Glioma biopsies Classification Using Raman Spectroscopy and Machine Learning Models on Fresh Tissue Samples
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. Sample Preparation and Analysis
2.3. Raman Spectroscopic Acquisition
2.4. Data Processing
2.5. Statistical Analysis
3. Results
3.1. Classification Features and Algorithms
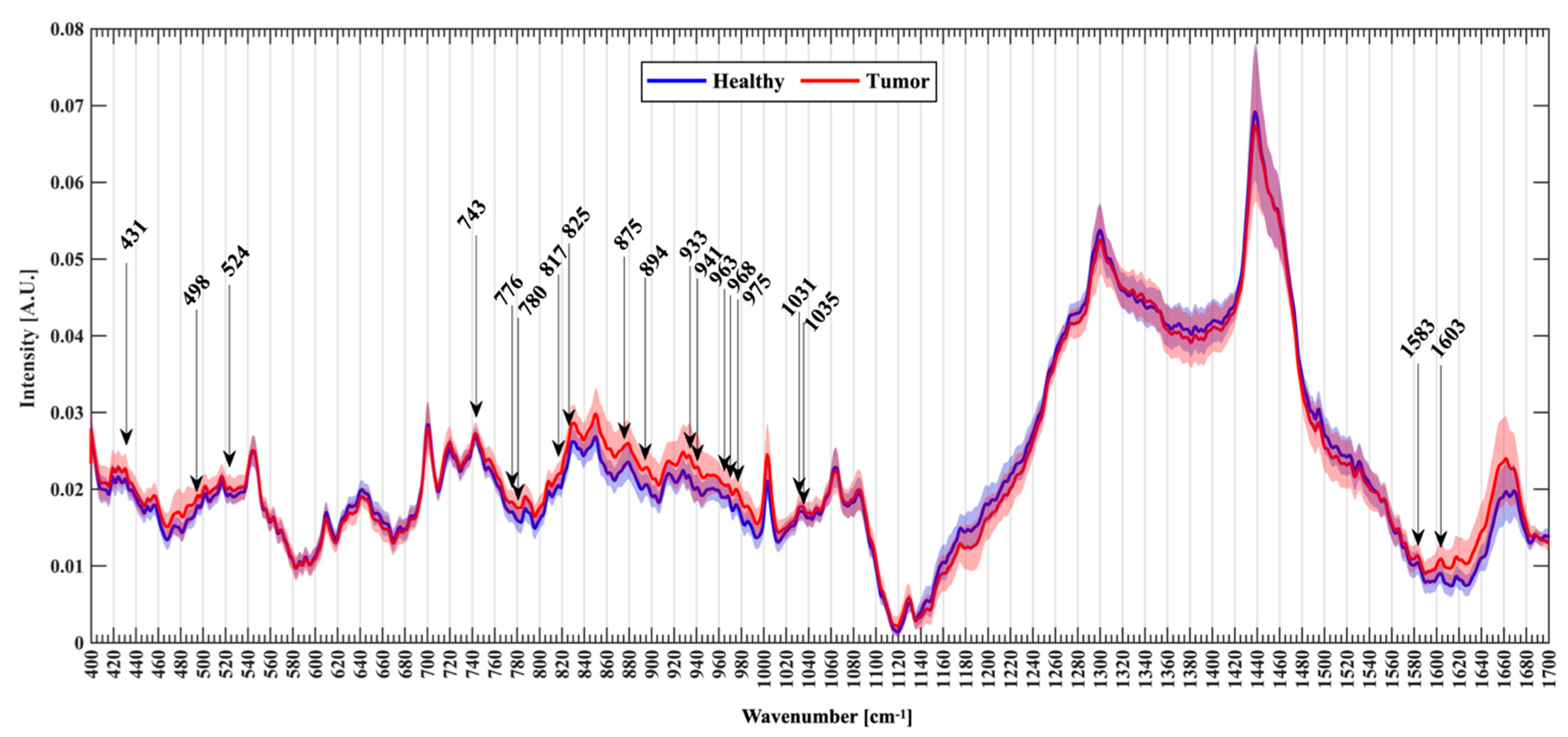
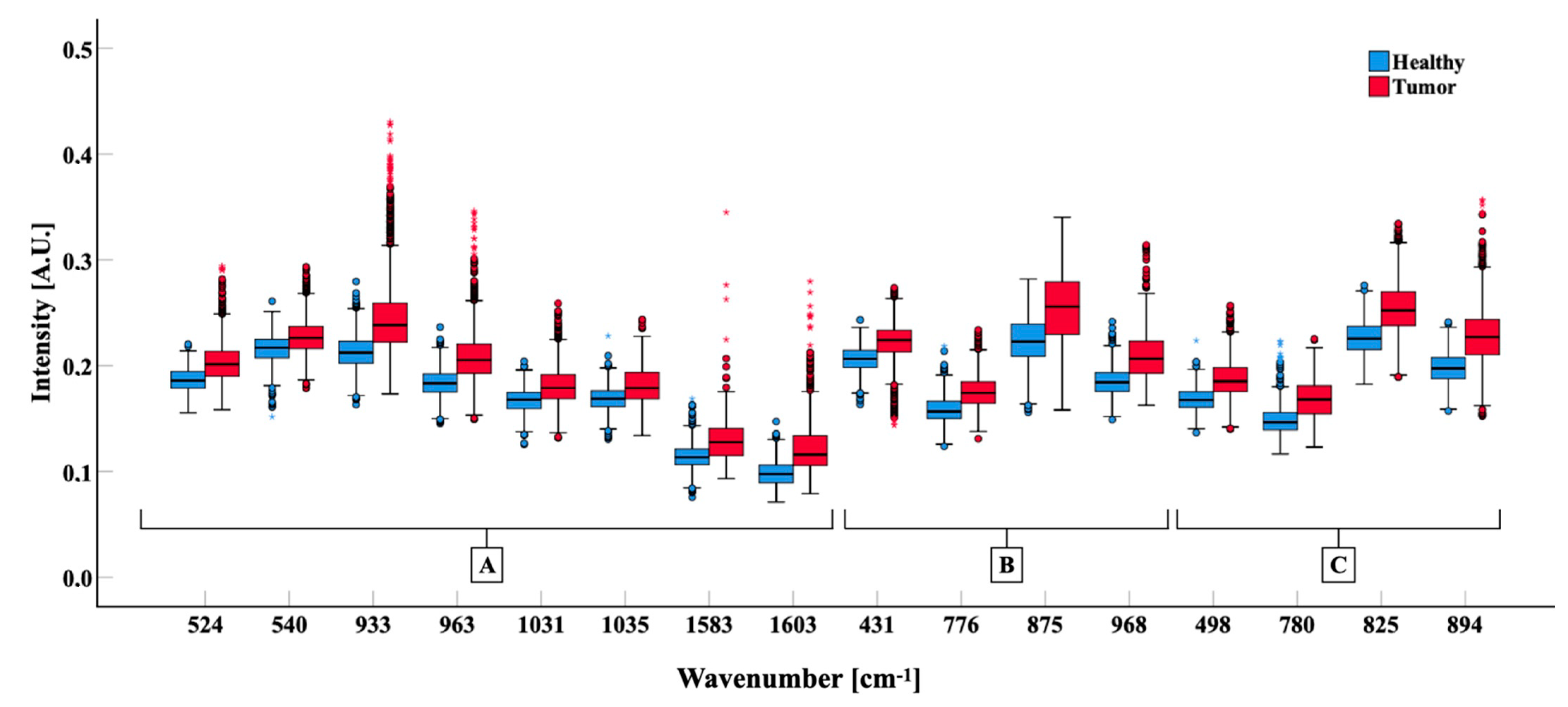
3.2. Spectral Analysis
3.3. Novel Raman Shifts
4. Discussion
4.1. Raman Spectroscopy with Fresh Tissue Samples in an Ex-Vivo Scenario
4.2. Characterization of Raman Shifts
4.3. Diagnostic Performance of Machine Learning Classification Models
4.4. Study Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Riva, M.; Lopci, E.; Gay, L.G.; Nibali, M.C.; Rossi, M.; Sciortino, T.; Castellano, A.; Bello, L. Advancing Imaging to Enhance Surgery: From Image to Information Guidance. Neurosurg. Clin. North Am. 2020, 32, 31–46. [Google Scholar] [CrossRef]
- Sanai, N.; Polley, M.-Y.; McDermott, M.W.; Parsa, A.T.; Berger, M.S. An extent of resection threshold for newly diagnosed glioblastomas. J. Neurosurg. 2011, 3–8. [Google Scholar] [CrossRef]
- Smith, J.S.; Chang, E.F.; Lamborn, K.R.; Chang, S.M.; Prados, M.D.; Cha, S.; Tihan, T.; Vandenberg, S.; McDermott, M.W.; Berger, M.S. Role of extent of resection in the long-term outcome of low-grade hemispheric gliomas. J. Clin. Oncol. 2008, 26, 1338–1345. [Google Scholar] [CrossRef]
- Talos, I.-F.; Zou, K.H.; Ohno-Machado, L.; Bhagwat, J.G.; Kikinis, R.; Black, P.M.; Jolesz, F.A. Supratentorial low-grade glioma resectability: Statistical predictive analysis based on anatomic MR features and tumor characteristics. Radiology 2006, 239, 506–513. [Google Scholar] [CrossRef]
- Livermore, L.J.; Isabelle, M.; Bell, I.M.; Edgar, O.; Voets, N.L.; Stacey, R.; Ansorge, O.; Vallance, C.; Plaha, P. Raman spectroscopy to differentiate between fresh tissue samples of glioma and normal brain: A comparison with 5-ALA–induced fluorescence-guided surgery. J. Neurosurg. 2020, 1–11. [Google Scholar] [CrossRef]
- Riva, M.; Hennersperger, C.; Milletari, F.; Katouzian, A.; Pessina, F.; Gutierrez-Becker, B.; Castellano, A.; Navab, N.; Bello, L. 3D intra-operative ultrasound and MR image guidance: Pursuing an ultrasound-based management of brainshift to enhance neuronavigation. Int. J. Comput. Assist. Radiol. Surg. 2017. [Google Scholar] [CrossRef]
- Cameron, J.M.; Conn, J.J.A.; Rinaldi, C.; Sala, A.; Brennan, P.M.; Jenkinson, M.D.; Caldwell, H.; Cinque, G.; Syed, K.; Butler, H.J.; et al. Interrogation of IDH1 Status in Gliomas by Fourier Transform Infrared Spectroscopy. Cancers 2020, 12, 3682. [Google Scholar] [CrossRef]
- Gajjar, K.; Heppenstall, L.D.; Pang, W.; Ashton, K.M.; Trevisan, J.; Patel, I.I.; Llabjani, V.; Stringfellow, H.F.; Martin-Hirsch, P.L.; Dawson, T.; et al. Diagnostic segregation of human brain tumours using Fourier-transform infrared and/or Raman spectroscopy coupled with discriminant analysis. Anal. Methods 2013, 5, 89–102. [Google Scholar] [CrossRef]
- Meyer, T.; Bergner, N.; Bielecki, C.; Krafft, C.; Akimov, D.; Romeike, B.F.M.; Reichart, R.; Kalff, R.; Dietzek, B.; Popp, J. Nonlinear microscopy, infrared, and Raman microspectroscopy for brain tumor analysis. J. Biomed. Opt. 2011, 16, 021113. [Google Scholar] [CrossRef] [PubMed]
- Ji, M.; Orringer, D.A.; Freudiger, C.W.; Ramkissoon, S.; Liu, X.; Lau, D.; Golby, A.J.; Norton, I.; Hayashi, M.; Agar, N.Y.R.; et al. Rapid, label-free detection of brain tumors with stimulated Raman scattering microscopy. Sci. Transl. Med. 2013, 5, 201ra119. [Google Scholar] [CrossRef]
- Jermyn, M.; Mok, K.; Mercier, J.; Desroches, J.; Pichette, J.; Saint-Arnaud, K.; Bernstein, L.; Guiot, M.-C.C.; Petrecca, K.; Leblond, F. Intraoperative brain cancer detection with Raman spectroscopy in humans. Sci. Transl. Med. 2015, 7, 274ra19. [Google Scholar] [CrossRef]
- Kircher, M.F.; de la Zerda, A.; Jokerst, J.V.; Zavaleta, C.L.; Kempen, P.J.; Mittra, E.; Pitter, K.; Huang, R.; Campos, C.; Habte, F.; et al. A brain tumor molecular imaging strategy using a new triple-modality MRI-photoacoustic-Raman nanoparticle. Nat. Med. 2012, 18, 829–834. [Google Scholar] [CrossRef] [PubMed]
- Kong, K.; Kendall, C.; Stone, N.; Notingher, I. Raman spectroscopy for medical diagnostics—From in-vitro biofluid assays to in-vivo cancer detection. Adv. Drug Deliv. Rev. 2015, 89, 121–134. [Google Scholar] [CrossRef] [PubMed]
- Surmacki, J.; Brozek-Pluska, B.; Kordek, R.; Abramczyk, H. The lipid-reactive oxygen species phenotype of breast cancer. Raman spectroscopy and mapping, PCA and PLSDA for invasive ductal carcinoma and invasive lobular carcinoma. Molecular tumorigenic mechanisms beyond Warburg effect. Analyst 2015, 140, 2121–2133. [Google Scholar] [CrossRef]
- Crow, P.; Ritchie, A.; Wright, M.; Kendall, C.; Stone, N. Evaluation of Raman spectroscopy to provide a real time, optical method for discrimination between normal and abnormal tissue in the prostate. Eur. Urol. Suppl. 2002, 1, 92. [Google Scholar] [CrossRef]
- Krishna, C.M.; Sockalingum, G.D.; Venteo, L.; Bhat, R.A.; Kushtagi, P.; Pluot, M.; Manfait, M. Evaluation of the suitability of ex vivo handled ovarian tissues for optical diagnosis by Raman microspectroscopy. Biopolymers 2005, 79, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Jermyn, M.; Desroches, J.; Mercier, J.; Tremblay, M.-A.; St-Arnaud, K.; Guiot, M.-C.; Petrecca, K.; Leblond, F. Neural networks improve brain cancer detection with Raman spectroscopy in the presence of operating room light artifacts. J. Biomed. Opt. 2016, 21, 094002. [Google Scholar] [CrossRef]
- Lemoine, É.; Dallaire, F.; Yadav, R.; Agarwal, R.; Kadoury, S.; Trudel, D.; Guiot, M.C.; Petrecca, K.; Leblond, F. Feature engineering applied to intraoperative: In vivo Raman spectroscopy sheds light on molecular processes in brain cancer: A retrospective study of 65 patients. Analyst 2019, 144, 6517–6532. [Google Scholar] [CrossRef]
- Kalkanis, S.N.; Kast, R.E.; Rosenblum, M.L.; Mikkelsen, T.; Yurgelevic, S.M.; Nelson, K.M.; Raghunathan, A.; Poisson, L.M.; Auner, G.W. Raman spectroscopy to distinguish grey matter, necrosis, and glioblastoma multiforme in frozen tissue sections. J. Neurooncol. 2014, 116, 477–485. [Google Scholar] [CrossRef]
- Rossi, M.; Gay, L.; Ambrogi, F.; Nibali, M.C.; Sciortino, T.; Puglisi, G.; Leonetti, A.; Mocellini, C.; Caroli, M.; Cordera, S.; et al. Association of Supratotal Resection with Progression-Free Survival, Malignant Transformation, and Overall Survival in Lower-Grade Gliomas. Neuro. Oncol. 2020. [Google Scholar] [CrossRef]
- Riva, M.; Hiepe, P.; Frommert, M.; Divenuto, I.; Gay, L.G.; Sciortino, T.; Nibali, M.C.; Rossi, M.; Pessina, F.; Bello, L. Intraoperative computed tomography and finite element modelling for multimodal image fusion in brain surgery. Oper. Neurosurg. 2020, 18, 531–541. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef]
- Eiseman, E.; Bloom, G.; Brower, J.; Clancy, N.; Olmsted, S.S. Case Studies of Existing Human Tissue Repositories “Best Practices” for a Biospecimen Resource for the Genomic and Proteomic Era; Rand Corporation: Santa Monica, CA, USA, 2013; ISBN 2325-7776 2325-7792. [Google Scholar]
- Barman, I.; Kong, C.-R.; Singh, G.P.; Dasari, R.R. Effect of photobleaching on calibration model development in biological Raman spectroscopy. J. Biomed. Opt. 2011, 16, 011004. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Henson, M.J. A practical algorithm to remove cosmic spikes in Raman imaging data for pharmaceutical applications. Appl. Spectrosc. 2007, 61, 1015–1020. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Lui, H.; McLean, D.I.; Zeng, H. Automated autofluorescence background subtraction algorithm for biomedical Raman spectroscopy. Appl. Spectrosc. 2007, 61, 1225–1232. [Google Scholar] [CrossRef]
- Banerjee, H.N.; Zhang, L. Deciphering the finger Prints of Brain Cancer Astrocytoma in comparison to Astrocytes by using near infrared Raman Spectroscopy. Mol. Cell. Biochem. 2007, 295, 237–240. [Google Scholar] [CrossRef]
- Krafft, C.; Sobottka, S.B.; Schackert, G.; Salzer, R. Raman and infrared spectroscopic mapping of human primary intracranial tumors: A comparative study. J. Raman Spectrosc. 2006, 37, 367–375. [Google Scholar] [CrossRef]
- Movasaghi, Z.; Rehman, S.; Rehman, I.U. Raman Spectroscopy of Biological Tissues. Appl. Spectrosc. Rev. 2007, 42, 493–541. [Google Scholar] [CrossRef]
- Brusatori, M.; Auner, G.; Noh, T.; Scarpace, L.; Broadbent, B.; Kalkanis, S.N. Intraoperative Raman Spectroscopy. Neurosurg. Clin. N. Am. 2017, 28, 633–652. [Google Scholar] [CrossRef] [PubMed]
- Bergner, N.; Krafft, C.; Geiger, K.D.; Kirsch, M.; Schackert, G.; Popp, J. Unsupervised unmixing of Raman microspectroscopic images for morphochemical analysis of non-dried brain tumor specimens. Proc. Anal. Bioanal. Chem. 2012, 403, 719–725. [Google Scholar] [CrossRef]
- Köhler, M.; MacHill, S.; Salzer, R.; Krafft, C. Characterization of lipid extracts from brain tissue and tumors using Raman spectroscopy and mass spectrometry. Anal. Bioanal. Chem. 2009, 393. [Google Scholar] [CrossRef]
- Krafft, C.; Shapoval, L.; Sobottka, S.B.; Geiger, K.D.; Schackert, G.; Salzer, R. Identification of primary tumors of brain metastases by SIMCA classification of IR spectroscopic images. Biochim. Biophys. Acta Biomembr. 2006, 1758, 883–891. [Google Scholar] [CrossRef]
- Koljenović, S.; Schut, T.B.; Vincent, A.; Kros, J.M.; Puppels, G.J. Detection of meningioma in dura mater by Raman spectroscopy. Anal. Chem. 2005, 77, 7958–7965. [Google Scholar] [CrossRef]
- Kast, R.E.; Auner, G.W.; Rosenblum, M.L.; Mikkelsen, T.; Yurgelevic, S.M.; Raghunathan, A.; Poisson, L.M.; Kalkanis, S.N. Raman molecular imaging of brain frozen tissue sections. J. Neurooncol. 2014, 120, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Beljebbar, A.; Dukic, S.; Amharref, N.; Manfait, M. Ex vivo and in vivo diagnosis of C6 glioblastoma development by Raman spectroscopy coupled to a microprobe. Anal. Bioanal. Chem. 2010, 398, 477–487. [Google Scholar] [CrossRef]
- Aydin, O.; Altaş, M.; Kahraman, M.; Bayrak, O.F.; Culha, M. Differentiation of healthy brain tissue and tumors using surface-enhanced Raman scattering. Appl. Spectrosc. 2009, 63, 1095–1100. [Google Scholar] [CrossRef] [PubMed]
- Tanahashi, K.; Natsume, A.; Ohka, F.; Momota, H.; Kato, A.; Motomura, K.; Watabe, N.; Muraishi, S.; Nakahara, H.; Saito, Y.; et al. Assessment of tumor cells in a mouse model of diffuse infiltrative glioma by Raman spectroscopy. Biomed Res. Int. 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Liu, C.-H.; Sun, Y.; Pu, Y.; Boydston-White, S.; Liu, Y.; Alfano, R.R. Human brain cancer studied by resonance Raman spectroscopy. J. Biomed. Opt. 2012, 17, 116021. [Google Scholar] [CrossRef] [PubMed]
- Krafft, C.; Sobottka, S.B.; Schackert, G.; Salzer, R. Near infrared Raman spectroscopic mapping of native brain tissue and intracranial tumors. Analyst 2005, 130, 1070–1077. [Google Scholar] [CrossRef] [PubMed]
- Bury, D.; Morais, C.L.M.; Martin, F.L.; Lima, K.M.G.; Ashton, K.M.; Baker, M.J.; Dawson, T.P. Discrimination of fresh frozen non-tumour and tumour brain tissue using spectrochemical analyses and a classification model. Br. J. Neurosurg. 2020, 34, 40–45. [Google Scholar] [CrossRef]
- Auner, A.W.; Kast, R.E.; Rabah, R.; Poulik, J.M.; Klein, M.D. Conclusions and data analysis: A 6-year study of Raman spectroscopy of solid tumors at a major pediatric institute. Pediatr. Surg. Int. 2013, 29, 129–140. [Google Scholar] [CrossRef]
- Beleites, C.; Geiger, K.; Kirsch, M.; Sobottka, S.B.; Schackert, G.; Salzer, R. Raman spectroscopic grading of astrocytoma tissues: Using soft reference information. Anal. Bioanal. Chem. 2011, 400, 2801–2816. [Google Scholar] [CrossRef]
- Aguiar, R.P.; Silveira, L.; Falcão, E.T.; Pacheco, M.T.T.; Zângaro, R.A.; Pasqualucci, C.A. Discriminating neoplastic and normal brain tissues in vitro through raman spectroscopy: A principal components analysis classification model. Photomed. Laser Surg. 2013, 31, 595–604. [Google Scholar] [CrossRef]
- Galli, R.; Meinhardt, M.; Koch, E.; Schackert, G.; Steiner, G.; Kirsch, M.; Uckermann, O. Rapid Label-Free Analysis of Brain Tumor Biopsies by Near Infrared Raman and Fluorescence Spectroscopy—A Study of 209 Patients. Front. Oncol. 2019, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Anna, I.; Bartosz, P.; Lech, P.; Halina, A. Novel strategies of Raman imaging for brain tumor research. Oncotarget 2017, 8, 85290–85310. [Google Scholar] [CrossRef]
- Kast, R.; Auner, G.; Yurgelevic, S.; Broadbent, B.; Raghunathan, A.; Poisson, L.M.; Mikkelsen, T.; Rosenblum, M.L.; Kalkanis, S.N. Identification of regions of normal grey matter and white matter from pathologic glioblastoma and necrosis in frozen sections using Raman imaging. J. Neurooncol. 2015, 125, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.M.; Bonnier, F.; Tfayli, A.; Lambkin, H.; Flynn, K.; McDonagh, V.; Healy, C.; Clive Lee, T.; Lyng, F.M.; Byrne, H.J. Raman spectroscopic analysis of human skin tissue sections ex-vivo: Evaluation of the effects of tissue processing and dewaxing. Med. Opt. 2013, 18, 061202. [Google Scholar] [CrossRef]
- Hollon, T.; Lewis, S.; Freudiger, C.W.; Sunney Xie, X.; Orringer, D.A. Improving the accuracy of brain tumor surgery via Raman-based technology. Neurosurg. Focus 2016. [Google Scholar] [CrossRef] [PubMed]
- Livermore, L.J.; Isabelle, M.; Bell, I.M.; Scott, C.; Walsby-Tickle, J.; Gannon, J.; Plaha, P.; Vallance, C.; Ansorge, O. Rapid intraoperative molecular genetic classification of gliomas using Raman spectroscopy. Neuro Oncol. Adv. 2019, 1. [Google Scholar] [CrossRef] [PubMed]
- Hendricks, B.K.; Sanai, N.; Stummer, W. Fluorescence-guided surgery with aminolevulinic acid for low-grade gliomas. J. Neurooncol. 2019, 141, 13–18. [Google Scholar] [CrossRef] [PubMed]
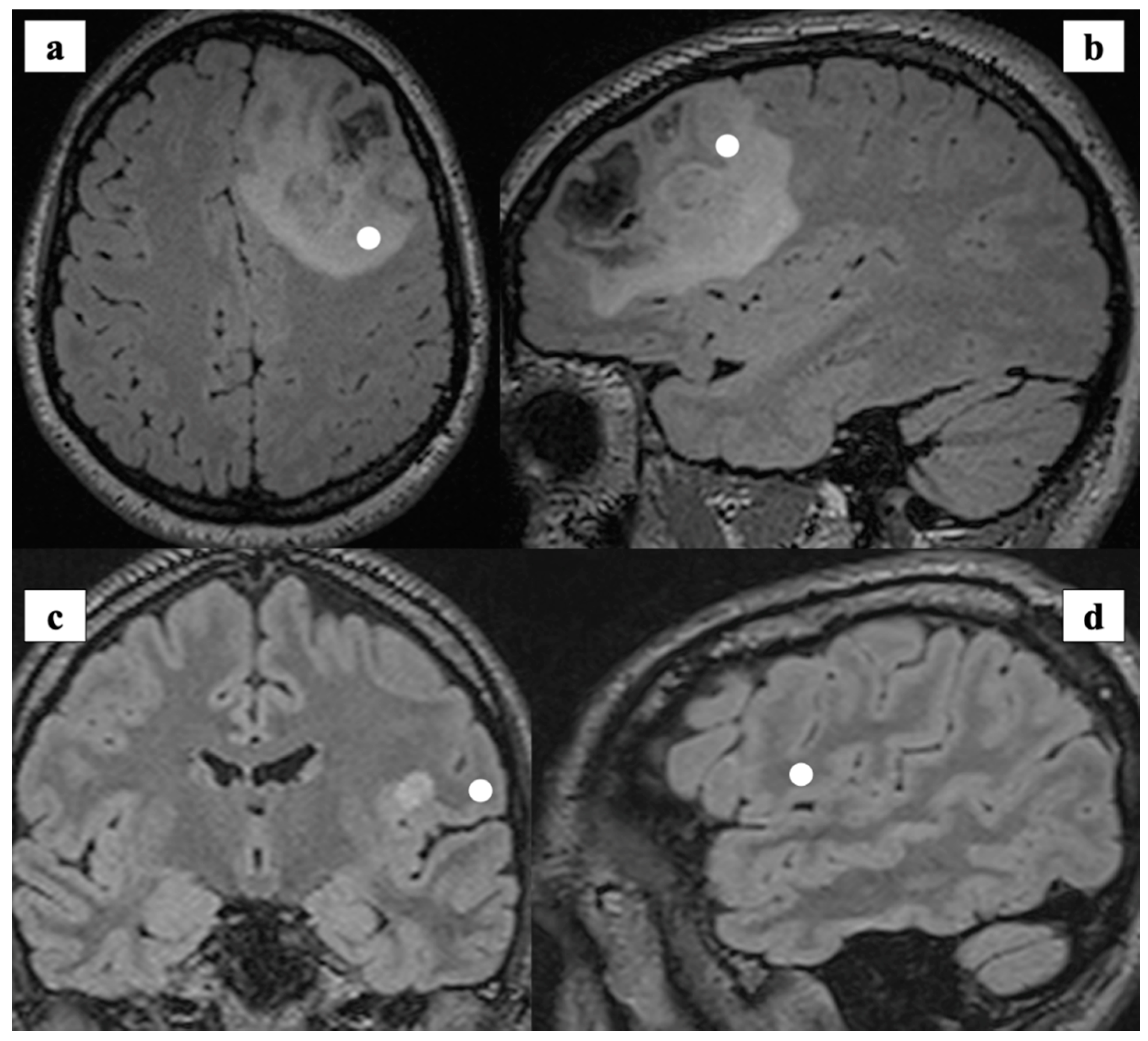
| n° of Samples | n° of Spectra | |
| Glioma tissue | 38 | 2073 |
| Grade II | ||
| Astrocytoma, WHO grade II (IDH-mutant) | 0 (0) | - |
| Oligodendroglioma, WHO grade II (IDH-mutant) | 4 (10.5) | 220 |
| Grade III | ||
| Astrocytoma, WHO grade III (IDH-mutant) | 5 (13.2) | 259 |
| Oligodendroglioma, WHO grade III (IDH-mutant) | 12 (31.6) | 642 |
| Grade IV | ||
| Glioblastoma, WHO grade IV (IDH wild-type) | 14 (36.8) | 773 |
| Glioblastoma, WHO grade IV (IDH mutant) | 3 (7.9) | 179 |
| Normal Tissue | 25 | 1377 |
| Total | 63 | 3450 |
| Raman Shift (s−1) | Proposed Assignments |
|---|---|
| 431 * | Cholesterol/cholesterol ester |
| 450 | Ring torsion of phenyl |
| 457 | Proteins and cholesterol |
| 460 | Undefined |
| 474 | Glycogen and polysaccharides |
| 478 | Polysaccharides |
| 495 | Undefined |
| 498 * | Nucleic acids/nucleotides |
| 502 | Undefined |
| 517 | Undefined |
| 524 * | S-S disulfide stretching in proteins |
| 540 | n(S-S) trans-gauche-trans(amino acid cysteine) |
| 743 * | Heme (blood) |
| 754 | Symmetric breathing of tryptophan |
| 776 * | Phosphatidylinositol |
| 780 * | Uracil based ring breathing mode |
| 808 | Undefined |
| 817 * | C-C stretching/collagen assignment |
| 825 * | Phosphodiester |
| 826 | Tyr, proline |
| 837 | Undefined |
| 850 | Glycogen (high-grade tumors) |
| 853 | Glycogen (high-grade tumors) |
| 875 * | Choline and phospholipids |
| 880 | Tryptophan, d(ring) |
| 881 | Hydroxyproline and tryptophan (collagen); sterol ring stretch of cholesterol; asymmetric stretching of choline |
| 883 | ρ (CH2) (protein assignment) |
| 893/4 * | Phosphodiester (nucleic acids) |
| 903 | Undefined |
| 916 | Undefined |
| 927 | Undefined |
| 928 | Amino acids proline & valine (protein band) |
| 933 * | Proline, hydroxyproline |
| 934 | C-C backbone (collagen assignment) |
| 941 * | Glycogen |
| 950 | single bond stretching vibrations for the amino acids proline and valine and polysaccharides |
| 954 | Undefined |
| 958 | Stretching vibrations of PO4 in hydroxyapatite |
| 963 * | Protein assignments |
| 968 * | Lipids |
| 975 * and 977 | Tricalcium phosphate Ca3(PO4) calcification seen in schwannoma and necrosis |
| 1003 | Ring breathing mode of phenylalanine of protein |
| 1030 | Phenylalanine (protein assignment) and collagen |
| 1031 * | Phenylalanine (protein assignment) |
| 1035 * | Collagen |
| 1036 | Undefined |
| 1122 | Glycogen |
| 1337 | Aliphatic amino acids, including tryptophan, nucleic acids. Glycogen in necrosis |
| 1342 | Aliphatic amino acids, including tryptophan, nucleic acids. Glycogen |
| 1578 | C-C stretch of protein, nucleic acids |
| 1581 | C-C stretch of protein, nucleic acids |
| 1583 * | C-C stretch of protein, phenylalanine, nucleic acids |
| 1603 * | Phenylalanine & tyrosine/Oxygenated haemoglobin |
| 1603 | Cytosine, tyrosine, phenylalanine |
| 1614 | Aromatic amino acids (protein); Tyrosine e proline |
| 1616 | C-C stretching mode of tyrosine & tryptophan |
| 1657 | Amide I (a-helix) (protein) |
| 1659 | Amide I (a-helix) |
| 1660 | Amide I band |
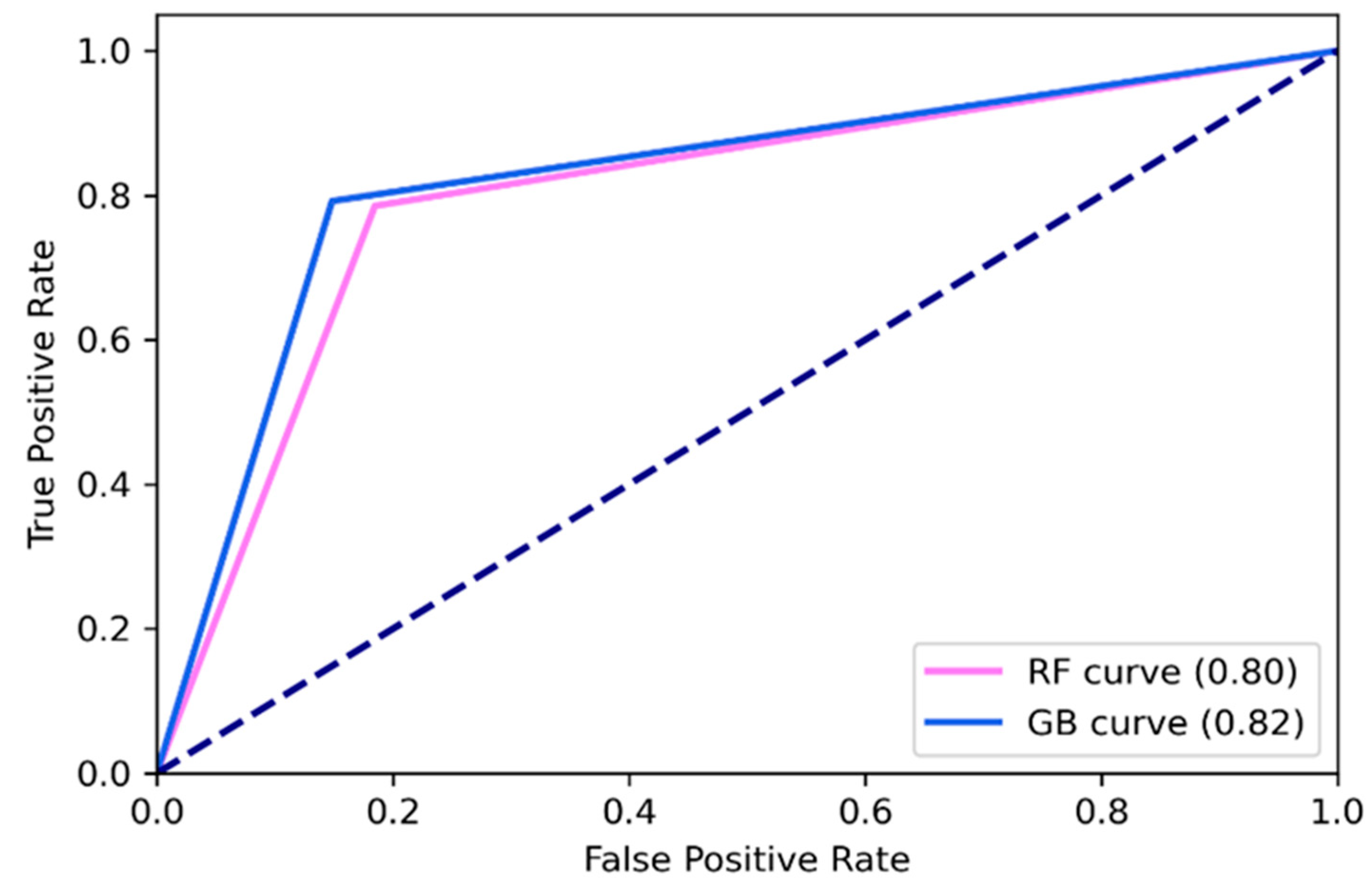
| Performance Metrics | Random Forest | Gradient Boosting |
|---|---|---|
| Accuracy | 0.80 | 0.83 |
| Precision | 0.79 | 0.82 |
| Recall | 0.80 | 0.82 |
| F1-score | 0.80 | 0.82 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Riva, M.; Sciortino, T.; Secoli, R.; D’Amico, E.; Moccia, S.; Fernandes, B.; Conti Nibali, M.; Gay, L.; Rossi, M.; De Momi, E.; et al. Glioma biopsies Classification Using Raman Spectroscopy and Machine Learning Models on Fresh Tissue Samples. Cancers 2021, 13, 1073. https://doi.org/10.3390/cancers13051073
Riva M, Sciortino T, Secoli R, D’Amico E, Moccia S, Fernandes B, Conti Nibali M, Gay L, Rossi M, De Momi E, et al. Glioma biopsies Classification Using Raman Spectroscopy and Machine Learning Models on Fresh Tissue Samples. Cancers. 2021; 13(5):1073. https://doi.org/10.3390/cancers13051073
Chicago/Turabian StyleRiva, Marco, Tommaso Sciortino, Riccardo Secoli, Ester D’Amico, Sara Moccia, Bethania Fernandes, Marco Conti Nibali, Lorenzo Gay, Marco Rossi, Elena De Momi, and et al. 2021. "Glioma biopsies Classification Using Raman Spectroscopy and Machine Learning Models on Fresh Tissue Samples" Cancers 13, no. 5: 1073. https://doi.org/10.3390/cancers13051073
APA StyleRiva, M., Sciortino, T., Secoli, R., D’Amico, E., Moccia, S., Fernandes, B., Conti Nibali, M., Gay, L., Rossi, M., De Momi, E., & Bello, L. (2021). Glioma biopsies Classification Using Raman Spectroscopy and Machine Learning Models on Fresh Tissue Samples. Cancers, 13(5), 1073. https://doi.org/10.3390/cancers13051073








