Grading Evolution and Contemporary Prognostic Biomarkers of Clinically Significant Prostate Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Historical Development of Prostate Cancer Grading
3. Contemporary Practices in Gleason Grading–ISUP Grade Groups
4. Quantitative Gleason and Artificial Intelligence
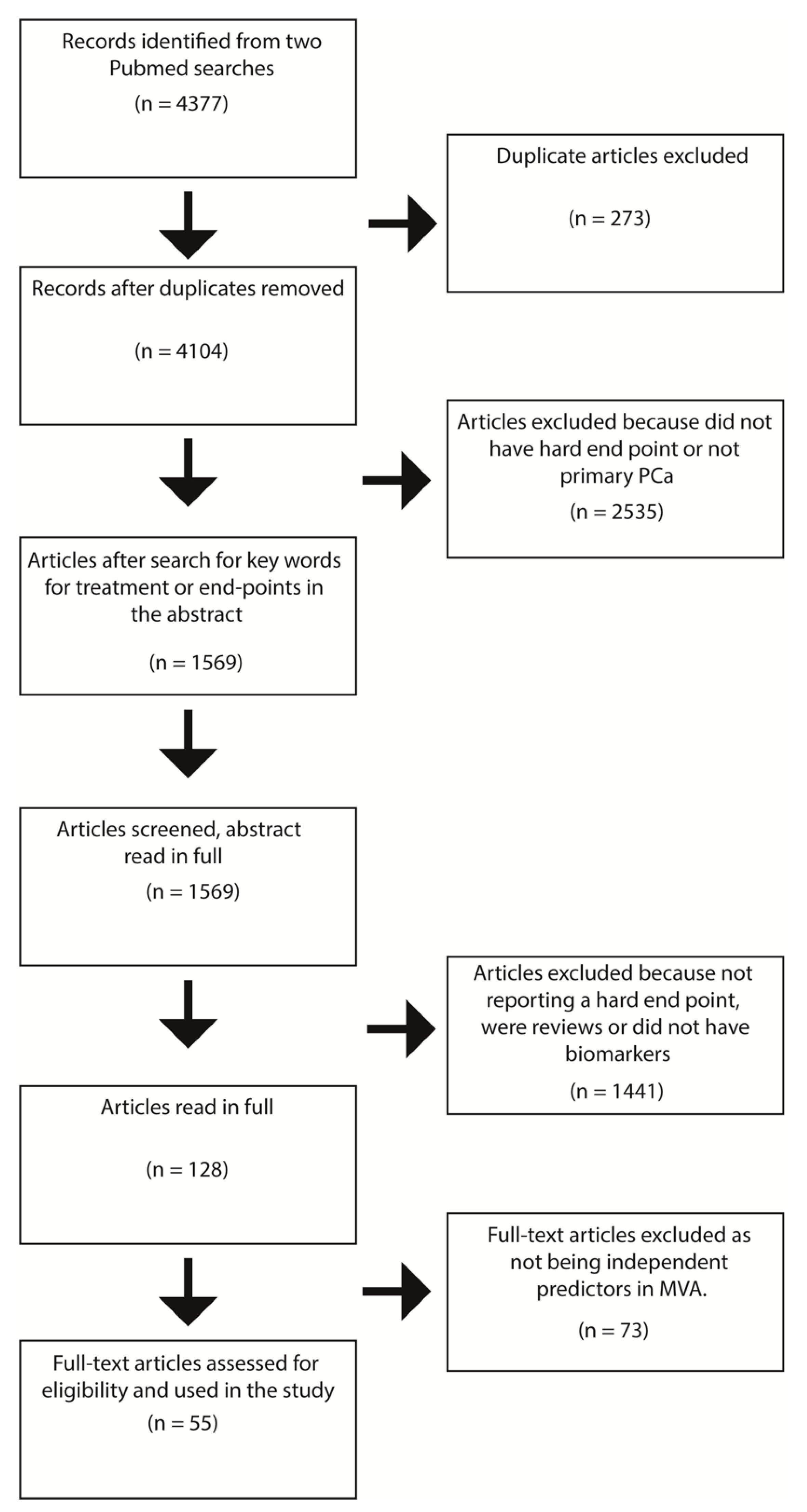
5. Systematic Review of Biomarkers Related to Clinically Relevant Endpoints
5.1. Results
5.1.1. Biopsy-Based Biomarkers—Radiation Therapy and Radical Prostatectomy
5.1.2. RP Specimen-Based Markers
5.1.3. Peripheral Blood Markers
5.1.4. Active Surveillance
6. Discussion
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
Search Terms Used in PUBMED
References
- Visser, W.C.H.; de Jong, H.; Melchers, W.J.G.; Mulders, P.F.A.; Schalken, J.A. Commercialized Blood-, Urinary- and Tissue-Based Biomarker Tests for Prostate Cancer Diagnosis and Prognosis. Cancers 2020, 12, 3790. [Google Scholar] [CrossRef] [PubMed]
- Adams, J. The case of scirrhous of the prostate gland with corresponding affliction of the lymphatic glands in the lumbar region and in the pelvis. Lancet 1853, 1, 393. [Google Scholar]
- Broders, A.C. The grading of carcinoma. Minn. Med. 1925, 8, 1730–1925. [Google Scholar]
- Shelley, H.S.; Auerbach, S.H.; Classen, K.L.; Marks, C.H.; Wiederanders, R.E. Carcinoma of the prostate: A new system of classification. AMA Arch. Surg. 1958, 77, 751–756. [Google Scholar] [CrossRef]
- Gleason, D.F. Classification of prostatic carcinomas. Cancer Chemother. Rep. 1966, 50, 125–128. [Google Scholar]
- Gleason, D.F.; Mellinger, G.T. Prediction of prognosis for prostatic adenocarcinoma by combined histological grading and clinical staging. J. Urol. 1974, 111, 58–64. [Google Scholar] [CrossRef]
- Albertsen, P.C.; Hanley, J.A.; Gleason, D.F.; Barry, M.J. Competing risk analysis of men aged 55 to 74 years at diagnosis managed conservatively for clinically localized prostate cancer. JAMA 1998, 280, 975–980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milonas, D.; Venclovas, Z.; Muilwijk, T.; Jievaltas, M.; Joniau, S. External validation of Memorial Sloan Kettering Cancer Center nomogram and prediction of optimal candidate for lymph node dissection in clinically localized prostate cancer. Cent. Eur. J. Urol. 2020, 73, 19–25. [Google Scholar] [CrossRef]
- Cooperberg, M.R.; Pasta, D.J.; Elkin, E.P.; Litwin, M.S.; Latini, D.M.; Du Chane, J.; Carroll, P.R. The University of California, San Francisco Cancer of the Prostate Risk Assessment score: A straightforward and reliable preoperative predictor of disease recurrence after radical prostatectomy. J. Urol. 2005, 173, 1938–1942. [Google Scholar] [CrossRef] [Green Version]
- Eifler, J.B.; Feng, Z.; Lin, B.M.; Partin, M.T.; Humphreys, E.B.; Han, M.; Epstein, J.I.; Walsh, P.C.; Trock, B.J.; Partin, A.W. An updated prostate cancer staging nomogram (Partin tables) based on cases from 2006 to 2011. BJU Int. 2013, 111, 22–29. [Google Scholar] [CrossRef]
- Hernandez, D.J.; Nielsen, M.E.; Han, M.; Partin, A.W. Contemporary evaluation of the D’amico risk classification of prostate cancer. Urology 2007, 70, 931–935. [Google Scholar] [CrossRef]
- Zelic, R.; Garmo, H.; Zugna, D.; Stattin, P.; Richiardi, L.; Akre, O.; Pettersson, A. Predicting Prostate Cancer Death with Different Pretreatment Risk Stratification Tools: A Head-to-head Comparison in a Nationwide Cohort Study. Eur. Urol. 2020, 77, 180–188. [Google Scholar] [CrossRef]
- Epstein, J.I. An update of the Gleason grading system. J. Urol. 2010, 183, 433–440. [Google Scholar] [CrossRef]
- Gordetsky, J.; Epstein, J. Grading of prostatic adenocarcinoma: Current state and prognostic implications. Diagn. Pathol. 2016, 11, 25. [Google Scholar] [CrossRef] [Green Version]
- Epstein, J.I.; Egevad, L.; Amin, M.B.; Delahunt, B.; Srigley, J.R.; Humphrey, P.A. The 2014 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma. Am. J. Surg. Pathol. 2016, 40, 244–252. [Google Scholar] [CrossRef]
- Ross, H.M.; Kryvenko, O.N.; Cowan, J.E.; Simko, J.P.; Wheeler, T.M.; Epstein, J.I. Do adenocarcinomas of the prostate with Gleason score (GS) ≤6 have the potential to metastasize to lymph nodes? Am. J. Surg. Pathol. 2012, 36, 1346–1352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walsh, P.C. The Gleason Grading System: A Complete Guide for Pathologists and Clinicians; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013. [Google Scholar]
- Kweldam, C.F.; Wildhagen, M.F.; Steyerberg, E.W.; Bangma, C.H.; van der Kwast, T.H.; van Leenders, G.J.L.H. Cribriform growth is highly predictive for postoperative metastasis and disease-specific death in Gleason score 7 prostate cancer. Mod. Pathol. 2015, 28, 457–464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Leenders, G.J.L.H.; van der Kwast, T.H.; Grignon, D.J.; Evans, A.J.; Kristiansen, G.; Kweldam, C.F.; Litjens, G.; McKenney, J.K.; Melamed, J.; Mottet, N.; et al. The 2019 International Society of Urological Pathology (ISUP) Consensus Conference on Grading of Prostatic Carcinoma. Am. J. Surg. Pathol. 2020, 44, e87–e99. [Google Scholar] [CrossRef]
- Smith, S.C.; Gandhi, J.S.; Moch, H.; Aron, M.; Compérat, E.; Paner, G.P.; McKenney, J.K.; Amin, M.B. Similarities and Differences in the 2019 ISUP and GUPS Recommendations on Prostate Cancer Grading: A Guide for Practicing Pathologists. Adv. Anat. Pathol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Sauter, G.; Steurer, S.; Clauditz, T.S.; Krech, T.; Wittmer, C.; Lutz, F.; Lennartz, M.; Janssen, T.; Hakimi, N.; Simon, R.; et al. Clinical Utility of Quantitative Gleason Grading in Prostate Biopsies and Prostatectomy Specimens. Eur. Urol. 2016, 69, 592–598. [Google Scholar] [CrossRef]
- Sauter, G.; Clauditz, T.; Steurer, S.; Wittmer, C.; Büscheck, F.; Krech, T.; Lutz, F.; Lennartz, M.; Harms, L.; Lawrenz, L.; et al. Integrating Tertiary Gleason 5 Patterns into Quantitative Gleason Grading in Prostate Biopsies and Prostatectomy Specimens. Eur. Urol. 2018, 73, 674–683. [Google Scholar] [CrossRef]
- Nagpal, K.; Foote, D.; Tan, F.; Liu, Y.; Chen, P.-H.C.; Steiner, D.F.; Manoj, N.; Olson, N.; Smith, J.L.; Mohtashamian, A.; et al. Development and Validation of a Deep Learning Algorithm for Gleason Grading of Prostate Cancer From Biopsy Specimens. JAMA Oncol. 2020, 6, 1372. [Google Scholar] [CrossRef] [PubMed]
- Goldenberg, S.L.; Nir, G.; Salcudean, S.E. A new era: Artificial intelligence and machine learning in prostate cancer. Nat. Rev. Urol. 2019, 16, 391–403. [Google Scholar] [CrossRef]
- Ström, P.; Kartasalo, K.; Olsson, H.; Solorzano, L.; Delahunt, B.; Berney, D.M.; Bostwick, D.G.; Evans, A.J.; Grignon, D.J.; Humphrey, P.A.; et al. Artificial intelligence for diagnosis and grading of prostate cancer in biopsies: A population-based, diagnostic study. Lancet Oncol. 2020, 21, 222–232. [Google Scholar] [CrossRef]
- Pantanowitz, L.; Quiroga-Garza, G.M.; Bien, L.; Heled, R.; Laifenfeld, D.; Linhart, C.; Sandbank, J.; Shach, A.A.; Shalev, V.; Vecsler, M.; et al. An artificial intelligence algorithm for prostate cancer diagnosis in whole slide images of core needle biopsies: A blinded clinical validation and deployment study. Lancet Digital Health 2020, 2, e407–e416. [Google Scholar] [CrossRef]
- Chandramouli, S.; Leo, P.; Lee, G.; Elliott, R.; Davis, C.; Zhu, G.; Fu, P.; Epstein, J.I.; Veltri, R.; Madabhushi, A. Computer Extracted Features from Initial H&E Tissue Biopsies Predict Disease Progression for Prostate Cancer Patients on Active Surveillance. Cancers 2020, 12, 2708. [Google Scholar] [CrossRef]
- Tollefson, M.K.; Karnes, R.J.; Kwon, E.D.; Lohse, C.M.; Rangel, L.J.; Mynderse, L.A.; Cheville, J.C.; Sebo, T.J. Prostate cancer Ki-67 (MIB-1) expression, perineural invasion, and gleason score as biopsy-based predictors of prostate cancer mortality: The Mayo model. Mayo Clin. Proc. 2014, 89, 308–318. [Google Scholar] [CrossRef]
- Verhoven, B.; Yan, Y.; Ritter, M.; Khor, L.-Y.; Hammond, E.; Jones, C.; Amin, M.; Bahary, J.-P.; Zeitzer, K.; Pollack, A. Ki-67 is an independent predictor of metastasis and cause-specific mortality for prostate cancer patients treated on Radiation Therapy Oncology Group (RTOG) 94-08. Int. J. Radiat. Oncol. Biol. Phys. 2013, 86, 317–323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pollack, A.; Dignam, J.J.; Diaz, D.A.; Wu, Q.; Stoyanova, R.; Bae, K.; Dicker, A.P.; Sandler, H.; Hanks, G.E.; Feng, F.Y. A tissue biomarker-based model that identifies patients with a high risk of distant metastasis and differential survival by length of androgen deprivation therapy in RTOG protocol 92-02. Clin. Cancer Res. 2014, 20, 6379–6388. [Google Scholar] [CrossRef] [Green Version]
- Krauss, D.J.; Hayek, S.; Amin, M.; Ye, H.; Kestin, L.L.; Zadora, S.; Vicini, F.A.; Cotant, M.; Brabbins, D.S.; Ghilezan, M.I.; et al. Prognostic significance of neuroendocrine differentiation in patients with Gleason score 8-10 prostate cancer treated with primary radiotherapy. Int. J. Radiat. Oncol. Biol. Phys. 2011, 81, e119–e125. [Google Scholar] [CrossRef]
- Cattrini, C.; Rubagotti, A.; Nuzzo, P.V.; Zinoli, L.; Salvi, S.; Boccardo, S.; Perachino, M.; Cerbone, L.; Vallome, G.; Latocca, M.M.; et al. Overexpression of Periostin in Tumor Biopsy Samples Is Associated With Prostate Cancer Phenotype and Clinical Outcome. Clin. Genitourin. Cancer 2018, 16, e1257–e1265. [Google Scholar] [CrossRef]
- Jacobs, C.; Tumati, V.; Kapur, P.; Yan, J.; Xie, X.-J.; Hannan, R.; Hsieh, J.-T.; Kim, D.W.N.; Saha, D. Pretreatment biopsy analysis of DAB2IP identifies subpopulation of high-risk prostate cancer patients with worse survival following radiation therapy. Cancer Med. 2015, 4, 1844–1852. [Google Scholar] [CrossRef] [PubMed]
- Kammerer-Jacquet, S.-F.; Ahmad, A.; Møller, H.; Sandu, H.; Scardino, P.; Soosay, G.; Beltran, L.; Cuzick, J.; Berney, D.M. Ki-67 is an independent predictor of prostate cancer death in routine needle biopsy samples: Proving utility for routine assessments. Mod. Pathol. 2019, 32, 1303–1309. [Google Scholar] [CrossRef]
- Megas, G.; Chrisofos, M.; Anastasiou, I.; Tsitlidou, A.; Choreftaki, T.; Deliveliotis, C. Estrogen receptor (α and β) but not androgen receptor expression is correlated with recurrence, progression and survival in post prostatectomy T3N0M0 locally advanced prostate cancer in an urban Greek population. Asian J. Androl. 2015, 17, 98–105. [Google Scholar] [CrossRef]
- Grindstad, T.; Skjefstad, K.; Andersen, S.; Ness, N.; Nordby, Y.; Al-Saad, S.; Fismen, S.; Donnem, T.; Khanehkenari, M.R.; Busund, L.-T.; et al. Estrogen receptors α and β and aromatase as independent predictors for prostate cancer outcome. Sci. Rep. 2016, 6, 33114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujimura, T.; Takahashi, S.; Urano, T.; Tanaka, T.; Zhang, W.; Azuma, K.; Takayama, K.; Obinata, D.; Murata, T.; Horie-Inoue, K.; et al. Clinical significance of steroid and xenobiotic receptor and its targeted gene CYP3A4 in human prostate cancer. Cancer Sci. 2012, 103, 176–180. [Google Scholar] [CrossRef]
- Quinn, D.I.; Stricker, P.D.; Kench, J.G.; Grogan, J.; Haynes, A.-M.; Henshall, S.M.; Grygiel, J.J.; Delprado, W.; Turner, J.J.; Horvath, L.G.; et al. p53 nuclear accumulation as an early indicator of lethal prostate cancer. Br. J. Cancer 2019, 121, 578–583. [Google Scholar] [CrossRef] [PubMed]
- Jiao, L.; Shen, D.; Liu, G.; Jia, J.; Geng, J.; Wang, H.; Sun, Y. PPM1D as a novel biomarker for prostate cancer after radical prostatectomy. Anticancer Res. 2014, 34, 2919–2925. [Google Scholar]
- Diao, Y.; Wu, D.; Dai, Z.; Kang, H.; Wang, Z.; Wang, X. Prognostic value of transformer 2β expression in prostate cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 6967–6973. [Google Scholar]
- Mortezavi, A.; Salemi, S.; Rupp, N.J.; Rüschoff, J.H.; Hermanns, T.; Poyet, C.; Randazzo, M.; Simon, H.-U.; Moch, H.; Sulser, T.; et al. Negative LC3b immunoreactivity in cancer cells is an independent prognostic predictor of prostate cancer specific death. Oncotarget 2017, 8, 31765–31774. [Google Scholar] [CrossRef] [Green Version]
- Staibano, S.; Mascolo, M.; Di Benedetto, M.; Vecchione, M.L.; Ilardi, G.; Di Lorenzo, G.; Autorino, R.; Salerno, V.; Morena, A.; Rocco, A.; et al. BAG3 protein delocalisation in prostate carcinoma. Tumour Biol. 2010, 31, 461–469. [Google Scholar] [CrossRef] [Green Version]
- Tradonsky, A.; Rubin, T.; Beck, R.; Ring, B.; Seitz, R.; Mair, S. A search for reliable molecular markers of prognosis in prostate cancer: A study of 240 cases. Am. J. Clin. Pathol. 2012, 137, 918–930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grosset, A.-A.; Ouellet, V.; Caron, C.; Fragoso, G.; Barrès, V.; Delvoye, N.; Latour, M.; Aprikian, A.; Bergeron, A.; Chevalier, S.; et al. Validation of the prognostic value of NF-κB p65 in prostate cancer: A retrospective study using a large multi-institutional cohort of the Canadian Prostate Cancer Biomarker Network. PLoS Med. 2019, 16, e1002847. [Google Scholar] [CrossRef] [PubMed]
- Ness, N.; Andersen, S.; Khanehkenari, M.R.; Nordbakken, C.V.; Valkov, A.; Paulsen, E.-E.; Nordby, Y.; Bremnes, R.M.; Donnem, T.; Busund, L.-T.; et al. The prognostic role of immune checkpoint markers programmed cell death protein 1 (PD-1) and programmed death ligand 1 (PD-L1) in a large, multicenter prostate cancer cohort. Oncotarget 2017, 8, 26789–26801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fleischmann, A.; Rocha, C.; Saxer-Sekulic, N.; Zlobec, I.; Sauter, G.; Thalmann, G.N. High CD10 expression in lymph node metastases from surgically treated prostate cancer independently predicts early death. Virchows Arch. 2011, 458, 741–748. [Google Scholar] [CrossRef] [PubMed]
- Nonsrijun, N.; Mitchai, J.; Brown, K.; Leksomboon, R.; Tuamsuk, P. Overexpression of matrix metalloproteinase 11 in Thai prostatic adenocarcinoma is associated with poor survival. Asian Pac. J. Cancer Prev. 2013, 14, 3331–3335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamid, A.A.; Gray, K.P.; Huang, Y.; Bowden, M.; Pomerantz, M.; Loda, M.; Sweeney, C.J. Loss of PTEN Expression Detected by Fluorescence Immunohistochemistry Predicts Lethal Prostate Cancer in Men Treated with Prostatectomy. Eur. Urol. Oncol. 2019, 2, 475–482. [Google Scholar] [CrossRef] [PubMed]
- Lahdensuo, K.; Erickson, A.; Saarinen, I.; Seikkula, H.; Lundin, J.; Lundin, M.; Nordling, S.; Bützow, A.; Vasarainen, H.; Boström, P.J.; et al. Loss of PTEN expression in ERG-negative prostate cancer predicts secondary therapies and leads to shorter disease-specific survival time after radical prostatectomy. Mod. Pathol. 2016, 29, 1565–1574. [Google Scholar] [CrossRef] [Green Version]
- Lin, Z.-Y.; Chen, G.; Zhang, Y.-Q.; He, H.-C.; Liang, Y.-X.; Ye, J.-H.; Liang, Y.-K.; Mo, R.-J.; Lu, J.-M.; Zhuo, Y.-J.; et al. MicroRNA-30d promotes angiogenesis and tumor growth via MYPT1/c-JUN/VEGFA pathway and predicts aggressive outcome in prostate cancer. Mol. Cancer 2017, 16, 48. [Google Scholar] [CrossRef] [Green Version]
- Nordby, Y.; Andersen, S.; Richardsen, E.; Ness, N.; Al-Saad, S.; Melbø-Jørgensen, C.; Patel, H.R.H.; Dønnem, T.; Busund, L.-T.; Bremnes, R.M. Stromal expression of VEGF-A and VEGFR-2 in prostate tissue is associated with biochemical and clinical recurrence after radical prostatectomy. Prostate 2015, 75, 1682–1693. [Google Scholar] [CrossRef]
- Borkowetz, A.; Froehner, M.; Rauner, M.; Conrad, S.; Erdmann, K.; Mayr, T.; Datta, K.; Hofbauer, L.C.; Baretton, G.B.; Wirth, M.; et al. Neuropilin-2 is an independent prognostic factor for shorter cancer-specific survival in patients with acinar adenocarcinoma of the prostate. Int. J. Cancer 2020, 146, 2619–2627. [Google Scholar] [CrossRef] [Green Version]
- Liu, R.; Yang, K.; Meng, C.; Zhang, Z.; Xu, Y. Vasculogenic mimicry is a marker of poor prognosis in prostate cancer. Cancer Biol. Ther. 2012, 13, 527–533. [Google Scholar] [CrossRef]
- Nordby, Y.; Richardsen, E.; Rakaee, M.; Ness, N.; Donnem, T.; Patel, H.R.H.; Busund, L.-T.; Bremnes, R.M.; Andersen, S. High expression of PDGFR-β in prostate cancer stroma is independently associated with clinical and biochemical prostate cancer recurrence. Sci. Rep. 2017, 7, 43378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, J.; Wang, M.; Wang, Z.; Liu, X. Overexpression of Pleomorphic Adenoma Gene-Like 2 Is a Novel Poor Prognostic Marker of Prostate Cancer. PLoS ONE 2016, 11, e0158667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Guo, F.; Gao, X.; Wu, Y. Golgi phosphoprotein 3 expression predicts poor prognosis in patients with prostate cancer undergoing radical prostatectomy. Mol. Med. Rep. 2015, 12, 1298–1304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tretiakova, M.S.; Wei, W.; Boyer, H.D.; Newcomb, L.F.; Hawley, S.; Auman, H.; Vakar-Lopez, F.; McKenney, J.K.; Fazli, L.; Simko, J.; et al. Prognostic value of Ki67 in localized prostate carcinoma: A multi-institutional study of >1000 prostatectomies. Prostate Cancer Prostatic Dis. 2016, 19, 264–270. [Google Scholar] [CrossRef] [Green Version]
- Haldrup, C.; Lynnerup, A.-S.; Storebjerg, T.M.; Vang, S.; Wild, P.; Visakorpi, T.; Arsov, C.; Schulz, W.A.; Lindberg, J.; Grönberg, H.; et al. Large-scale evaluation of SLC18A2 in prostate cancer reveals diagnostic and prognostic biomarker potential at three molecular levels. Mol. Oncol. 2016, 10, 825–837. [Google Scholar] [CrossRef]
- Rynkiewicz, N.K.; Fedele, C.G.; Chiam, K.; Gupta, R.; Kench, J.G.; Ooms, L.M.; McLean, C.A.; Giles, G.G.; Horvath, L.G.; Mitchell, C.A. INPP4B is highly expressed in prostate intermediate cells and its loss of expression in prostate carcinoma predicts for recurrence and poor long term survival. Prostate 2015, 75, 92–102. [Google Scholar] [CrossRef]
- Genitsch, V.; Zlobec, I.; Thalmann, G.N.; Fleischmann, A. MUC1 is upregulated in advanced prostate cancer and is an independent prognostic factor. Prostate Cancer Prostatic Dis. 2016, 19, 242–247. [Google Scholar] [CrossRef]
- Hammarsten, P.; Dahl Scherdin, T.; Hägglöf, C.; Andersson, P.; Wikström, P.; Stattin, P.; Egevad, L.; Granfors, T.; Bergh, A. High Caveolin-1 Expression in Tumor Stroma Is Associated with a Favourable Outcome in Prostate Cancer Patients Managed by Watchful Waiting. PLoS ONE 2016, 11, e0164016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, P.L.; Martin, N.E.; Choeurng, V.; Palmer-Aronsten, B.; Kolisnik, T.; Beard, C.J.; Orio, P.F.; Nezolosky, M.D.; Chen, Y.-W.; Shin, H.; et al. Utilization of biopsy-based genomic classifier to predict distant metastasis after definitive radiation and short-course ADT for intermediate and high-risk prostate cancer. Prostate Cancer Prostatic Dis. 2017, 20, 186–192. [Google Scholar] [CrossRef] [Green Version]
- Van Den Eeden, S.K.; Lu, R.; Zhang, N.; Quesenberry, C.P., Jr.; Shan, J.; Han, J.S.; Tsiatis, A.C.; Leimpeter, A.D.; Lawrence, H.J.; Febbo, P.G.; et al. A Biopsy-based 17-gene Genomic Prostate Score as a Predictor of Metastases and Prostate Cancer Death in Surgically Treated Men with Clinically Localized Disease. Eur. Urol. 2018, 73, 129–138. [Google Scholar] [CrossRef]
- Zeng, W.; Sun, H.; Meng, F.; Liu, Z.; Xiong, J.; Zhou, S.; Li, F.; Hu, J.; Hu, Z.; Liu, Z. Nuclear C-MYC expression level is associated with disease progression and potentially predictive of two year overall survival in prostate cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 1878–1888. [Google Scholar]
- Castro, E.; Goh, C.; Leongamornlert, D.; Saunders, E.; Tymrakiewicz, M.; Dadaev, T.; Govindasami, K.; Guy, M.; Ellis, S.; Frost, D.; et al. Effect of BRCA Mutations on Metastatic Relapse and Cause-specific Survival After Radical Treatment for Localised Prostate Cancer. Eur. Urol. 2015, 68, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Cooperberg, M.R.; Davicioni, E.; Crisan, A.; Jenkins, R.B.; Ghadessi, M.; Karnes, R.J. Combined value of validated clinical and genomic risk stratification tools for predicting prostate cancer mortality in a high-risk prostatectomy cohort. Eur. Urol. 2015, 67, 326–333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ross, A.E.; Johnson, M.H.; Yousefi, K.; Davicioni, E.; Netto, G.J.; Marchionni, L.; Fedor, H.L.; Glavaris, S.; Choeurng, V.; Buerki, C.; et al. Tissue-based Genomics Augments Post-prostatectomy Risk Stratification in a Natural History Cohort of Intermediate- and High-Risk Men. Eur. Urol. 2016, 69, 157–165. [Google Scholar] [CrossRef]
- Zhao, S.G.; Jackson, W.C.; Kothari, V.; Schipper, M.J.; Erho, N.; Evans, J.R.; Speers, C.; Hamstra, D.A.; Niknafs, Y.S.; Nguyen, P.L.; et al. High-throughput transcriptomic analysis nominates proteasomal genes as age-specific biomarkers and therapeutic targets in prostate cancer. Prostate Cancer Prostatic Dis. 2015, 18, 229–236. [Google Scholar] [CrossRef]
- Zhao, S.G.; Evans, J.R.; Kothari, V.; Sun, G.; Larm, A.; Mondine, V.; Schaeffer, E.M.; Ross, A.E.; Klein, E.A.; Den, R.B.; et al. The Landscape of Prognostic Outlier Genes in High-Risk Prostate Cancer. Clin. Cancer Res. 2016, 22, 1777–1786. [Google Scholar] [CrossRef] [Green Version]
- Moen, L.V.; Ramberg, H.; Zhao, S.; Grytli, H.H.; Sveen, A.; Berge, V.; Skotheim, R.I.; Taskén, K.A.; Skålhegg, B.S. Observed correlation between the expression levels of catalytic subunit, Cβ2, of cyclic adenosine monophosphate-dependent protein kinase and prostate cancer aggressiveness. Urol. Oncol. 2017, 35, 111.e1–111.e8. [Google Scholar] [CrossRef]
- Evans, J.R.; Zhao, S.G.; Chang, S.L.; Tomlins, S.A.; Erho, N.; Sboner, A.; Schiewer, M.J.; Spratt, D.E.; Kothari, V.; Klein, E.A.; et al. Patient-Level DNA Damage and Repair Pathway Profiles and Prognosis After Prostatectomy for High-Risk Prostate Cancer. JAMA Oncol. 2016, 2, 471–480. [Google Scholar] [CrossRef]
- Hu, B.R.; Fairey, A.S.; Madhav, A.; Yang, D.; Li, M.; Groshen, S.; Stephens, C.; Kim, P.H.; Virk, N.; Wang, L.; et al. AXIN2 expression predicts prostate cancer recurrence and regulates invasion and tumor growth. Prostate 2016, 76, 597–608. [Google Scholar] [CrossRef]
- Schmidt, L.; Fredsøe, J.; Kristensen, H.; Strand, S.H.; Rasmussen, A.; Høyer, S.; Borre, M.; Mouritzen, P.; Ørntoft, T.; Sørensen, K.D. Training and validation of a novel 4-miRNA ratio model (MiCaP) for prediction of postoperative outcome in prostate cancer patients. Ann. Oncol. 2018, 29, 2003–2009. [Google Scholar] [CrossRef]
- Richardsen, E.; Andersen, S.; Al-Saad, S.; Rakaee, M.; Nordby, Y.; Pedersen, M.I.; Ness, N.; Ingebriktsen, L.M.; Fassina, A.; Taskén, K.A.; et al. Low Expression of miR-424-3p is Highly Correlated with Clinical Failure in Prostate Cancer. Sci. Rep. 2019, 9, 10662. [Google Scholar] [CrossRef]
- Laursen, E.B.; Fredsøe, J.; Schmidt, L.; Strand, S.H.; Kristensen, H.; Rasmussen, A.K.I.; Daugaard, T.F.; Mouritzen, P.; Høyer, S.; Kristensen, G.; et al. Elevated miR-615-3p Expression Predicts Adverse Clinical Outcome and Promotes Proliferation and Migration of Prostate Cancer Cells. Am. J. Pathol. 2019, 189, 2377–2388. [Google Scholar] [CrossRef] [Green Version]
- Troyer, D.A.; Jamaspishvili, T.; Wei, W.; Feng, Z.; Good, J.; Hawley, S.; Fazli, L.; McKenney, J.K.; Simko, J.; Hurtado-Coll, A.; et al. A multicenter study shows PTEN deletion is strongly associated with seminal vesicle involvement and extracapsular extension in localized prostate cancer. Prostate 2015, 75, 1206–1215. [Google Scholar] [CrossRef] [Green Version]
- Thurner, E.-M.; Krenn-Pilko, S.; Langsenlehner, U.; Stojakovic, T.; Pichler, M.; Gerger, A.; Kapp, K.S.; Langsenlehner, T. The association of an elevated plasma fibrinogen level with cancer-specific and overall survival in prostate cancer patients. World J. Urol. 2015, 33, 1467–1473. [Google Scholar] [CrossRef]
- Renner, W.; Krenn-Pilko, S.; Gruber, H.-J.; Herrmann, M.; Langsenlehner, T. Relative telomere length and prostate cancer mortality. Prostate Cancer Prostatic Dis. 2018, 21, 579–583. [Google Scholar] [CrossRef]
- Lévesque, E.; Laverdière, I.; Audet-Walsh, E.; Caron, P.; Rouleau, M.; Fradet, Y.; Lacombe, L.; Guillemette, C. Steroidogenic germline polymorphism predictors of prostate cancer progression in the estradiol pathway. Clin. Cancer Res. 2014, 20, 2971–2983. [Google Scholar] [CrossRef] [Green Version]
- Schoenfeld, J.D.; Margalit, D.N.; Kasperzyk, J.L.; Shui, I.M.; Rider, J.R.; Epstein, M.M.; Meisner, A.; Kenfield, S.A.; Martin, N.E.; Nguyen, P.L.; et al. A single nucleotide polymorphism in inflammatory gene RNASEL predicts outcome after radiation therapy for localized prostate cancer. Clin. Cancer Res. 2013, 19, 1612–1619. [Google Scholar] [CrossRef] [Green Version]
- Szarvas, T.; Tschirdewahn, S.; Niedworok, C.; Kramer, G.; Sevcenco, S.; Reis, H.; Shariat, S.F.; Rübben, H.; vom Dorp, F. Prognostic value of tissue and circulating levels of IMP3 in prostate cancer. Int. J. Cancer 2014, 135, 1596–1604. [Google Scholar] [CrossRef]
- Bishoff, J.T.; Freedland, S.J.; Gerber, L.; Tennstedt, P.; Reid, J.; Welbourn, W.; Graefen, M.; Sangale, Z.; Tikishvili, E.; Park, J.; et al. Prognostic utility of the cell cycle progression score generated from biopsy in men treated with prostatectomy. J. Urol. 2014, 192, 409–414. [Google Scholar] [CrossRef]
- Nguyen, P.L.; Haddad, Z.; Ross, A.E.; Martin, N.E.; Deheshi, S.; Lam, L.L.C.; Chelliserry, J.; Tosoian, J.J.; Lotan, T.L.; Spratt, D.E.; et al. Ability of a Genomic Classifier to Predict Metastasis and Prostate Cancer-specific Mortality after Radiation or Surgery based on Needle Biopsy Specimens. Eur. Urol. 2017, 72, 845–852. [Google Scholar] [CrossRef] [Green Version]
- Morra, L.; Moch, H. Periostin expression and epithelial-mesenchymal transition in cancer: A review and an update. Virchows Arch. 2011, 459, 465–475. [Google Scholar] [CrossRef] [Green Version]
- Tomlins, S.A.; Rhodes, D.R.; Perner, S.; Dhanasekaran, S.M.; Mehra, R.; Sun, X.-W.; Varambally, S.; Cao, X.; Tchinda, J.; Kuefer, R.; et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 2005, 310, 644–648. [Google Scholar] [CrossRef]
- Na, R.; Zheng, S.L.; Han, M.; Yu, H.; Jiang, D.; Shah, S.; Ewing, C.M.; Zhang, L.; Novakovic, K.; Petkewicz, J.; et al. Germline Mutations in ATM and BRCA1/2 Distinguish Risk for Lethal and Indolent Prostate Cancer and are Associated with Early Age at Death. Eur. Urol. 2017, 71, 740–747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, W.; Xie, C.C.; Thomas, C.Y.; Kim, S.-T.; Lindberg, J.; Egevad, L.; Wang, Z.; Zhang, Z.; Sun, J.; Sun, J.; et al. Genetic markers associated with early cancer-specific mortality following prostatectomy. Cancer 2013, 119, 2405–2412. [Google Scholar] [CrossRef] [PubMed]
- Rüegger, S.; Großhans, H. MicroRNA turnover: When, how, and why. Trends Biochem. Sci. 2012, 37, 436–446. [Google Scholar] [CrossRef] [PubMed]
- Pettersson, A.; Lis, R.T.; Meisner, A.; Flavin, R.; Stack, E.C.; Fiorentino, M.; Finn, S.; Graff, R.E.; Penney, K.L.; Rider, J.R.; et al. Modification of the association between obesity and lethal prostate cancer by TMPRSS2:ERG. J. Natl. Cancer Inst. 2013, 105, 1881–1890. [Google Scholar] [CrossRef] [Green Version]
- Peng, K.; Bai, Y.; Zhu, Q.; Hu, B.; Xu, Y. Targeting VEGF-neuropilin interactions: A promising antitumor strategy. Drug Discov. Today 2019, 24, 656–664. [Google Scholar] [CrossRef]
- Scott, K.L.; Kabbarah, O.; Liang, M.-C.; Ivanova, E.; Anagnostou, V.; Wu, J.; Dhakal, S.; Wu, M.; Chen, S.; Feinberg, T.; et al. GOLPH3 modulates mTOR signalling and rapamycin sensitivity in cancer. Nature 2009, 459, 1085–1090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pascale, M.; Aversa, C.; Barbazza, R.; Marongiu, B.; Siracusano, S.; Stoffel, F.; Sulfaro, S.; Roggero, E.; Bonin, S.; Stanta, G. The proliferation marker Ki67, but not neuroendocrine expression, is an independent factor in the prediction of prognosis of primary prostate cancer patients. Radiol. Oncol. 2016, 50, 313–320. [Google Scholar] [CrossRef] [Green Version]
- Hodgson, M.C.; Shao, L.-J.; Frolov, A.; Li, R.; Peterson, L.E.; Ayala, G.; Ittmann, M.M.; Weigel, N.L.; Agoulnik, I.U. Decreased expression and androgen regulation of the tumor suppressor gene INPP4B in prostate cancer. Cancer Res. 2011, 71, 572–582. [Google Scholar] [CrossRef] [Green Version]
- Rajabi, H.; Ahmad, R.; Jin, C.; Joshi, M.D.; Guha, M.; Alam, M.; Kharbanda, S.; Kufe, D. MUC1-C oncoprotein confers androgen-independent growth of human prostate cancer cells. Prostate 2012, 72, 1659–1668. [Google Scholar] [CrossRef] [Green Version]
- Bokhorst, L.P.; Valdagni, R.; Rannikko, A.; Kakehi, Y.; Pickles, T.; Bangma, C.H.; Roobol, M.J.; PRIAS study group. A Decade of Active Surveillance in the PRIAS Study: An Update and Evaluation of the Criteria Used to Recommend a Switch to Active Treatment. Eur. Urol. 2016, 70, 954–960. [Google Scholar] [CrossRef] [Green Version]
- Lokman, U.; Erickson, A.M.; Vasarainen, H.; Rannikko, A.S.; Mirtti, T. PTEN Loss but Not ERG Expression in Diagnostic Biopsies Is Associated with Increased Risk of Progression and Adverse Surgical Findings in Men with Prostate Cancer on Active Surveillance. Eur. Urol. Focus 2018, 4, 867–873. [Google Scholar] [CrossRef] [Green Version]
- Hammarsten, P.; Josefsson, A.; Thysell, E.; Lundholm, M.; Hägglöf, C.; Iglesias-Gato, D.; Flores-Morales, A.; Stattin, P.; Egevad, L.; Granfors, T.; et al. Immunoreactivity for prostate specific antigen and Ki67 differentiates subgroups of prostate cancer related to outcome. Mod. Pathol. 2019, 32, 1310–1319. [Google Scholar] [CrossRef]
- Fisher, G.; Yang, Z.H.; Kudahetti, S.; Møller, H.; Scardino, P.; Cuzick, J.; Berney, D.M. Transatlantic Prostate Group Prognostic value of Ki-67 for prostate cancer death in a conservatively managed cohort. Br. J. Cancer 2013, 108, 271–277. [Google Scholar] [CrossRef] [Green Version]
- Lobo, J.; Rodrigues, Â.; Antunes, L.; Graça, I.; Ramalho-Carvalho, J.; Vieira, F.Q.; Martins, A.T.; Oliveira, J.; Jerónimo, C.; Henrique, R. High immunoexpression of Ki67, EZH2, and SMYD3 in diagnostic prostate biopsies independently predicts outcome in patients with prostate cancer. Urol. Oncol. 2018, 36, 161.e7–161.e17. [Google Scholar] [CrossRef]
- Mirtti, T.; Leiby, B.E.; Abdulghani, J.; Aaltonen, E.; Pavela, M.; Mamtani, A.; Alanen, K.; Egevad, L.; Granfors, T.; Josefsson, A.; et al. Nuclear Stat5a/b predicts early recurrence and prostate cancer-specific death in patients treated by radical prostatectomy. Hum. Pathol. 2013, 44, 310–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thorgeirsson, T.; Jordahl, K.M.; Flavin, R.; Epstein, M.M.; Fiorentino, M.; Andersson, S.-O.; Andren, O.; Rider, J.R.; Mosquera, J.M.; Ingoldsby, H.; et al. Intracellular location of BRCA2 protein expression and prostate cancer progression in the Swedish Watchful Waiting Cohort. Carcinogenesis 2016, 37, 262–268. [Google Scholar] [CrossRef] [Green Version]
- Cuzick, J.; Berney, D.M.; Fisher, G.; Mesher, D.; Møller, H.; Reid, J.E.; Perry, M.; Park, J.; Younus, A.; Gutin, A.; et al. Prognostic value of a cell cycle progression signature for prostate cancer death in a conservatively managed needle biopsy cohort. Br. J. Cancer 2012, 106, 1095–1099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.L.; Li, P.; Huang, H.-C.; Deheshi, S.; Marti, T.; Knudsen, B.; Abou-Ouf, H.; Alam, R.; Lotan, T.L.; Lam, L.L.C.; et al. Validation of the Decipher Test for predicting adverse pathology in candidates for prostate cancer active surveillance. Prostate Cancer Prostatic Dis. 2019, 22, 399–405. [Google Scholar] [CrossRef] [Green Version]
- Herlemann, A.; Huang, H.-C.; Alam, R.; Tosoian, J.J.; Kim, H.L.; Klein, E.A.; Simko, J.P.; Chan, J.M.; Lane, B.R.; Davis, J.W.; et al. Decipher identifies men with otherwise clinically favorable-intermediate risk disease who may not be good candidates for active surveillance. Prostate Cancer Prostatic Dis. 2020, 23, 136–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kornberg, Z.; Cowan, J.E.; Westphalen, A.C.; Cooperberg, M.R.; Chan, J.M.; Zhao, S.; Shinohara, K.; Carroll, P.R. Genomic Prostate Score, PI-RADSTM version 2 and Progression in Men with Prostate Cancer on Active Surveillance. J. Urol. 2019, 201, 300–307. [Google Scholar] [CrossRef]
- Cooperberg, M.R.; Carroll, P.R.; Dall’Era, M.A.; Davies, B.J.; Davis, J.W.; Eggener, S.E.; Feng, F.Y.; Lin, D.W.; Morgan, T.M.; Morgans, A.K.; et al. The State of the Science on Prostate Cancer Biomarkers: The San Francisco Consensus Statement. Eur. Urol. 2019, 76, 268–272. [Google Scholar] [CrossRef] [PubMed]
- Lotan, T.L.; Tomlins, S.A.; Bismar, T.A.; Van der Kwast, T.H.; Grignon, D.; Egevad, L.; Kristiansen, G.; Pritchard, C.C.; Rubin, M.A.; Bubendorf, L. Report From the International Society of Urological Pathology (ISUP) Consultation Conference on Molecular Pathology of Urogenital Cancers. I. Molecular Biomarkers in Prostate Cancer. Am. J. Surg. Pathol. 2020, 44, e15–e29. [Google Scholar] [CrossRef]
- Eggener, S.E.; Rumble, R.B.; Armstrong, A.J.; Morgan, T.M.; Crispino, T.; Cornford, P.; van der Kwast, T.; Grignon, D.J.; Rai, A.J.; Agarwal, N.; et al. Molecular Biomarkers in Localized Prostate Cancer: ASCO Guideline. J. Clin. Oncol. 2020, 38, 1474–1494. [Google Scholar] [CrossRef]
- Jhun, M.A.; Geybels, M.S.; Wright, J.L.; Kolb, S.; April, C.; Bibikova, M.; Ostrander, E.A.; Fan, J.-B.; Feng, Z.; Stanford, J.L. Gene expression signature of Gleason score is associated with prostate cancer outcomes in a radical prostatectomy cohort. Oncotarget 2017, 8, 43035–43047. [Google Scholar] [CrossRef] [PubMed]
- Penney, K.L.; Sinnott, J.A.; Fall, K.; Pawitan, Y.; Hoshida, Y.; Kraft, P.; Stark, J.R.; Fiorentino, M.; Perner, S.; Finn, S.; et al. mRNA expression signature of Gleason grade predicts lethal prostate cancer. J. Clin. Oncol. 2011, 29, 2391–2396. [Google Scholar] [CrossRef] [Green Version]
- Rubicz, R.; Zhao, S.; Wright, J.L.; Coleman, I.; Grasso, C.; Geybels, M.S.; Leonardson, A.; Kolb, S.; April, C.; Bibikova, M.; et al. Gene expression panel predicts metastatic-lethal prostate cancer outcomes in men diagnosed with clinically localized prostate cancer. Mol. Oncol. 2017, 11, 140–150. [Google Scholar] [CrossRef] [Green Version]
- Sinnott, J.A.; Peisch, S.F.; Tyekucheva, S.; Gerke, T.; Lis, R.; Rider, J.R.; Fiorentino, M.; Stampfer, M.J.; Mucci, L.A.; Loda, M.; et al. Prognostic Utility of a New mRNA Expression Signature of Gleason Score. Clin. Cancer Res. 2017, 23, 81–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woo, S.; Cho, J.Y.; Ku, J.H.; Kim, S.Y.; Kim, S.H. Prostate cancer-specific mortality after radical prostatectomy: Value of preoperative MRI. Acta Radiol. 2016, 57, 1006–1013. [Google Scholar] [CrossRef] [PubMed]

| Source Material, Analysis | Ref. No. | First Author | Year | Primary Therapy | Biomarkers | Additional Analysis Info | Outcome | Correlation |
|---|---|---|---|---|---|---|---|---|
| Biopsy, IHC | [28] | Tollefson | 2014 | RP | Ki-67 | MFS, DSS | Positive | |
| [29] | Verhoven | 2013 | RT + ADT | Ki-67 | DSS, MFS | Positive | ||
| [30] | Pollack | 2015 | RT + ADT | Ki-67, MDM2, p16, Cox-2 | MFS | Negative/Positive | ||
| [31] | Krauss | 2011 | RT + ADT | Chromogranin A (CgA) | MFS, DSS | Positive | ||
| [32] | Cattrini | 2019 | RP, RT, ADT | POSTN | OS, MFS * | Positive | ||
| [33] | Jacobs | 2016 | RT + ADT | EZH2 | MFS | Negative | ||
| [34] | Kammerer-Jacquet | 2020 | AS | Ki-67 | DSS | Positive | ||
| RP, IHC | [35] | Megas | 2016 | RP, RT, ADT | ER(α), ER(β) | OS, MFS * | Positive/Negative | |
| [36] | Grindstad | 2018 | RP | ER(α), Aromatase | MFS *, DSS | Negative | ||
| [37] | Fujimura | 2012 | RP, RT, ADT | SXR, CYP3A4 | RP and Western blot | DSS | Negative | |
| [38] | Quinn | 2020 | RP, RT, ADT | p53 | MFS, DSS | Positive | ||
| [39] | Jiao | 2014 | RP | PPM1D | OS | Positive | ||
| [40] | Diao | 2016 | RP | Tra2β | OS | Positive | ||
| [41] | Mortezavi | 2018 | RP, ADT | LC3b | DSS | Negative | ||
| [42] | Staibano | 2010 | RP | BAG3 | MFS | Positive | ||
| [43] | Tradonsky | 2012 | RP | Hey2 | MFS | Positive | ||
| [44] | Grosset | 2019 | RP | NF-κB p65 | MFS, DSS | Positive | ||
| [45] | Ness | 2018 | RP | PD-1+ stromal lymphocytes | MFS * | Positive | ||
| [46] | Fleischmann | 2011 | RP + ADT | CD10 | LN+ patients only | OS | Positive | |
| [47] | Nonsrijun | 2015 | RP | MMP-11 | DSS | Positive | ||
| [48] | Hamid | 2020 | RP, RT, ADT | PTEN | OS and MFS ** | Negative | ||
| [49] | Lahdensuo | 2018 | RP | ERG, PTEN, | DSS | Negative | ||
| [50] | Lin | 2017 | RP | MYPT1 | OS | Negative | ||
| [51] | Nordby | 2015 | RP | VEGFR-2 | MFS * | Positive | ||
| [52] | Borkowetz | 2020 | RP | NRP2 | DSS | Positive | ||
| [53] | Liu | 2012 | RP | Vasculogenic mimicry (VM) | OS, MFS * | Positive | ||
| [54] | Nordby | 2018 | RP, ADT, RT | PDGFR-β | MFS * | Positive | ||
| [55] | Guo | 2017 | RP | PLAGL2 | OS | Positive | ||
| [56] | Zhang | 2016 | RP | GOLPH3 | OS | Positive | ||
| [57] | Tretiakova | 2017 | RP | Ki67 | OS, DSS, MFS * | Negative | ||
| [58] | Haldrup | 2017 | RP | SLC18A2 | OS | Negative | ||
| [59] | Rynkiewicz | 2015 | RP, RT, ADT | INPP4B | MFS * | Negative | ||
| [60] | Genitsch | 2017 | RP | MUC1 | RP and LN Mets | DSS | Positive | |
| [61] | Hammarsten | 2017 | AS + TURP | Caveolin-1 | RP and TURP | DSS | Negative | |
| Tissue—other | [62] | Nguyen | 2018 | RP, RT + ADT | Decipher | exon microarray, Bx | MFS | Positive |
| [63] | Van Den Eden | 2018 | RP | Oncotype DX | RNA-PCR, Bx | MFS, DSS | Positive | |
| [64] | Zeng | 2016 | RP | TMPRSS2-ERG | RP and Bx, FISH | OS | Positive | |
| [65] | Castro | 2016 | RP/RT + ADT | BRCA1 and 2 | Mutational analysis, Bx and RP | DSS, MFS | Positive | |
| [66] | Cooperberg | 2015 | RP, RT + ADT | Decipher | RNA hybridisation, RP | DSS | Positive | |
| [67] | Ross | 2016 | RP | Decipher | RNA hybridisation, RP | MFS | Positive | |
| [68] | Zhao | 2016 | RP, RT | PSMB4, PSMB7, PSMD14, PSMB2, PSMD11 | RNA microarray hybridization, RP | MFS | Positive | |
| [69] | Zhao | 2016 | RP | NVL, SMC4, SQLE | qRT-PCR | MFS, OS | Negative | |
| [70] | Moen | 2018 | RP, RT, ADT | catalytic subunit Cβ2 | RNA nanostring, Bx | DSS | Positive | |
| [71] | Evans | 2016 | RP, RT, ADT | 17 genes, DDR pathway | GSEA, RP | OS, MFS | Positive | |
| [72] | Hu | 2016 | RP, RT, ADT | AXIN2 | qRT-PCR, RP | MFS | Negative | |
| [73] | Schmidt | 2019 | RP | 4-miRNA ratio model (MiCaP) | miRNA PCR, RP | DSS | Negative/Positive | |
| [74] | Richardsen | 2020 | RP | miR-424-3p | miRNA ISH, RP | MFS * | Negative | |
| [75] | Laursen | 2020 | RP | miR-615-3p | miRNA-PCR, RP | DSS | Positive | |
| [76] | Troyer | 2015 | RP | PTEN | FISH, RP | DSS * | Negative | |
| [53] | Liu | 2013 | RP | PTEN, MYC | SNP array analysis, RP | DSS | Negative/Positive | |
| Blood | [77] | Thurner | 2016 | RT + ADT | Plasma fibrinogen level | Fibrinogen assay | DSS, OS | Positive |
| [78] | Renner | 2019 | RT + ADT | Leukocyte relative telomere (RTL) | DNA-PCR | OS, DSS | Positive | |
| [79] | Lévesque | 2015 | RP | CYP1B1, COMT, and SULT2B1 (3 SNPs) | SNP genotyping | OS, MFS * | Positive/Negative | |
| [80] | Schoenfeld | 2013 | RP, RT + ADT | Ribunuclease-L (rs12757998) | SNP genotyping | DSS and MFS ** | Negative | |
| [81] | Szarvas | 2014 | RP + TURP | IMP3 | ELISA | DSS | Positive |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sopyllo, K.; Erickson, A.M.; Mirtti, T. Grading Evolution and Contemporary Prognostic Biomarkers of Clinically Significant Prostate Cancer. Cancers 2021, 13, 628. https://doi.org/10.3390/cancers13040628
Sopyllo K, Erickson AM, Mirtti T. Grading Evolution and Contemporary Prognostic Biomarkers of Clinically Significant Prostate Cancer. Cancers. 2021; 13(4):628. https://doi.org/10.3390/cancers13040628
Chicago/Turabian StyleSopyllo, Konrad, Andrew M. Erickson, and Tuomas Mirtti. 2021. "Grading Evolution and Contemporary Prognostic Biomarkers of Clinically Significant Prostate Cancer" Cancers 13, no. 4: 628. https://doi.org/10.3390/cancers13040628
APA StyleSopyllo, K., Erickson, A. M., & Mirtti, T. (2021). Grading Evolution and Contemporary Prognostic Biomarkers of Clinically Significant Prostate Cancer. Cancers, 13(4), 628. https://doi.org/10.3390/cancers13040628





