Argonaute Proteins Take Center Stage in Cancers
Abstract
Simple Summary
Abstract
1. Introduction
2. The Functional Role of AGO Proteins in RISC
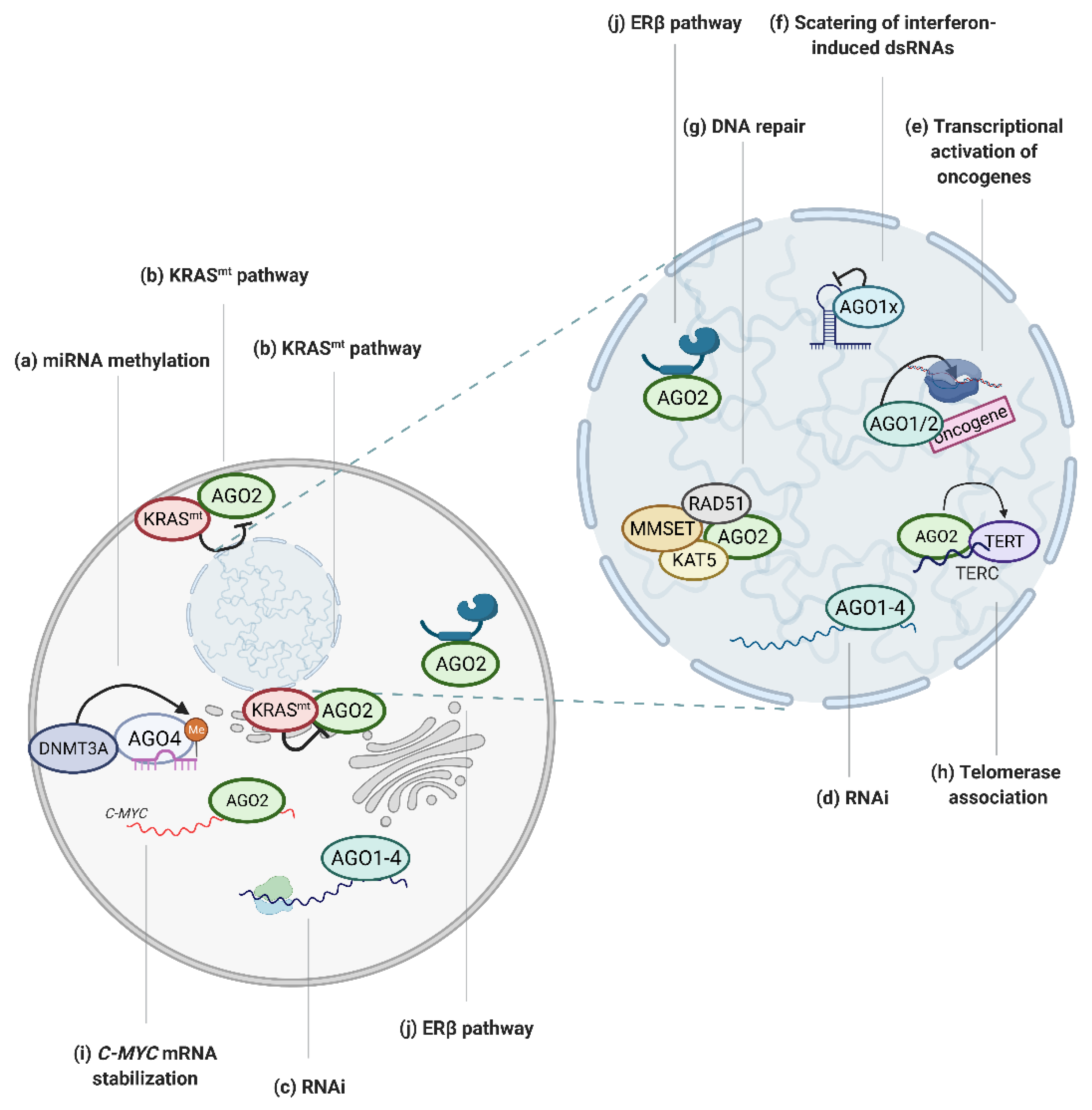
3. The Involvement of AGO Proteins in Tumor Associated Processes
3.1. Regulation of AGO:miRNAs by Post-Transcriptional Modifications
3.2. Regulation of AGOs by Protein/RNA Co-Factors
3.3. AGOs in the Nucleus Affect Cancer Progression
3.4. Regulation of DNA Integrity
3.5. Cellular Differentiation
3.6. Angiogenesis
3.7. Motility and Metastasis
3.8. Tumor-Promoting and Anti-Cancer Functions of the AGOs
4. AGO Proteins as Potential Biomarkers
4.1. The Prognostic Value of AGO Protein Expression in Cancer
4.2. The Biomarker Potential of Modifications of AGO Proteins
4.3. The Biomarker Potential of Genetic Variations of AGO Proteins
5. AGO Proteins as Potential Therapeutic Applications
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef]
- Kobayashi, H.; Tomari, Y. RISC assembly: Coordination between small RNAs and Argonaute proteins. Biochim. Biophys. Acta 2016, 1859, 71–81. [Google Scholar] [CrossRef]
- Niaz, S. The AGO proteins: An overview. Biol. Chem. 2018, 399, 525–547. [Google Scholar] [CrossRef]
- Catalanotto, C.; Cogoni, C.; Zardo, G. MicroRNA in Control of Gene Expression: An Overview of Nuclear Functions. Int. J. Mol. Sci. 2016, 17. [Google Scholar] [CrossRef]
- Neilson, J.R.; Zheng, G.X.Y.; Burge, C.B.; Sharp, P.A. Dynamic regulation of miRNA expression in ordered stages of cellular development. Genes Dev. 2007, 21, 578–589. [Google Scholar] [CrossRef]
- Muller, M.; Fazi, F.; Ciaudo, C. Argonaute Proteins: From Structure to Function in Development and Pathological Cell Fate Determination. Front. Cell Dev. Biol. 2019, 7, 360. [Google Scholar] [CrossRef] [PubMed]
- Völler, D.; Linck, L.; Bruckmann, A.; Hauptmann, J.; Deutzmann, R.; Meister, G.; Bosserhoff, A.K. Argonaute Family Protein Expression in Normal Tissue and Cancer Entities. PLoS ONE 2016, 11, e0161165. [Google Scholar] [CrossRef] [PubMed]
- Valdmanis, P.N.; Gu, S.; Schüermann, N.; Sethupathy, P.; Grimm, D.; Kay, M.A. Expression determinants of mammalian argonaute proteins in mediating gene silencing. Nucleic Acids Res. 2012, 40, 3704–3713. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Boroviak, T.; Stirparo, G.G.; Dietmann, S.; Hernando-Herraez, I.; Mohammed, H.; Reik, W.; Smith, A.; Sasaki, E.; Nichols, J.; Bertone, P. Single cell transcriptome analysis of human, marmoset and mouse embryos reveals common and divergent features of preimplantation development. Development 2018, 145. [Google Scholar] [CrossRef]
- Vishnoi, A.; Rani, S. MiRNA Biogenesis and Regulation of Diseases: An Overview. Methods Mol. Biol. 2017, 1509, 1–10. [Google Scholar] [CrossRef]
- Sandiford, O.A.; Moore, C.A.; Du, J.; Boulad, M.; Gergues, M.; Eltouky, H.; Rameshwar, P. Human Aging and Cancer: Role of miRNA in Tumor Microenvironment. Adv. Exp. Med. Biol. 2018, 1056, 137–152. [Google Scholar] [CrossRef]
- Lee, Y.S.; Dutta, A. MicroRNAs in cancer. Annu. Rev. Pathol. 2009, 4, 199–227. [Google Scholar] [CrossRef] [PubMed]
- Di Leva, G.; Garofalo, M.; Croce, C.M. MicroRNAs in cancer. Annu. Rev. Pathol. 2014, 9, 287–314. [Google Scholar] [CrossRef]
- Acunzo, M.; Romano, G.; Wernicke, D.; Croce, C.M. MicroRNA and cancer--a brief overview. Adv. Biol. Regul. 2015, 57, 1–9. [Google Scholar] [CrossRef]
- Mei, Q.; Li, X.; Guo, M.; Fu, X.; Han, W. The miRNA network: Micro-regulator of cell signaling in cancer. Expert Rev. Anticancer Ther. 2014, 14, 1515–1527. [Google Scholar] [CrossRef] [PubMed]
- Svoronos, A.A.; Engelman, D.M.; Slack, F.J. OncomiR or Tumor Suppressor? The Duplicity of MicroRNAs in Cancer. Cancer Res. 2016, 76, 3666–3670. [Google Scholar] [CrossRef]
- Tan, G.S.; Garchow, B.G.; Liu, X.; Yeung, J.; Morris, J.P.; Cuellar, T.L.; McManus, M.T.; Kiriakidou, M. Expanded RNA-binding activities of mammalian Argonaute 2. Nucleic Acids Res. 2009, 37, 7533–7545. [Google Scholar] [CrossRef]
- Huang, V.; Zheng, J.; Qi, Z.; Wang, J.; Place, R.F.; Yu, J.; Li, H.; Li, L.-C. Ago1 Interacts with RNA Polymerase II and Binds to the Promoters of Actively Transcribed Genes in Human Cancer Cells. PLoS Genet. 2013, 9, e1003821. [Google Scholar] [CrossRef]
- Pin-on, P.; Aporntewan, C.; Siriluksana, J.; Bhummaphan, N.; Chanvorachote, P.; Mutirangura, A. Targeting high transcriptional control activity of long mononucleotide A-T repeats in cancer by Argonaute 1. Gene 2019, 699, 54–61. [Google Scholar] [CrossRef]
- Huang, V.; Li, L.-C. Demystifying the nuclear function of Argonaute proteins. RNA Biol. 2014, 11, 18–24. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, L.; Liu, Z.; Zhou, J.; Pan, Q.; Fan, J.; Zang, R.; Wang, L. AGO1 may influence the prognosis of hepatocellular carcinoma through TGF-β pathway. Cell Death Dis. 2018, 9, 324. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Guimaraes, J.C.; Lanzafame, M.; Schmidt, A.; Syed, A.P.; Dimitriades, B.; Börsch, A.; Ghosh, S.; Mittal, N.; Montavon, T.; et al. Prevention of dsRNA-induced interferon signaling by AGO1x is linked to breast cancer cell proliferation. EMBO J. 2020, 39. [Google Scholar] [CrossRef]
- Zardo, G.; Ciolfi, A.; Vian, L.; Starnes, L.M.; Billi, M.; Racanicchi, S.; Maresca, C.; Fazi, F.; Travaglini, L.; Noguera, N.; et al. Polycombs and microRNA-223 regulate human granulopoiesis by transcriptional control of target gene expression. Blood 2012, 119, 4034–4046. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Lai, T.-C.; Jan, Y.-H.; Lin, F.-M.; Wang, W.-C.; Xiao, H.; Wang, Y.-T.; Sun, W.; Cui, X.; Li, Y.-S.; et al. Hypoxia-responsive miRNAs target argonaute 1 to promote angiogenesis. J. Clin. Investig. 2013, 123, 1057–1067. [Google Scholar] [CrossRef]
- Bellissimo, T.; Tito, C.; Ganci, F.; Sacconi, A.; Masciarelli, S.; Di Martino, G.; Porta, N.; Cirenza, M.; Sorci, M.; De Angelis, L.; et al. Argonaute 2 drives miR-145-5p-dependent gene expression program in breast cancer cells. Cell Death Dis. 2019, 10, 17. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, B.; Chen, X.; Li, W.; Dong, P. AGO2 involves the malignant phenotypes and FAK/PI3K/AKT signaling pathway in hypopharyngeal-derived FaDu cells. Oncotarget 2017, 8, 54735–54746. [Google Scholar] [CrossRef]
- Zhang, K.; Pomyen, Y.; Barry, A.E.; Martin, S.P.; Khatib, S.; Knight, L.; Forgues, M.; Dominguez, D.A.; Parhar, R.; Shah, A.P.; et al. AGO2 Mediates MYC mRNA Stability in Hepatocellular Carcinoma. Mol. Cancer Res. 2020, 18, 612–622. [Google Scholar] [CrossRef]
- Shankar, S.; Pitchiaya, S.; Malik, R.; Kothari, V.; Hosono, Y.; Yocum, A.K.; Gundlapalli, H.; White, Y.; Firestone, A.; Cao, X.; et al. KRAS Engages AGO2 to Enhance Cellular Transformation. Cell Rep. 2016, 14, 1448–1461. [Google Scholar] [CrossRef]
- Shankar, S.; Tien, J.C.-Y.; Siebenaler, R.F.; Chugh, S.; Dommeti, V.L.; Zelenka-Wang, S.; Wang, X.-M.; Apel, I.J.; Waninger, J.; Eyunni, S.; et al. An essential role for Argonaute 2 in EGFR-KRAS signaling in pancreatic cancer development. Nat. Commun. 2020, 11, 2817. [Google Scholar] [CrossRef]
- Zhang, X.; Graves, P.; Zeng, Y. Overexpression of human Argonaute2 inhibits cell and tumor growth. Biochim. Biophys. Acta 2013, 1830, 2553–2561. [Google Scholar] [CrossRef]
- Tarallo, R.; Giurato, G.; Bruno, G.; Ravo, M.; Rizzo, F.; Salvati, A.; Ricciardi, L.; Marchese, G.; Cordella, A.; Rocco, T.; et al. The nuclear receptor ERβ engages AGO2 in regulation of gene transcription, RNA splicing and RISC loading. Genome Biol. 2017, 18, 189. [Google Scholar] [CrossRef] [PubMed]
- Iosue, I.; Quaranta, R.; Masciarelli, S.; Fontemaggi, G.; Batassa, E.M.; Bertolami, C.; Ottone, T.; Divona, M.; Salvatori, B.; Padula, F.; et al. Argonaute 2 sustains the gene expression program driving human monocytic differentiation of acute myeloid leukemia cells. Cell Death Dis. 2013, 4, e926. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Wei, W.; Li, M.-M.; Wu, Y.-S.; Ba, Z.; Jin, K.-X.; Li, M.-M.; Liao, Y.-Q.; Adhikari, S.; Chong, Z.; et al. Ago2 facilitates Rad51 recruitment and DNA double-strand break repair by homologous recombination. Cell Res. 2014, 24, 532–541. [Google Scholar] [CrossRef]
- Wang, Q.; Goldstein, M. Small RNAs Recruit Chromatin-Modifying Enzymes MMSET and Tip60 to Reconfigure Damaged DNA upon Double-Strand Break and Facilitate Repair. Cancer Res. 2016, 76, 1904–1915. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.-l.; Huang, Y.; Li, L.-f.; Zhu, H.-l.; Gao, H.-x.; Liu, H.; Lv, S.-q.; Xu, Z.-h.; Zheng, L.-n.; Liu, T.; et al. Argonaute 2 promotes angiogenesis via the PTEN/VEGF signaling pathway in human hepatocellular carcinoma. Acta Pharmacol. Sin. 2015, 36, 1237–1245. [Google Scholar] [CrossRef]
- Wu, S.; Yu, W.; Qu, X.; Wang, R.; Xu, J.; Zhang, Q.; Xu, J.; Li, J.; Chen, L. Argonaute 2 promotes myeloma angiogenesis via microRNA dysregulation. J. Hematol. Oncol. 2014, 7, 40. [Google Scholar] [CrossRef] [PubMed]
- Laudadio, I.; Orso, F.; Azzalin, G.; Calabrò, C.; Berardinelli, F.; Coluzzi, E.; Gioiosa, S.; Taverna, D.; Sgura, A.; Carissimi, C.; et al. AGO2 promotes telomerase activity and interaction between the telomerase components TERT and TERC. EMBO Rep. 2019, 20. [Google Scholar] [CrossRef]
- Cheray, M.; Etcheverry, A.; Jacques, C.; Pacaud, R.; Bougras-Cartron, G.; Aubry, M.; Denoual, F.; Peterlongo, P.; Nadaradjane, A.; Briand, J.; et al. Cytosine methylation of mature microRNAs inhibits their functions and is associated with poor prognosis in glioblastoma multiforme. Mol. Cancer 2020, 19, 36. [Google Scholar] [CrossRef] [PubMed]
- Potenza, N.; Papa, U.; Russo, A. Differential expression of Dicer and Argonaute genes duringthe differentiation of human neuroblastoma cells. Cell Biol. Int. 2009, 33, 734–738. [Google Scholar] [CrossRef]
- Meister, G. Argonaute proteins: Functional insights and emerging roles. Nat. Rev. Genet. 2013, 14, 447–459. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Trombly, M.I.; Chen, J.; Wang, X. Essential and overlapping functions for mammalian Argonautes in microRNA silencing. Genes Dev. 2009, 23, 304–317. [Google Scholar] [CrossRef] [PubMed]
- Park, M.S.; Phan, H.-D.; Busch, F.; Hinckley, S.H.; Brackbill, J.A.; Wysocki, V.H.; Nakanishi, K. Human Argonaute3 has slicer activity. Nucleic Acids Res. 2017, 45, 11867–11877. [Google Scholar] [CrossRef]
- Park, M.S.; Sim, G.; Kehling, A.C.; Nakanishi, K. Human Argonaute2 and Argonaute3 are catalytically activated by different lengths of guide RNA. Proc. Natl. Acad Sci. USA 2020, 117, 28576–28578. [Google Scholar] [CrossRef] [PubMed]
- Hammond, S.M. An overview of microRNAs. Adv. Drug Deliv. Rev. 2015, 87, 3–14. [Google Scholar] [CrossRef]
- Gebert, L.F.R.; MacRae, I.J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 2019, 20, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Azlan, A.; Dzaki, N.; Azzam, G. Argonaute: The executor of small RNA function. J. Genet. Genom. 2016, 43, 481–494. [Google Scholar] [CrossRef] [PubMed]
- Sarshad, A.A.; Juan, A.H.; Muler, A.I.C.; Anastasakis, D.G.; Wang, X.; Genzor, P.; Feng, X.; Tsai, P.-F.; Sun, H.-W.; Haase, A.D.; et al. Argonaute-miRNA Complexes Silence Target mRNAs in the Nucleus of Mammalian Stem Cells. Mol. Cell 2018, 71, 1040–1050. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Xia, W.; Khotskaya, Y.B.; Huo, L.; Nakanishi, K.; Lim, S.-O.; Du, Y.; Wang, Y.; Chang, W.-C.; Chen, C.-H.; et al. EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nature 2013, 497, 383–387. [Google Scholar] [CrossRef] [PubMed]
- Golden, R.J.; Chen, B.; Li, T.; Braun, J.; Manjunath, H.; Chen, X.; Wu, J.; Schmid, V.; Chang, T.-C.; Kopp, F.; et al. An Argonaute phosphorylation cycle promotes microRNA-mediated silencing. Nature 2017, 542, 197–202. [Google Scholar] [CrossRef]
- Quévillon Huberdeau, M.; Zeitler, D.M.; Hauptmann, J.; Bruckmann, A.; Fressigné, L.; Danner, J.; Piquet, S.; Strieder, N.; Engelmann, J.C.; Jannot, G.; et al. Phosphorylation of Argonaute proteins affects mRNA binding and is essential for microRNA-guided gene silencing in vivo. EMBO J. 2017, 36, 2088–2106. [Google Scholar] [CrossRef]
- Ribezzo, F.; Snoeren, I.A.M.; Ziegler, S.; Stoelben, J.; Olofsen, P.A.; Henic, A.; Ferreira, M.V.; Chen, S.; Stalmann, U.S.A.; Buesche, G.; et al. Rps14, Csnk1a1 and miRNA145/miRNA146a deficiency cooperate in the clinical phenotype and activation of the innate immune system in the 5q- syndrome. Leukemia 2019, 33, 1759–1772. [Google Scholar] [CrossRef] [PubMed]
- Järås, M.; Miller, P.G.; Chu, L.P.; Puram, R.V.; Fink, E.C.; Schneider, R.K.; Al-Shahrour, F.; Peña, P.; Breyfogle, L.J.; Hartwell, K.A.; et al. Csnk1a1 inhibition has p53-dependent therapeutic efficacy in acute myeloid leukemia. J. Exp. Med. 2014, 211, 605–612. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, Y.; Dou, J.; Guo, Y.; He, J.; Li, L.; Liu, X.; Chen, R.; Deng, R.; Huang, J.; et al. Acetylation of AGO2 promotes cancer progression by increasing oncogenic miR-19b biogenesis. Oncogene 2019, 38, 1410–1431. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Sankala, H.; Zhang, X.; Graves, P.R. Phosphorylation of Argonaute 2 at serine-387 facilitates its localization to processing bodies. Biochem. J. 2008, 413, 429–436. [Google Scholar] [CrossRef]
- Horman, S.R.; Janas, M.M.; Litterst, C.; Wang, B.; MacRae, I.J.; Sever, M.J.; Morrissey, D.V.; Graves, P.; Luo, B.; Umesalma, S.; et al. Akt-Mediated Phosphorylation of Argonaute 2 Downregulates Cleavage and Upregulates Translational Repression of MicroRNA Targets. Mol. Cell 2013, 50, 356–367. [Google Scholar] [CrossRef] [PubMed]
- McKenzie, A.J.; Hoshino, D.; Hong, N.H.; Cha, D.J.; Franklin, J.L.; Coffey, R.J.; Patton, J.G.; Weaver, A.M. KRAS-MEK Signaling Controls Ago2 Sorting into Exosomes. Cell Rep. 2016, 15, 978–987. [Google Scholar] [CrossRef] [PubMed]
- Erson-Bensan, A.E.; Begik, O. m6A Modification and Implications for microRNAs. Microrna 2017, 6, 97–101. [Google Scholar] [CrossRef]
- Zhang, J.; Jin, H.; Liu, H.; lv, S.; Wang, B.; Wang, R.; Liu, H.; Ding, M.; Yang, Y.; Li, L.; et al. MiRNA-99a directly regulates AGO2 through translational repression in hepatocellular carcinoma. Oncogenesis 2014, 3, e97. [Google Scholar] [CrossRef]
- Tan, Y.; Zhang, B.; Wu, T.; Skogerbo, G.; Zhu, X.; Guo, X.; He, S.; Chen, R. Transcriptional inhibition of Hoxd4 expression by noncoding RNAs in human breast cancer cells. BMC Mol. Biol. 2009, 10, 12. [Google Scholar] [CrossRef]
- Xu, Q.; Hou, Y.-X.; Langlais, P.; Erickson, P.; Zhu, J.; Shi, C.-X.; Luo, M.; Zhu, Y.; Xu, Y.; Mandarino, L.J.; et al. Expression of the cereblon binding protein argonaute 2 plays an important role for multiple myeloma cell growth and survival. BMC Cancer 2016, 16, 297. [Google Scholar] [CrossRef]
- Zeng, Y.; Cullen, B.R. RNA interference in human cells is restricted to the cytoplasm. RNA 2002, 8, 855–860. [Google Scholar] [CrossRef]
- Vickers, T.A.; Koo, S.; Bennett, C.F.; Crooke, S.T.; Dean, N.M.; Baker, B.F. Efficient reduction of target RNAs by small interfering RNA and RNase H-dependent antisense agents. A comparative analysis. J. Biol. Chem. 2003, 278, 7108–7118. [Google Scholar] [CrossRef] [PubMed]
- Gagnon, K.T.; Li, L.; Chu, Y.; Janowski, B.A.; Corey, D.R. RNAi factors are present and active in human cell nuclei. Cell Rep. 2014, 6, 211–221. [Google Scholar] [CrossRef]
- Ohrt, T.; Mütze, J.; Staroske, W.; Weinmann, L.; Höck, J.; Crell, K.; Meister, G.; Schwille, P. Fluorescence correlation spectroscopy and fluorescence cross-correlation spectroscopy reveal the cytoplasmic origination of loaded nuclear RISC in vivo in human cells. Nucleic Acids Res. 2008, 36, 6439–6449. [Google Scholar] [CrossRef]
- Morris, K.V. Small Interfering RNA-Induced Transcriptional Gene Silencing in Human Cells. Science 2004, 305, 1289–1292. [Google Scholar] [CrossRef]
- Suzuki, K.; Shijuuku, T.; Fukamachi, T.; Zaunders, J.; Guillemin, G.; Cooper, D.; Kelleher, A. Prolonged transcriptional silencing and CpG methylation induced by siRNAs targeted to the HIV-1 promoter region. J. RNAi Gene Silencing 2005, 1, 66–78. [Google Scholar] [PubMed]
- Yamagishi, M.; Ishida, T.; Miyake, A.; Cooper, D.A.; Kelleher, A.D.; Suzuki, K.; Watanabe, T. Retroviral delivery of promoter-targeted shRNA induces long-term silencing of HIV-1 transcription. Microbes Infec. 2009, 11, 500–508. [Google Scholar] [CrossRef]
- Park, H.K.; Min, B.Y.; Kim, N.Y.; Jang, E.S.; Shin, C.M.; Park, Y.S.; Hwang, J.-H.; Jeong, S.-H.; Kim, N.; Lee, D.H.; et al. Short hairpin RNA induces methylation of hepatitis B virus covalently closed circular DNA in human hepatoma cells. Biochem. Biophys. Res. Commun. 2013, 436, 152–155. [Google Scholar] [CrossRef] [PubMed]
- Ting, A.H.; Schuebel, K.E.; Herman, J.G.; Baylin, S.B. Short double-stranded RNA induces transcriptional gene silencing in human cancer cells in the absence of DNA methylation. Nat. Genet. 2005, 37, 906–910. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, P.G.; Santoso, S.; Adams, C.; Anest, V.; Morris, K.V. Promoter targeted small RNAs induce long-term transcriptional gene silencing in human cells. Nucleic Acids Res. 2009, 37, 2984–2995. [Google Scholar] [CrossRef]
- Kim, D.H.; Villeneuve, L.M.; Morris, K.V.; Rossi, J.J. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat. Struct. Mol. Biol. 2006, 13, 793–797. [Google Scholar] [CrossRef]
- Janowski, B.A.; Younger, S.T.; Hardy, D.B.; Ram, R.; Huffman, K.E.; Corey, D.R. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat. Chem. Biol. 2007, 3, 166–173. [Google Scholar] [CrossRef]
- Li, L.C.; Okino, S.T.; Zhao, H.; Pookot, D.; Place, R.F.; Urakami, S.; Enokida, H.; Dahiya, R. Small dsRNAs induce transcriptional activation in human cells. Proc. Natl. Acad Sci. USA 2006, 103, 17337–17342. [Google Scholar] [CrossRef] [PubMed]
- Yue, X.; Schwartz, J.C.; Chu, Y.; Younger, S.T.; Gagnon, K.T.; Elbashir, S.; Janowski, B.A.; Corey, D.R. Transcriptional regulation by small RNAs at sequences downstream from 3′ gene termini. Nat. Chem. Biol. 2010, 6, 621–629. [Google Scholar] [CrossRef]
- Turunen, M.P.; Lehtola, T.; Heinonen, S.E.; Assefa, G.S.; Korpisalo, P.; Girnary, R.; Glass, C.K.; Väisänen, S.; Ylä-Herttuala, S. Efficient Regulation of VEGF Expression by Promoter-Targeted Lentiviral shRNAs Based on Epigenetic Mechanism: A Novel Example of Epigenetherapy. Circ. Res. 2009, 105, 604–609. [Google Scholar] [CrossRef] [PubMed]
- Yokota, A.; Hiramoto, M.; Hino, H.; Tokuhisa, M.; Miyazaki, M.; Kazama, H.; Takano, N.; Miyazawa, K. Sequestosome 1 (p62) accumulation in breast cancer cells suppresses progesterone receptor expression via argonaute 2. Biochem. Biophys. Res. Commun. 2020, 531, 256–263. [Google Scholar] [CrossRef]
- Jackson, S.P.; Bartek, J. The DNA-damage response in human biology and disease. Nature 2009, 461, 1071–1078. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Ba, Z.; Gao, M.; Wu, Y.; Ma, Y.; Amiard, S.; White, C.I.; Danielsen, R.; Jannie, M.; Yang, Y.-G.; et al. A Role for Small RNAs in DNA Double-Strand Break Repair. Cell 2012, 149, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Krell, J.; Stebbing, J.; Carissimi, C.; Dabrowska, A.F.; de Giorgio, A.; Frampton, A.E.; Harding, V.; Fulci, V.; Macino, G.; Colombo, T.; et al. TP53 regulates miRNA association with AGO2 to remodel the miRNA–mRNA interaction network. Genome Res. 2016, 26, 331–341. [Google Scholar] [CrossRef] [PubMed]
- Zvereva, M.I.; Shcherbakova, D.M.; Dontsova, O.A. Telomerase: Structure, functions, and activity regulation. Biochemistry 2010, 75, 1563–1583. [Google Scholar] [CrossRef]
- Artandi, S.E.; DePinho, R.A. Telomeres and telomerase in cancer. Carcinogenesis 2010, 31, 9–18. [Google Scholar] [CrossRef]
- Ngondo, R.P.; Cirera-Salinas, D.; Yu, J.; Wischnewski, H.; Bodak, M.; Vandormael-Pournin, S.; Geiselmann, A.; Wettstein, R.; Luitz, J.; Cohen-Tannoudji, M.; et al. Argonaute 2 Is Required for Extra-embryonic Endoderm Differentiation of Mouse Embryonic Stem Cells. Stem Cell Rep. 2018, 10, 461–476. [Google Scholar] [CrossRef] [PubMed]
- Dueck, A.; Meister, G. Assembly and function of small RNA-argonaute protein complexes. Biol. Chem. 2014, 395, 611–629. [Google Scholar] [CrossRef] [PubMed]
- Pugh, C.W.; Ratcliffe, P.J. Regulation of angiogenesis by hypoxia: Role of the HIF system. Nat. Med. 2003, 9, 677–684. [Google Scholar] [CrossRef]
- Asai, T.; Suzuki, Y.; Matsushita, S.; Yonezawa, S.; Yokota, J.; Katanasaka, Y.; Ishida, T.; Dewa, T.; Kiwada, H.; Nango, M.; et al. Disappearance of the angiogenic potential of endothelial cells caused by Argonaute2 knockdown. Biochem. Biophys. Res. Commun. 2008, 368, 243–248. [Google Scholar] [CrossRef]
- Hatanaka, K.; Shimizu, K.; Asai, T.; Oku, N. Antineovascular gene therapy by Ago2 knockdown. Yakugaku Zasshi 2008, 128, 1567–1575. [Google Scholar] [CrossRef][Green Version]
- Hale, A.; Lee, C.; Annis, S.; Min, P.-K.; Pande, R.; Creager, M.A.; Julian, C.G.; Moore, L.G.; Mitsialis, S.A.; Hwang, S.J.; et al. An Argonaute 2 switch regulates circulating miR-210 to coordinate hypoxic adaptation across cells. Biochim. Biophys. Acta 2014, 1843, 2528–2542. [Google Scholar] [CrossRef]
- Qi, H.H.; Ongusaha, P.P.; Myllyharju, J.; Cheng, D.; Pakkanen, O.; Shi, Y.; Lee, S.W.; Peng, J.; Shi, Y. Prolyl 4-hydroxylation regulates Argonaute 2 stability. Nature 2008, 455, 421–424. [Google Scholar] [CrossRef] [PubMed]
- Bavelloni, A.; Ramazzotti, G.; Poli, A.; Piazzi, M.; Focaccia, E.; Blalock, W.; Faenza, I. MiRNA-210: A Current Overview. Anticancer Res. 2017, 37, 6511–6521. [Google Scholar] [CrossRef]
- Wu, C.; So, J.; Davis-Dusenbery, B.N.; Qi, H.H.; Bloch, D.B.; Shi, Y.; Lagna, G.; Hata, A. Hypoxia Potentiates MicroRNA-Mediated Gene Silencing through Posttranslational Modification of Argonaute2. Mol. Cell. Biol. 2011, 31, 4760–4774. [Google Scholar] [CrossRef]
- Saitoh, M. Involvement of partial EMT in cancer progression. J. Biochem. 2018, 164, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Cheng, N.; Li, Y.; Han, Z.-G. Argonaute2 promotes tumor metastasis by way of up-regulating focal adhesion kinase expression in hepatocellular carcinoma. Hepatology 2013, 57, 1906–1918. [Google Scholar] [CrossRef] [PubMed]
- Tilley, A.M.C.; Howard, C.M.; Sridharan, S.; Subramaniyan, B.; Bearss, N.R.; Alkhalili, S.; Raman, D. The CXCR4-Dependent LASP1-Ago2 Interaction in Triple-Negative Breast Cancer. Cancers 2020, 12, 2455. [Google Scholar] [CrossRef]
- MacFarlane, L.-A.; Gu, Y.; Casson, A.G.; Murphy, P.R. Regulation of Fibroblast Growth Factor-2 by an Endogenous Antisense RNA and by Argonaute-2. Mol. Endocrinol. 2010, 24, 800–812. [Google Scholar] [CrossRef]
- Vaksman, O.; Hetland, T.E.; Trope, C.G.; Reich, R.; Davidson, B. Argonaute, Dicer, and Drosha are up-regulated along tumor progression in serous ovarian carcinoma. Hum. Pathol. 2012, 43, 2062–2069. [Google Scholar] [CrossRef] [PubMed]
- Conger, A.; Martin, E.; Yan, T.; Rhodes, L.; Hoang, V.; La, J.; Anbalagan, M.; Burks, H.; Rowan, B.; Nephew, K.; et al. Argonaute 2 Expression Correlates with a Luminal B Breast Cancer Subtype and Induces Estrogen Receptor Alpha Isoform Variation. ncRNA 2016, 2, 8. [Google Scholar] [CrossRef] [PubMed]
- Casey, M.C.; Prakash, A.; Holian, E.; McGuire, A.; Kalinina, O.; Shalaby, A.; Curran, C.; Webber, M.; Callagy, G.; Bourke, E.; et al. Quantifying Argonaute 2 (Ago2) expression to stratify breast cancer. BMC Cancer 2019, 19, 712. [Google Scholar] [CrossRef]
- Yang, H.; Dinney, C.P.; Ye, Y.; Zhu, Y.; Grossman, H.B.; Wu, X. Evaluation of Genetic Variants in MicroRNA-Related Genes and Risk of Bladder Cancer. Cancer Res. 2008, 68, 2530–2537. [Google Scholar] [CrossRef]
- Rabien, A.; Ratert, N.; Högner, A.; Erbersdobler, A.; Jung, K.; Ecke, T.H.; Kilic, E. Diagnostic and Prognostic Potential of MicroRNA Maturation Regulators Drosha, AGO1 and AGO2 in Urothelial Carcinomas of the Bladder. Int. J. Mol. Sci. 2018, 19. [Google Scholar] [CrossRef] [PubMed]
- Papachristou, D.J.; Korpetinou, A.; Giannopoulou, E.; Antonacopoulou, A.G.; Papadaki, H.; Grivas, P.; Scopa, C.D.; Kalofonos, H.P. Expression of the ribonucleases Drosha, Dicer, and Ago2 in colorectal carcinomas. Virchows Arch. 2011, 459, 431–440. [Google Scholar] [CrossRef]
- Zhang, J.; Fan, X.-S.; Wang, C.-X.; Liu, B.; Li, Q.; Zhou, X.-J. Up-regulation of Ago2 expression in gastric carcinoma. Med. Oncol. 2013, 30, 628. [Google Scholar] [CrossRef] [PubMed]
- Feng, B.; Hu, P.; Lu, S.-J.; Chen, J.-B.; Ge, R.-L. Increased argonaute 2 expression in gliomas and its association with tumor progression and poor prognosis. Asian Pac. J. Cancer Prev. 2014, 15, 4079–4083. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Yu, C.; Gao, H.; Li, Y. Argonaute proteins: Potential biomarkers for human colon cancer. BMC Cancer 2010, 10, 38. [Google Scholar] [CrossRef]
- Chang, S.S.; Smith, I.; Glazer, C.; Hennessey, P.; Califano, J.A. EIF2C is overexpressed and amplified in head and neck squamous cell carcinoma. ORL J. Otorhinolaryngol. Relat. Spec. 2010, 72, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Sand, M.; Skrygan, M.; Georgas, D.; Arenz, C.; Gambichler, T.; Sand, D.; Altmeyer, P.; Bechara, F.G. Expression levels of the microRNA maturing microprocessor complex component DGCR8 and the RNA-induced silencing complex (RISC) components argonaute-1, argonaute-2, PACT, TARBP1, and TARBP2 in epithelial skin cancer. Mol. Carcinog. 2012, 51, 916–922. [Google Scholar] [CrossRef]
- Piroozian, F.; Bagheri Varkiyani, H.; Koolivand, M.; Ansari, M.; Afsa, M.; AtashAbParvar, A.; MalekZadeh, K. The impact of variations in transcription of DICER and AGO2 on exacerbation of childhood B-cell lineage acute lymphoblastic leukaemia. Int. J. Exp. Pathol. 2019, 100, 184–191. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.S.; Min, H.; Ha, J.Y.; Kim, B.H.; Choi, M.S.; Kim, S. Dysregulation of the miRNA biogenesis components DICER1, DROSHA, DGCR8 and AGO2 in clear cell renal cell carcinoma in both a Korean cohort and the cancer genome atlas kidney clear cell carcinoma cohort. Oncol. Lett. 2019, 18, 4337–4345. [Google Scholar] [CrossRef]
- Kitagawa, N.; Ojima, H.; Shirakihara, T.; Shimizu, H.; Kokubu, A.; Urushidate, T.; Totoki, Y.; Kosuge, T.; Miyagawa, S.; Shibata, T. Downregulation of the microRNA biogenesis components and its association with poor prognosis in hepatocellular carcinoma. Cancer Sci. 2013, 104, 543–551. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Meng, J.; Zhai, Y.; Zhang, H.; Yu, L.; Wang, Z.; Zhang, X.; Cao, P.; Chen, X.; Han, Y.; et al. Argonaute 2 and nasopharyngeal carcinoma: A genetic association study and functional analysis. BMC Cancer 2015, 15, 862. [Google Scholar] [CrossRef][Green Version]
- Sung, H.; Jeon, S.; Lee, K.-M.; Han, S.; Song, M.; Choi, J.-Y.; Park, S.K.; Yoo, K.-Y.; Noh, D.-Y.; Ahn, S.-H.; et al. Common genetic polymorphisms of microRNA biogenesis pathway genes and breast cancer survival. BMC Cancer 2012, 12, 195. [Google Scholar] [CrossRef]
- Bermisheva, M.A.; Takhirova, Z.R.; Gilyazova, I.R.; Khusnutdinova, E.K. MicroRNA Biogenesis Pathway Gene Polymorphisms Are Associated with Breast Cancer Risk. Russ. J. Genet. 2018, 54, 568–575. [Google Scholar] [CrossRef]
- Fawzy, M.S.; Toraih, E.A.; Alelwani, W.; Kattan, S.W.; Alnajeebi, A.M.; Hassan, R. The prognostic value of microRNA-biogenesis genes Argonaute 1 and 2 variants in breast cancer patients. Am. J. Transl. Res. 2020, 12, 1994–2006. [Google Scholar]
- Song, X.; Zhong, H.; Wu, Q.; Wang, M.; Zhou, J.; Zhou, Y.; Lu, X.; Ying, B. Association between SNPs in microRNA machinery genes and gastric cancer susceptibility, invasion, and metastasis in Chinese Han population. Oncotarget 2017, 8, 86435–86446. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Yin, Z.; Li, X.; Xia, L.; Zhou, B. Polymorphisms in GEMIN4 and AGO1 Genes Are Associated with the Risk of Lung Cancer: A Case-Control Study in Chinese Female Non-Smokers. Int. J. Environ. Res. Public Health 2016, 13. [Google Scholar] [CrossRef]
- Kim, J.-S.; Choi, Y.Y.; Jin, G.; Kang, H.-G.; Choi, J.-E.; Jeon, H.-S.; Lee, W.-K.; Kim, D.-S.; Kim, C.H.; Kim, Y.J.; et al. Association of a common AGO1 variant with lung cancer risk: A two-stage case-control study. Mol. Carcinog. 2010, 49, 913–921. [Google Scholar] [CrossRef]
- Horikawa, Y.; Wood, C.G.; Yang, H.; Zhao, H.; Ye, Y.; Gu, J.; Lin, J.; Habuchi, T.; Wu, X. Single nucleotide polymorphisms of microRNA machinery genes modify the risk of renal cell carcinoma. Clin. Cancer Res. 2008, 14, 7956–7962. [Google Scholar] [CrossRef]
- Permuth-Wey, J.; Kim, D.; Tsai, Y.-Y.; Lin, H.-Y.; Chen, Y.A.; Barnholtz-Sloan, J.; Birrer, M.J.; Bloom, G.; Chanock, S.J.; Chen, Z.; et al. LIN28B Polymorphisms Influence Susceptibility to Epithelial Ovarian Cancer. Cancer Res. 2011, 71, 3896–3903. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez-Camino, A.; Lopez-Lopez, E.; Martin-Guerrero, I.; Piñan, M.A.; Garcia-Miguel, P.; Sanchez-Toledo, J.; Carbone Bañeres, A.; Uriz, J.; Navajas, A.; Garcia-Orad, A. Noncoding RNA–related polymorphisms in pediatric acute lymphoblastic leukemia susceptibility. Pediatr. Res. 2014, 75, 767–773. [Google Scholar] [CrossRef]
- Martin-Guerrero, I.; Gutierrez-Camino, A.; Lopez-Lopez, E.; Bilbao-Aldaiturriaga, N.; Pombar-Gomez, M.; Ardanaz, M.; Garcia-Orad, A. Genetic Variants in MiRNA Processing Genes and Pre-MiRNAs Are Associated with the Risk of Chronic Lymphocytic Leukemia. PLoS ONE 2015, 10, e0118905. [Google Scholar] [CrossRef]
- Peckham-Gregory, E.C.; Thapa, D.R.; Martinson, J.; Duggal, P.; Penugonda, S.; Bream, J.H.; Chang, P.-Y.; Dandekar, S.; Chang, S.-C.; Detels, R.; et al. MicroRNA-related polymorphisms and non-Hodgkin lymphoma susceptibility in the Multicenter AIDS Cohort Study. Cancer Epidemiol. 2016, 45, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Nikolić, Z.; Savić Pavićević, D.; Vučić, N.; Cerović, S.; Vukotić, V.; Brajušković, G. Genetic variants in RNA-induced silencing complex genes and prostate cancer. World J. Urol. 2017, 35, 613–624. [Google Scholar] [CrossRef]
- Dobrijević, Z.; Matijašević, S.; Savić-Pavićević, D.; Brajušković, G. Association between genetic variants in genes encoding Argonaute proteins and cancer risk: A meta-analysis. Pathol. Res. Pract. 2020, 216, 152906. [Google Scholar] [CrossRef]
- Hu, B.; Zhong, L.; Weng, Y.; Peng, L.; Huang, Y.; Zhao, Y.; Liang, X.-J. Therapeutic siRNA: State of the art. Signal. Transduct. Target. Ther. 2020, 5, 101. [Google Scholar] [CrossRef] [PubMed]
- Hager, S.; Fittler, F.J.; Wagner, E.; Bros, M. Nucleic Acid-Based Approaches for Tumor Therapy. Cells 2020, 9. [Google Scholar] [CrossRef] [PubMed]
- Watashi, K.; Yeung, M.L.; Starost, M.F.; Hosmane, R.S.; Jeang, K.-T. Identification of Small Molecules That Suppress MicroRNA Function and Reverse Tumorigenesis. J. Biol. Chem. 2010, 285, 24707–24716. [Google Scholar] [CrossRef] [PubMed]
- Masciarelli, S.; Quaranta, R.; Iosue, I.; Colotti, G.; Padula, F.; Varchi, G.; Fazi, F.; Del Rio, A. A Small-Molecule Targeting the MicroRNA Binding Domain of Argonaute 2 improves the Retinoic Acid Differentiation Response of the Acute Promyelocytic Leukemia Cell Line NB4. ACS Chem. Biol. 2014, 9, 1674–1679. [Google Scholar] [CrossRef]
- Fuji, T.; Umeda, Y.; Nyuya, A.; Taniguchi, F.; Kawai, T.; Yasui, K.; Toshima, T.; Yoshida, K.; Fujiwara, T.; Goel, A.; et al. Detection of circulating microRNAs with Ago2 complexes to monitor the tumor dynamics of colorectal cancer patients during chemotherapy: Detection of circulating MicroRNAs. Int. J. Cancer 2019, 144, 2169–2180. [Google Scholar] [CrossRef]
- Unal, O.; Akkoc, Y.; Kocak, M.; Nalbat, E.; Dogan-Ekici, A.I.; Yagci Acar, H.; Gozuacik, D. Treatment of breast cancer with autophagy inhibitory microRNAs carried by AGO2-conjugated nanoparticles. J. Nanobiotechnol. 2020, 18, 65. [Google Scholar] [CrossRef]


| AGO | SNP | Cancer Type | Odd Ratio | Reference |
|---|---|---|---|---|
| AGO1 | rs595055 C/T | Breast cancer | Increased | [111] |
| rs636832 A/A | Breast cancer | Increased | [112] | |
| rs636832 AA + A | Gastric cancer | Decreased | [113] | |
| rs636832 A > G | Lung cancer | Decreased | [115] | |
| rs595961 AG | Lung cancer | Increased | [114] | |
| rs595961 AG + GG | Clear cell renal cell carcinoma | Decreased | [116] | |
 | ||||
| AGO2 | rs3928672 GA + AA | Nasopharyngeal carcinoma | Increased | [109] |
| rs11786030 A/G | Breast cancer | Increased | [110] | |
| rs2292779 C/G | Breast cancer | Increased | [110] | |
| rs2977490 G/G | Breast cancer | Increased | [112] | |
 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nowak, I.; Sarshad, A.A. Argonaute Proteins Take Center Stage in Cancers. Cancers 2021, 13, 788. https://doi.org/10.3390/cancers13040788
Nowak I, Sarshad AA. Argonaute Proteins Take Center Stage in Cancers. Cancers. 2021; 13(4):788. https://doi.org/10.3390/cancers13040788
Chicago/Turabian StyleNowak, Iwona, and Aishe A. Sarshad. 2021. "Argonaute Proteins Take Center Stage in Cancers" Cancers 13, no. 4: 788. https://doi.org/10.3390/cancers13040788
APA StyleNowak, I., & Sarshad, A. A. (2021). Argonaute Proteins Take Center Stage in Cancers. Cancers, 13(4), 788. https://doi.org/10.3390/cancers13040788






