Differential Effects of Trp53 Alterations in Murine Colorectal Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Mice
- i.
- Apcfl/fl (“Apc”);
- ii.
- Apcfl/fl/KrasLSL-G12D/+ (“AK”);
- iii.
- Apcfl/fl/KrasLSL-G12D/+ /Trp53fl/fl (“AKPfl”);
- iv.
- Apcfl/fl/KrasLSL-G12D/+ /Trp53LSL-R172H/+ (“AKPr”);
2.3. Animal Experiments
2.4. Migration Assay
2.5. Histology
2.6. Immunohistochemistry
2.7. Microsatellite Instability Testing
2.8. Next Generation Sequencing and Data Analysis
2.9. Statistics
3. Results
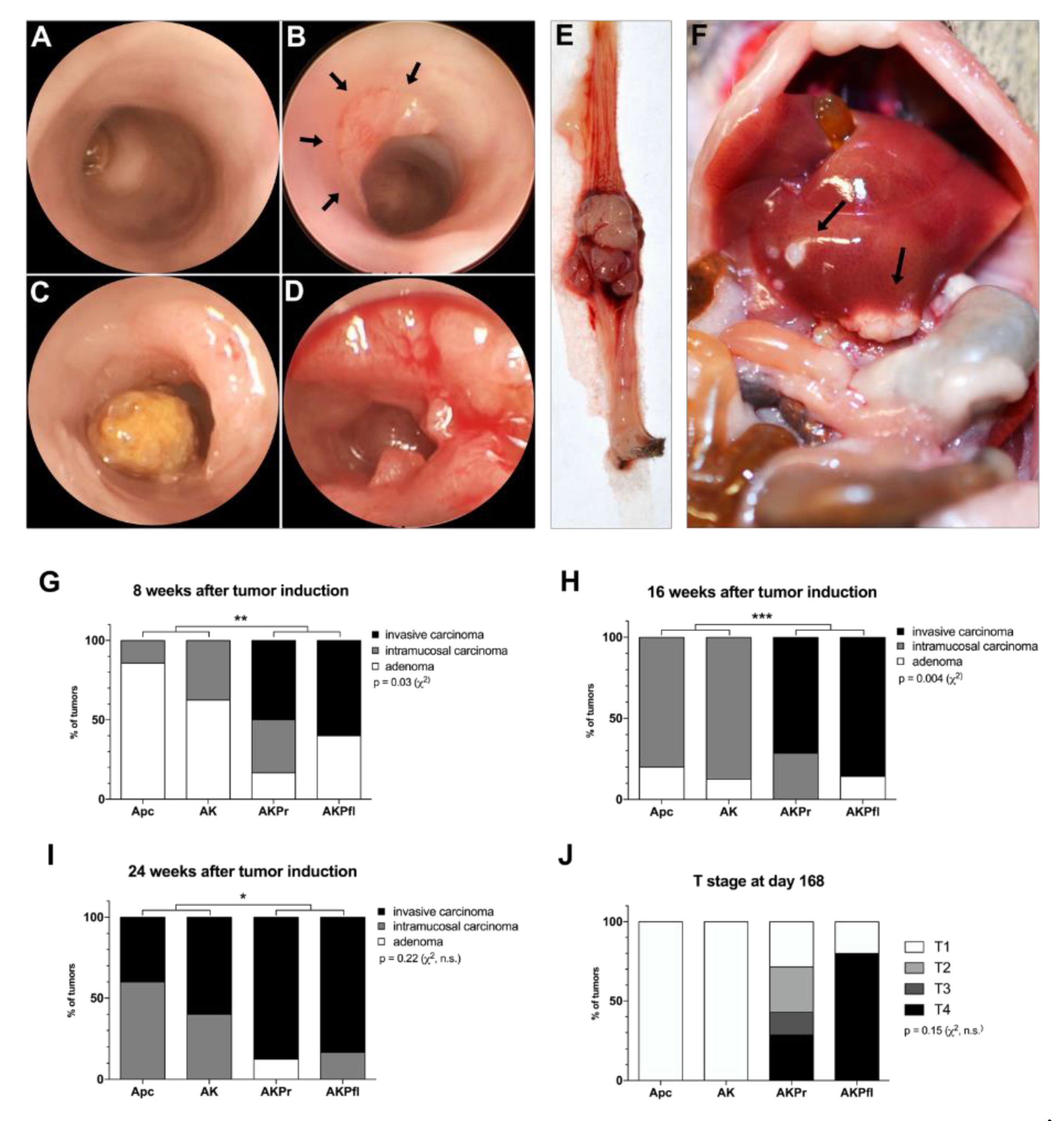
3.1. Sequential Addition of Mutations Accelerates Malignant Tumor Formation
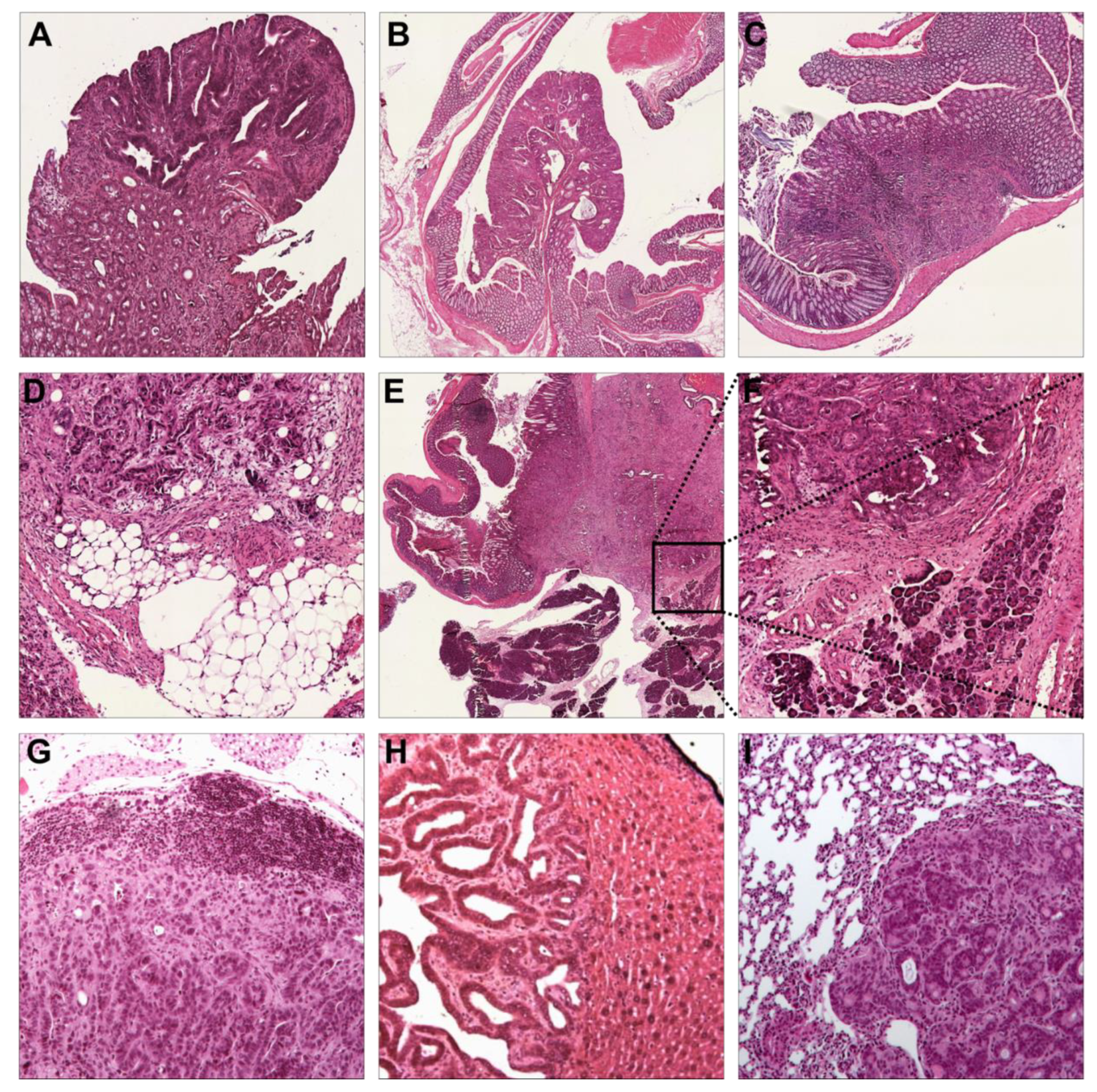
3.2. Immunohistochemistry Confirms Colorectal Origin of Primary Tumors and Metastases
3.3. Trp53 Alterations Accelerate Differentiation, Local Invasion, and Metastasis
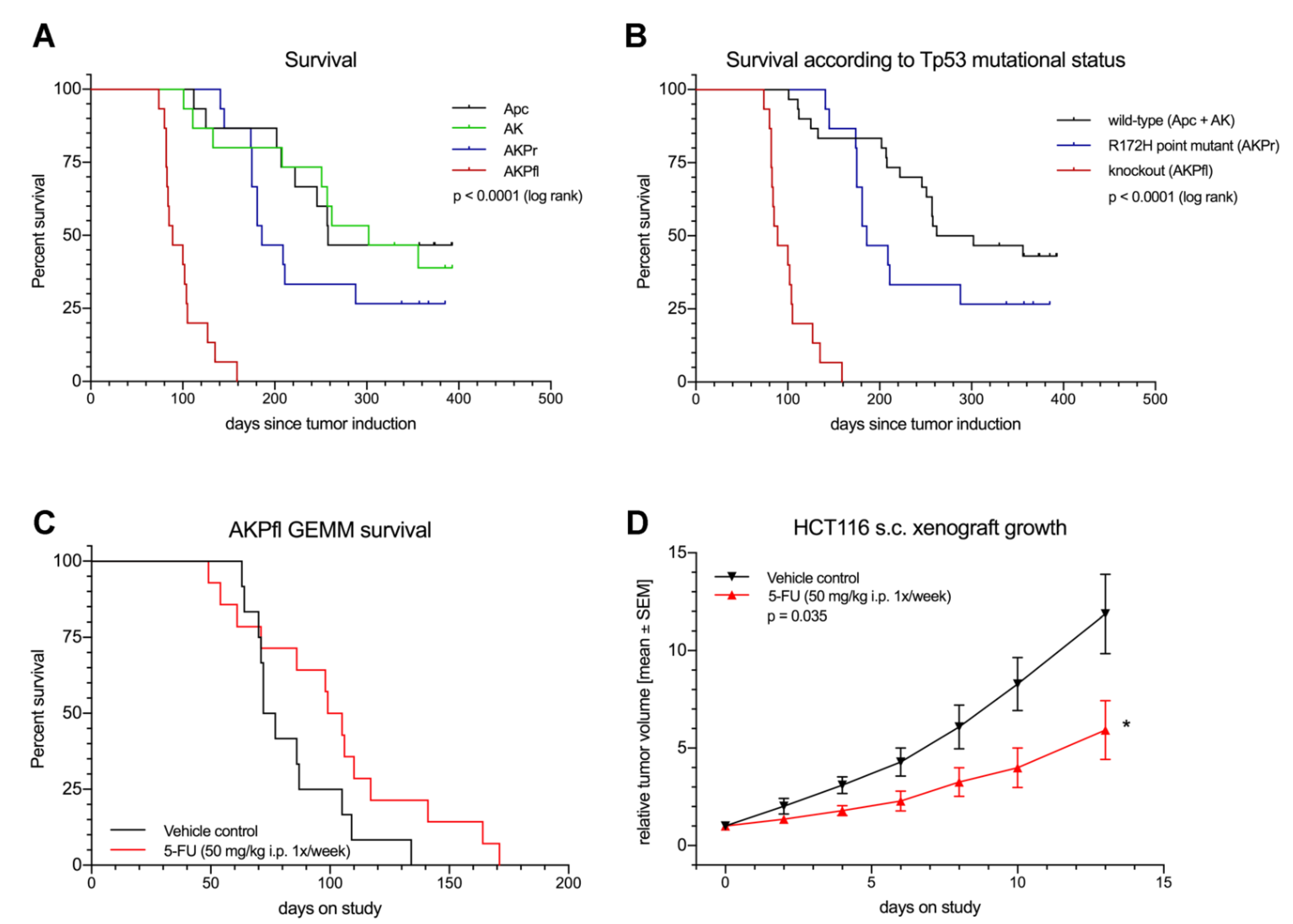
3.4. Trp53 Loss and Point Mutation Have Differential Effects on Survival and in Vitro Cell Migration
3.5. GEMM Tumors Mimic Human Response to Treatment
3.6. Mutations in Trp53 Destabilize the Genetic Integrity of Mouse Tumors
3.7. Many Key Mutations Are Shared between Human and Murine CRC
3.8. Differential Effects of Trp53 Mutation and Knockout on Tumor Biology
3.9. Copy Number Variation and Pathway Analyses Reveal Similarities between Human and Murine CRC
3.10. Spontaneous Development of Microsatellite Instability in Murine CRC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cancer Genome Atlas Network. Comprehensive Molecular Characterization of Human Colon and Rectal Cancer. Nature 2012, 487, 330–337. [Google Scholar] [CrossRef] [PubMed]
- Powell, S.M.; Zilz, N.; Beazer-Barclay, Y.; Bryan, T.M.; Hamilton, S.R.; Thibodeau, S.N.; Vogelstein, B.; Kinzler, K.W. APC Mutations Occur Early during Colorectal Tumorigenesis. Nature 1992, 359, 235–237. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.R.; Vogelstein, B. Genetic Alterations in the Adenoma—Carcinoma Sequence. Cancer 1992, 70, 1727–1731. [Google Scholar] [CrossRef]
- Felipe De Sousa, E.M.; Wang, X.; Jansen, M.; Fessler, E.; Trinh, A.; de Rooij, L.P.M.H.; de Jong, J.H.; de Boer, O.J.; van Leersum, R.; Bijlsma, M.F.; et al. Poor-Prognosis Colon Cancer Is Defined by a Molecularly Distinct Subtype and Develops from Serrated Precursor Lesions. Nat. Med. 2013, 19, 614–618. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Salas, N.; Dominguez, G.; Barderas, R.; Mendiola, M.; García-Albéniz, X.; Maurel, J.; Batlle, J.F. Clinical Relevance of Colorectal Cancer Molecular Subtypes. Crit. Rev. Oncol. Hematol. 2017, 109, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Schölch, S.; Rauber, C.; Tietz, A.; Rahbari, N.N.; Bork, U.; Schmidt, T.; Kahlert, C.; Haberkorn, U.; Tomai, M.A.; Lipson, K.E.; et al. Radiotherapy Combined with TLR7/8 Activation Induces Strong Immune Responses against Gastrointestinal Tumors. Oncotarget 2015, 6, 4663–4676. [Google Scholar] [CrossRef] [PubMed]
- Schölch, S.; Rauber, C.; Weitz, J.; Koch, M.; Huber, P.E. TLR Activation and Ionizing Radiation Induce Strong Immune Responses against Multiple Tumor Entities. Oncoimmunology 2015, 4, e1042201. [Google Scholar] [CrossRef]
- Van Noort, V.; Schölch, S.; Iskar, M.; Zeller, G.; Ostertag, K.; Schweitzer, C.; Werner, K.; Weitz, J.; Koch, M.; Bork, P. Novel Drug Candidates for the Treatment of Metastatic Colorectal Cancer through Global Inverse Gene-Expression Profiling. Cancer Res. 2014, 74, 5690–5699. [Google Scholar] [CrossRef]
- Kochall, S.; Thepkaysone, M.-L.; García, S.A.; Betzler, A.M.; Weitz, J.; Reissfelder, C.; Schölch, S. Isolation of Circulating Tumor Cells in an Orthotopic Mouse Model of Colorectal Cancer. J. Vis. Exp. 2017. [Google Scholar] [CrossRef]
- Schölch, S.; García, S.A.; Iwata, N.; Niemietz, T.; Betzler, A.M.; Nanduri, L.K.; Bork, U.; Kahlert, C.; Thepkaysone, M.-L.; Swiersy, A.; et al. Circulating Tumor Cells Exhibit Stem Cell Characteristics in an Orthotopic Mouse Model of Colorectal Cancer. Oncotarget 2016, 7, 27232–27242. [Google Scholar] [CrossRef]
- Sharpless, N.E.; Depinho, R.A. The Mighty Mouse: Genetically Engineered Mouse Models in Cancer Drug Development. Nat. Rev. Drug Discov. 2006, 5, 741–754. [Google Scholar] [CrossRef] [PubMed]
- Roper, J.; Hung, K.E. Priceless GEMMs: Genetically Engineered Mouse Models for Colorectal Cancer Drug Development. Trends Pharmacol. Sci. 2012, 33, 449–455. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Lima, A.; Molina, R.; Hamilton, P.; Clermont, A.C.; Devasthali, V.; Thompson, J.D.; Cheng, J.H.; Bou Reslan, H.; Ho, C.C.K.; et al. Assessing Therapeutic Responses in Kras Mutant Cancers Using Genetically Engineered Mouse Models. Nat. Biotechnol. 2010, 28, 585–593. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Murriel, C.L.; Johnson, L. Genetically Engineered Mouse Models: Closing the Gap between Preclinical Data and Trial Outcomes. Cancer Res. 2012, 72, 2695–2700. [Google Scholar] [CrossRef]
- Zhu, L.; Hissa, B.; Győrffy, B.; Jann, J.-C.; Yang, C.; Reissfelder, C.; Schölch, S. Characterization of Stem-like Circulating Tumor Cells in Pancreatic Cancer. Diagnostics 2020, 10, 305. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Kan, K.-J.; Grün, J.L.; Hissa, B.; Yang, C.; Győrffy, B.; Loges, S.; Reißfelder, C.; Schölch, S. GAS2L1 Is a Potential Biomarker of Circulating Tumor Cells in Pancreatic Cancer. Cancers 2020, 12, 3774. [Google Scholar] [CrossRef]
- Seidlitz, T.; Chen, Y.-T.; Uhlemann, H.; Schölch, S.; Kochall, S.; Merker, S.R.; Klimova, A.; Hennig, A.; Schweitzer, C.; Pape, K.; et al. Mouse Models of Human Gastric Cancer Subtypes With Stomach-Specific CreERT2-Mediated Pathway Alterations. Gastroenterology 2019, 157, 1599–1614.e2. [Google Scholar] [CrossRef]
- Shibata, H.; Toyama, K.; Shioya, H.; Ito, M.; Hirota, M.; Hasegawa, S.; Matsumoto, H.; Takano, H.; Akiyama, T.; Toyoshima, K.; et al. Rapid Colorectal Adenoma Formation Initiated by Conditional Targeting of the Apc Gene. Science 1997, 278, 120–123. [Google Scholar]
- Hung, K.E.; Maricevich, M.A.; Richard, L.G.; Chen, W.Y.; Richardson, M.P.; Kunin, A.; Bronson, R.T.; Mahmood, U.; Kucherlapati, R. Development of a Mouse Model for Sporadic and Metastatic Colon Tumors and Its Use in Assessing Drug Treatment. Proc. Natl. Acad. Sci. USA 2010, 107, 1565–1570. [Google Scholar] [CrossRef]
- Betzler, A.M.; Kochall, S.; Blickensdörfer, L.; Garcia, S.A.; Thepkaysone, M.-L.; Nanduri, L.K.; Muders, M.H.; Weitz, J.; Reissfelder, C.; Schölch, S. A Genetically Engineered Mouse Model of Sporadic Colorectal Cancer. J. Vis. Exp. 2017. [Google Scholar] [CrossRef]
- Kuraguchi, M.; Wang, X.-P.; Bronson, R.T.; Rothenberg, R.; Ohene-Baah, N.Y.; Lund, J.J.; Kucherlapati, M.; Maas, R.L.; Kucherlapati, R. Adenomatous Polyposis Coli (APC) Is Required for Normal Development of Skin and Thymus. PLoS Genet. 2006, 2, e146. [Google Scholar] [CrossRef] [PubMed]
- Johnson, L.; Mercer, K.; Greenbaum, D.; Bronson, R.T.; Crowley, D.; Tuveson, D.A.; Jacks, T. Somatic Activation of the K-Ras Oncogene Causes Early Onset Lung Cancer in Mice. Nature 2001, 410, 1111–1116. [Google Scholar] [CrossRef]
- Olive, K.P.; Tuveson, D.A.; Ruhe, Z.C.; Yin, B.; Willis, N.A.; Bronson, R.T.; Crowley, D.; Jacks, T. Mutant P53 Gain of Function in Two Mouse Models of Li-Fraumeni Syndrome. Cell 2004, 119, 847–860. [Google Scholar] [CrossRef] [PubMed]
- Marino, S.; Vooijs, M.; van Der Gulden, H.; Jonkers, J.; Berns, A. Induction of Medulloblastomas in P53-Null Mutant Mice by Somatic Inactivation of Rb in the External Granular Layer Cells of the Cerebellum. Genes Dev. 2000, 14, 994–1004. [Google Scholar]
- Boivin, G.P.; Washington, K.; Yang, K.; Ward, J.M.; Pretlow, T.P.; Russell, R.; Besselsen, D.G.; Godfrey, V.L.; Doetschman, T.; Dove, W.F.; et al. Pathology of Mouse Models of Intestinal Cancer: Consensus Report and Recommendations. Gastroenterology 2003, 124, 762–777. [Google Scholar] [CrossRef] [PubMed]
- Washington, M.K.; Powell, A.E.; Sullivan, R.; Sundberg, J.P.; Wright, N.; Coffey, R.J.; Dove, W.F. Pathology of Rodent Models of Intestinal Cancer: Progress Report and Recommendations. Gastroenterology 2013, 144, 705–717. [Google Scholar] [CrossRef]
- White, B.D.; Chien, A.J.; Dawson, D.W. Dysregulation of Wnt/β-Catenin Signaling in Gastrointestinal Cancers. Gastroenterology 2012, 142, 219–232. [Google Scholar] [CrossRef]
- Liu, Q.; Teh, M.; Ito, K.; Shah, N.; Ito, Y.; Yeoh, K.G. CDX2 Expression Is Progressively Decreased in Human Gastric Intestinal Metaplasia, Dysplasia and Cancer. Mod. Pathol. 2007, 20, 1286–1297. [Google Scholar] [CrossRef] [PubMed]
- Goh, A.M.; Coffill, C.R.; Lane, D.P. The Role of Mutant P53 in Human Cancer. J. Pathol. 2011, 223, 116–126. [Google Scholar] [CrossRef]
- He, J.; Pei, L.; Jiang, H.; Yang, W.; Chen, J.; Liang, H. Chemoresistance of Colorectal Cancer to 5-Fluorouracil Is Associated with Silencing of the BNIP3 Gene through Aberrant Methylation. J Cancer 2017, 8, 1187–1196. [Google Scholar] [CrossRef]
- Ahlquist, T.; Bottillo, I.; Danielsen, S.A.; Meling, G.I.; Rognum, T.O.; Lind, G.E.; Dallapiccola, B.; Lothe, R.A. RAS Signaling in Colorectal Carcinomas through Alteration of RAS, RAF, NF1, and/or RASSF1A. Neoplasia 2008, 10, 680–686. [Google Scholar] [CrossRef]
- Sinnott, R.; Winters, L.; Larson, B.; Mytsa, D.; Taus, P.; Cappell, K.M.; Whitehurst, A.W. Mechanisms Promoting Escape from Mitotic Stress-Induced Tumor Cell Death. Cancer Res. 2014, 74, 3857–3869. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Wang, Y.; Vikis, H.; Maciag, A.; Wang, D.; Lu, Y.; Liu, Y.; You, M. Candidate Lung Tumor Susceptibility Genes Identified through Whole-Genome Association Analyses in Inbred Mice. Nat. Genet. 2006, 38, 888–895. [Google Scholar] [CrossRef] [PubMed]
- Steinert, G.; Schölch, S.; Niemietz, T.; Iwata, N.; García, S.A.; Behrens, B.; Voigt, A.; Kloor, M.; Benner, A.; Bork, U.; et al. Immune Escape and Survival Mechanisms in Circulating Tumor Cells of Colorectal Cancer. Cancer Res. 2014, 74, 1694–1704. [Google Scholar] [CrossRef] [PubMed]
- Fabregat, A.; Sidiropoulos, K.; Garapati, P.; Gillespie, M.; Hausmann, K.; Haw, R.; Jassal, B.; Jupe, S.; Korninger, F.; McKay, S.; et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2016, 44, D481–D487. [Google Scholar] [CrossRef]
- Milacic, M.; Haw, R.; Rothfels, K.; Wu, G.; Croft, D.; Hermjakob, H.; D’Eustachio, P.; Stein, L. Annotating Cancer Variants and Anti-Cancer Therapeutics in Reactome. Cancers 2012, 4, 1180–1211. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and Integrative Analysis of Large Gene Lists Using DAVID Bioinformatics Resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Woerner, S.M.; Tosti, E.; Yuan, Y.P.; Kloor, M.; Bork, P.; Edelmann, W.; Gebert, J. Detection of Coding Microsatellite Frameshift Mutations in DNA Mismatch Repair-Deficient Mouse Intestinal Tumors. Mol. Carcinog. 2015, 54, 1376–1386. [Google Scholar] [CrossRef] [PubMed]
- Su, L.K.; Kinzler, K.W.; Vogelstein, B.; Preisinger, A.C.; Moser, A.R.; Luongo, C.; Gould, K.A.; Dove, W.F. Multiple Intestinal Neoplasia Caused by a Mutation in the Murine Homolog of the APC Gene. Science 1992, 256, 668–670. [Google Scholar] [CrossRef]
- Tetteh, P.W.; Kretzschmar, K.; Begthel, H.; van den Born, M.; Korving, J.; Morsink, F.; Farin, H.; van Es, J.H.; Offerhaus, G.J.A.; Clevers, H. Generation of an Inducible Colon-Specific Cre Enzyme Mouse Line for Colon Cancer Research. Proc. Natl. Acad. Sci. USA 2016, 113, 11859–11864. [Google Scholar] [CrossRef]
- Jackson, E.L.; Olive, K.P.; Tuveson, D.A.; Bronson, R.; Crowley, D.; Brown, M.; Jacks, T. The Differential Effects of Mutant P53 Alleles on Advanced Murine Lung Cancer. Cancer Res. 2005, 65, 10280–10288. [Google Scholar] [CrossRef] [PubMed]
- Deschoemaeker, S.; Di Conza, G.; Lilla, S.; Martín-Pérez, R.; Mennerich, D.; Boon, L.; Hendrikx, S.; Maddocks, O.D.K.; Marx, C.; Radhakrishnan, P.; et al. PHD1 Regulates P53-Mediated Colorectal Cancer Chemoresistance. EMBO Mol. Med. 2015, 7, 1350–1365. [Google Scholar] [CrossRef] [PubMed]
- Houghton, P.J.; Houghton, J.A. Evaluation of Single-Agent Therapy in Human Colorectal Tumour Xenografts. Br. J. Cancer 1978, 37, 833–840. [Google Scholar] [CrossRef]
- De Gramont, A.; Figer, A.; Seymour, M.; Homerin, M.; Hmissi, A.; Cassidy, J.; Boni, C.; Cortes-Funes, H.; Cervantes, A.; Freyer, G.; et al. Leucovorin and Fluorouracil with or without Oxaliplatin as First-Line Treatment in Advanced Colorectal Cancer. J. Clin. Oncol. 2000, 18, 2938–2947. [Google Scholar] [CrossRef] [PubMed]
- Coffee, E.M.; Faber, A.C.; Roper, J.; Sinnamon, M.J.; Goel, G.; Keung, L.; Wang, W.V.; Vecchione, L.; de Vriendt, V.; Weinstein, B.J.; et al. Concomitant BRAF and PI3K/MTOR Blockade Is Required for Effective Treatment of BRAF(V600E) Colorectal Cancer. Clin. Cancer Res. 2013, 19, 2688–2698. [Google Scholar] [CrossRef] [PubMed]


| A | |||||
| Genotype | Total | TCGA | % TCGA | TCGA >1% | % TCGA >1% |
| Apc | 133 | 8 | 6.0 | 6 | 4.5 |
| AK | 132 | 10 | 7.6 | 8 | 6.1 |
| AKPr | 2381 | 123 | 5.2 | 108 | 4.5 |
| AKPfl | 2479 | 127 | 5.1 | 103 | 4.2 |
| B | |||||
| Genotype | TCGA>1% Mutations | Shared | Unique | % Unique | % Overlap |
| AKPr | 108 | 41 | 67 | 62.04 | 37.96 |
| AKPfl | 103 | 41 | 62 | 61.19 | 39.81 |
| C | |||||
| Genotype | Affected pathways | Shared | Unique | % Unique | % Overlap |
| AKPr | 99 | 33 | 66 | 66.67 | 33.33 |
| AKPfl | 46 | 33 | 13 | 28.26 | 71.74 |
| Pathways | AKPfl | AKPr | Function |
|---|---|---|---|
| Adhesion | X | - | Adhesion |
| Hedgehog | X | - | Differentiation |
| DNA Repair * | X | X | DNA maintenance |
| Methylation | - | X | DNA methylation |
| Chromatin regulation | - | X | Gene expression |
| Nuclear transcription | X | X | Gene expression |
| Regulation of immune response | X | X | Immune modulation |
| Dap12 | - | X | Immune modulation, tumor cell survival |
| IL-2 | - | X | Immune modulation, identifying self and non self |
| Angiopoeitin | - | X | Migration, survival |
| IL-3, -5, GM-CSF | - | X | Migration, survival |
| Cell cycle regulation | X | - | Proliferation |
| Egfr * | X | X | Proliferation |
| Erbb | X | X | Proliferation |
| Fgfr1 | X | X | Proliferation |
| Igfr | - | X | Proliferation |
| Igfr1 * | X | X | Proliferation |
| IL-6 | X | - | Proliferation |
| Insulin pathway * | X | X | Proliferation |
| Proto-oncogene | - | X | Proliferation |
| Ptk6 * | X | X | Proliferation |
| Raf | - | X | Proliferation |
| Ras * | X | X | Proliferation |
| Ras family of oncogenes | X | X | Proliferation |
| Erk * | X | X | Proliferation, migration |
| Growth factor /receptor transcription | X | X | Proliferation, migration |
| Interleukins | X | X | Proliferation, migration |
| Mapk | X | X | Proliferation, migration |
| Notch * | - | X | Proliferation, migration |
| Notch * | X | - | Proliferation, migration |
| p38 | X | X | Proliferation, migration |
| Vegfr2 | - | X | Proliferation, migration |
| Scf-Kit | X | X | Proliferation, survival |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Betzler, A.M.; Nanduri, L.K.; Hissa, B.; Blickensdörfer, L.; Muders, M.H.; Roy, J.; Jesinghaus, M.; Steiger, K.; Weichert, W.; Kloor, M.; et al. Differential Effects of Trp53 Alterations in Murine Colorectal Cancer. Cancers 2021, 13, 808. https://doi.org/10.3390/cancers13040808
Betzler AM, Nanduri LK, Hissa B, Blickensdörfer L, Muders MH, Roy J, Jesinghaus M, Steiger K, Weichert W, Kloor M, et al. Differential Effects of Trp53 Alterations in Murine Colorectal Cancer. Cancers. 2021; 13(4):808. https://doi.org/10.3390/cancers13040808
Chicago/Turabian StyleBetzler, Alexander M., Lahiri K. Nanduri, Barbara Hissa, Linda Blickensdörfer, Michael H. Muders, Janine Roy, Moritz Jesinghaus, Katja Steiger, Wilko Weichert, Matthias Kloor, and et al. 2021. "Differential Effects of Trp53 Alterations in Murine Colorectal Cancer" Cancers 13, no. 4: 808. https://doi.org/10.3390/cancers13040808
APA StyleBetzler, A. M., Nanduri, L. K., Hissa, B., Blickensdörfer, L., Muders, M. H., Roy, J., Jesinghaus, M., Steiger, K., Weichert, W., Kloor, M., Klink, B., Schroeder, M., Mazzone, M., Weitz, J., Reissfelder, C., Rahbari, N. N., & Schölch, S. (2021). Differential Effects of Trp53 Alterations in Murine Colorectal Cancer. Cancers, 13(4), 808. https://doi.org/10.3390/cancers13040808











