B.-X.; Chu, L. Small Nucleolar RNAs: Insight into Their Function in Cancer. Front. Oncol. 2019, 9, 587. [Google Scholar] [CrossRef] [Green Version]
- Zhou, F.B.; Liu, Y.; Rohde, C.; Pauli, C.; Gerloff, D.; Köhn, M.; Misiak, D.; Bäumer, N.; Cui, C.; Göllner, S.; et al. AML1-ETO Requires Enhanced C/D box snoRNA/RNP Formation to Induce Self-Renewal and Leukaemia. Nat. Cell Biol. 2017, 19, 844–855. [Google Scholar] [CrossRef]
- McMahon, M.; Contreras, A.; Holm, M.; Uechi, T.; Forester, C.M.; Pang, X.; Jackson, C.; Calvert, M.E.; Chen, B.; Quigley, D.A.; et al. A Single H/ACA Small Nucleolar RNA Mediates Tumor Suppression Downstream of Oncogenic RAS. eLife 2019, 8, e48847. [Google Scholar] [CrossRef]
- Pacilli, A.; Ceccarelli, C.; Treré, D.; Montanaro, L. SnoRNA U50 Levels Are Regulated by Cell Proliferation and rRNA Transcription. Int. J. Mol. Sci. 2013, 14, 14923–14935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Han, L.; Wei, J.; Zhang, K.; Shi, Z.; Duan, R.; Li, S.; Zhou, X.; Pu, P.; Zhang, J.; et al. SNORD76, a Box C/D snoRNA, Acts as a Tumor Suppressor in Glioblastoma. Sci. Rep. 2015, 5, 8588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siprashvili, Z.; Webster, D.E.; Johnston, D.; Shenoy, R.M.; Ungewickell, A.J.; Bhaduri, A.; Flockhart, R.; Zarnegar, B.J.; Che, Y.; Meschi, F.; et al. The Noncoding RNAs SNORD50A and SNORD50B bind K-Ras and Are Recurrently Deleted in Human Cancer. Nat. Genet. 2016, 48, 53–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, X.-Y.; Guo, P.; Boyd, J.; Sun, X.; Li, Q.; Zhou, W.; Dong, J.-T. Implication of snoRNA U50 in Human Breast Cancer. J. Genet. Genom. 2009, 36, 447–454. [Google Scholar] [CrossRef] [Green Version]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [Green Version]
- McIntosh, J.R. Mitosis. Cold Spring Harb. Perspect. Biol. 2016, 8, a023218. [Google Scholar] [CrossRef]
- Batty, P.; Gerlich, D.W. Mitotic Chromosome Mechanics: How Cells Segregate Their Genome. Trends Cell Biol. 2019, 29, 717–726. [Google Scholar] [CrossRef] [Green Version]
- Behlke-Steinert, S.; Touat-Todeschini, L.; Skoufias, D.A.; Margolis, R.L. SMC5 and MMS21 Are Required for Chromosome Cohesion and Mitotic Progression. Cell Cycl. 2009, 8, 2211–2218. [Google Scholar] [CrossRef]
- Gallego-Paez, L.M.; Tanaka, H.; Bando, M.; Takahashi, M.; Nozaki, N.; Nakato, R.; Shirahige, K.; Hirota, T. Smc5/6-Mediated Regulation of Replication Progression Contributes to Chromosome Assembly during Mitosis in Human Cells. Mol. Biol. Cell 2014, 25, 302–317. [Google Scholar] [CrossRef]
- De La Fuente, R.; Baumann, C.; Viveiros, M.M. Role of ATRX in Chromatin Structure and Function: Implications for Chromosome Instability and Human Disease. Reproduction 2011, 142, 221–234. [Google Scholar] [CrossRef] [PubMed]
- Ciossani, G.; Overlack, K.; Petrovic, A.; Huis In ‘t Veld, P.J.; Koerner, C.; Wohlgemuth, S.; Maffini, S.; Musacchio, A. The Kinetochore Proteins CENP-E and CENP-F Directly and Specifically Interact with Distinct BUB Mitotic Checkpoint Ser/Thr Kinases. J. Biol. Chem. 2018, 293, 10084–10101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hans, F.; Dimitrov, S. Histone H3 Phosphorylation and Cell Division. Oncogene 2001, 20, 3021–3027. [Google Scholar] [CrossRef] [Green Version]
- Crosio, C.; Fimia, G.M.; Loury, R.; Kimura, M.; Okano, Y.; Zhou, H.Y.; Sen, S.; Allis, C.D.; Sassone-Corsi, P. Mitotic Phosphorylation of Histone H3: Spatio-Temporal Regulation by Mammalian Aurora Kinases. Mol. Cell. Biol. 2002, 22, 874–885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perez-Cadahia, B.; Drobic, B.; Davie, J.R. H3 Phosphorylation: Dual Role in Mitosis and Interphase. Biochem. Cell. Biol. 2009, 87, 695–709. [Google Scholar]
- Blajeski, A.L.; Phan, V.A.; Kottke, T.J.; Kaufmann, S.H. G(1) and G(2) Cell-Cycle Arrest Following Microtubule Depolymerization in Human Breast Cancer Cells. J. Clin. Investig. 2002, 110, 91–99. [Google Scholar] [CrossRef]
- Bhaud, Y.; Guillebault, D.; Lennon, J.; Defacque, H.; Soyer-Gobillard, M.O.; Moreau, H. Morphology and Behaviour of Dinoflagellate Chromosomes during the Cell Cycle and Mitosis. J. Cell Sci. 2000, 113, 1231–1239. [Google Scholar] [CrossRef] [PubMed]
- Nayyar, V.K.; Shire, K.; Frappier, L. Mitotic Chromosome Interactions of Epstein-Barr Nuclear Antigen 1 (EBNA1) and Human EBNA1-Binding Protein 2 (EBP2). J. Cell Sci. 2009, 122, 4341–4350. [Google Scholar] [CrossRef] [Green Version]
- Graber-Feesl, C.L.; Pederson, K.D.; Aney, K.J.; Shima, N. Mitotic DNA Synthesis Is Differentially Regulated between Cancer and Noncancerous Cells. Mol. Cancer Res. 2019, 17, 1687–1698. [Google Scholar] [CrossRef] [PubMed]
- Mei, Y.P.; Liao, J.P.; Shen, J.; Yu, L.; Liu, B.L.; Liu, L.; Li, R.Y.; Ji, L.; Dorsey, S.G.; Jiang, Z.R.; et al. Small Nucleolar RNA 42 Acts as an Oncogene in Lung Tumorigenesis. Oncogene 2012, 31, 2794–2804. [Google Scholar] [CrossRef] [PubMed]
- Shan, L.N.; Zhao, M.J.; Lu, Y.; Ning, H.J.; Yang, S.M.; Song, Y.G.; Chai, W.S.; Shi, X.B. CENPE Promotes Lung Adenocarcinoma Proliferation and Is Directly Regulated by FOXM1. Int. J. Oncol. 2019, 55, 257–266. [Google Scholar]
- Craske, B.; Welburn, J.P.I. Leaving No-One Behind: How CENP-E Facilitates Chromosome Alignment. Kinet. Chromosom. Segreg. 2020, 64, 313–324. [Google Scholar]
- Schafer-Hales, K.; Iaconelli, J.; Snyder, J.P.; Prussia, A.; Nettles, J.H.; El-Naggar, A.; Khuri, F.R.; Giannakakou, P.; Marcus, A.I. Farnesyl Transferase Inhibitors Impair Chromosomal Maintenance in Cell Lines and Human Tumors by Compromising CENP-E and CENP-F function. Mol. Cancer Ther. 2007, 6, 1317–1328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
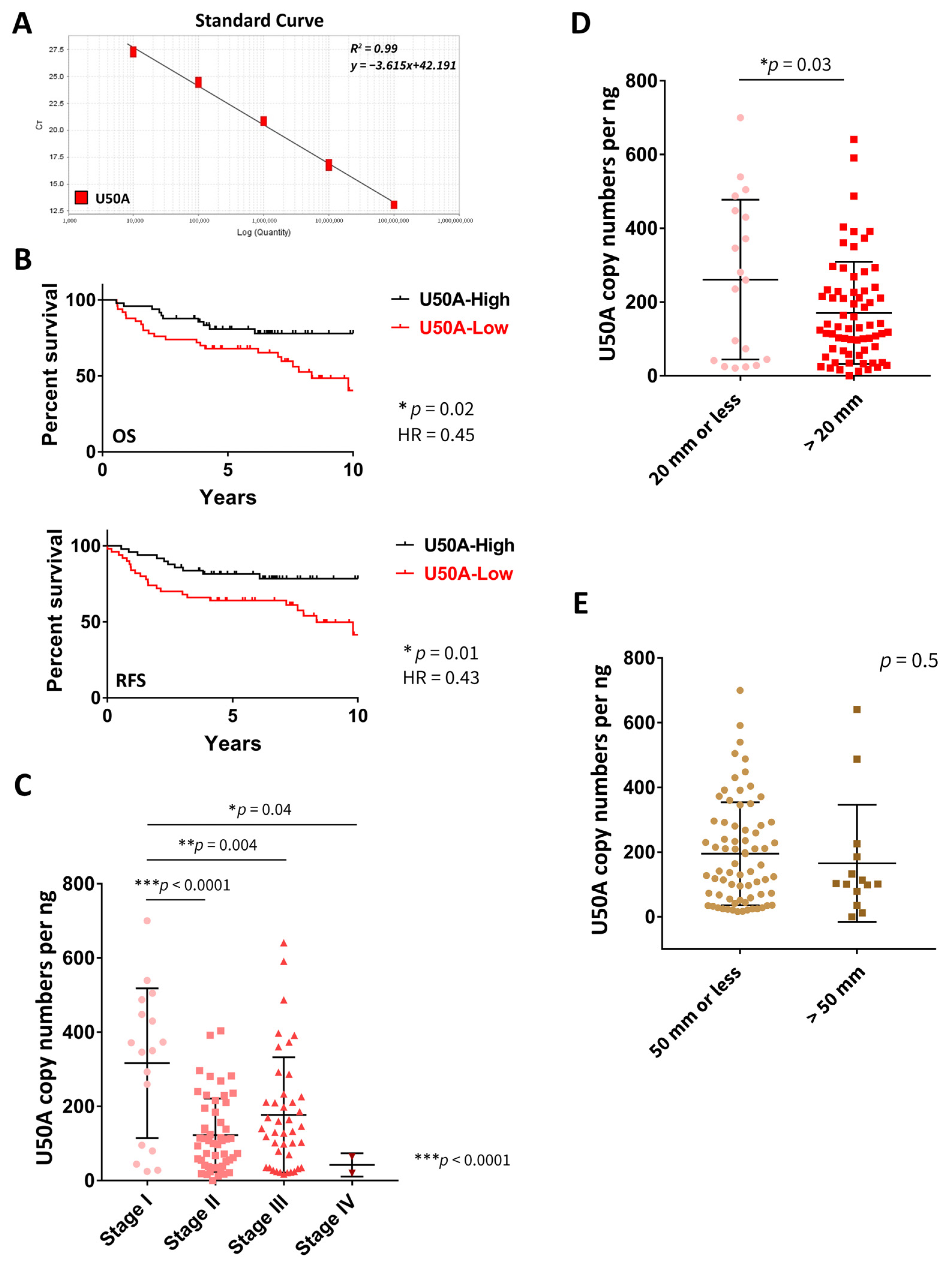




| Characteristics | No. of Patients (%) |
|---|---|
| Median age (range, years) | 50 (31–84) |
| Age ≤ 50 | 52 (46%) |
| Age > 50 | 62 (54%) |
| Pathologic characteristic | - |
| Invasive ductal carcinoma | 97 (85%) |
| Invasive lobular carcinoma | 1 (1%) |
| Others | 16 (14%) |
| Others: Mucinous carcinoma, papillary carcinoma, and invasive carcinoma | |
| Breast cancer subtype | - |
| Luminal A/B | 68 (60%) |
| HER2-enriched | 28 (24.6%) |
| Triple-negative | 18 (15.4%) |
| Variable | All Breast Cancer Patients in This Study | |||
|---|---|---|---|---|
| Overall Survival (OS) | Relapse-Free Survival (RFS) | |||
| p | HR | p | HR | |
| U50A | 0.02 * | 0.45 # | 0.01 * | 0.43 # |
| U50B | 0.32 | 0.7 | 0.36 | 0.72 |
| SNHG5 | 0.2 | 0.64 | 0.22 | 0.65 |
| All | Stage (Mean ± S.D.) | ||||
|---|---|---|---|---|---|
| Marker | I | II | III | IV | p |
| U50A | 316.3 ± 201.7 | 122.3 ± 98.98 | 177.1 ± 155.3 | 42.2 ± 31.4 | *** <0.0001 |
| U50B | 10.48 ± 17.28 | 6.38 ± 10.86 | 5.005 ± 7.4 | 1.705 ± 2.39 | 0.62 |
| SNHG5 | 36.04 ± 44.1 | 19.07 ± 38.69 | 60.72 ± 117.1 | 0.49 ± 0.67 | 0.07 |
| Selected Targets | Fold Change from RNA-Sequencing (pLUX-U50A/pLUX) |
|---|---|
| CENPE | 0.49 |
| CENPF | 0.58 |
| SMC5 | 0.63 |
| ATRX | 0.65 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.-N.; Wang, M.-Y.; Chen, Y.-T.; Kuo, Y.-L.; Chen, P.-S. Expression of SnoRNA U50A Is Associated with Better Prognosis and Prolonged Mitosis in Breast Cancer. Cancers 2021, 13, 6304. https://doi.org/10.3390/cancers13246304
Li J-N, Wang M-Y, Chen Y-T, Kuo Y-L, Chen P-S. Expression of SnoRNA U50A Is Associated with Better Prognosis and Prolonged Mitosis in Breast Cancer. Cancers. 2021; 13(24):6304. https://doi.org/10.3390/cancers13246304
Chicago/Turabian StyleLi, Jie-Ning, Ming-Yang Wang, Yi-Ting Chen, Yao-Lung Kuo, and Pai-Sheng Chen. 2021. "Expression of SnoRNA U50A Is Associated with Better Prognosis and Prolonged Mitosis in Breast Cancer" Cancers 13, no. 24: 6304. https://doi.org/10.3390/cancers13246304






