The Two-Faced Role of SIRT6 in Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Regulation of SIRT6 Expression and Activity
3. Role of SIRT6 in Cancer
3.1. Tumor Suppression Function
3.2. Tumor Promoter Function
4. Pharmacological Modulation of SIRT6
4.1. SIRT6 Activators
4.2. SIRT6 Inhibitors
5. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ADP | Adenosine diphosphate |
| AML | Acute myeloid leukemia; |
| AMP | Adenosine monophosphate |
| AP-1 | Activator protein 1 |
| ATP | Adenosine triphosphate |
| AMPK | AMP-activated protein kinase |
| Bax | Bcl-2 associated X protein |
| BER | Base excision repair |
| cAMP | Cyclic adenosine monophosphate |
| CHIP | Carboxyl terminus of Hsp70-Interacting protein |
| CREB | cAMP response element-binding protein |
| DSB | Double strand break |
| DLBCL | Diffuse large B-cell lymphoma |
| EC50 | Half maximal effective concentration |
| EMT | Epithelial-mesenchymal transition |
| FFA | Free fatty acid |
| HCC | Hepatocellular carcinoma |
| HDAC | Histone deacetylase |
| HIF- α | Hypoxia-inducible factor 1α |
| HP1 α | Heterochromatin protein 1α; |
| HR | Homologous recombination |
| IC50 | Half maximal inhibitory concentration |
| IGF | Insulin-like growth factor |
| IGFBP2 | Insulin-like growth factor-binding protein 2 |
| IL-6 | Interleukin-6 |
| IL-8 | Interleukin-8 |
| IPSCs | Induced pluripotent stem cells |
| L1 | Long interspersed element 1 |
| MAPK | Mitogen-activated protein kinase |
| MDM2 | Mouse double minute 2 homolog |
| miRNA | micro-RNA |
| MM | Multiple myeloma |
| NAD+ | Nicotinamide adenine dinucleotide |
| NHEJ | Non-homologous end-joining |
| NPC | Nasopharyngeal carcinoma |
| NSCLC | Non-small cell lung cancer |
| PARP1 | Poly [ADP-ribose] polymerase 1 |
| PCPB2 | Poly(C)-binding protein 2 |
| PCD | Programmed cell death |
| PDAC | Pancreatic ductal adenocarcinoma |
| PKA | Protein kinase A |
| PKM2 | Pyruvate kinase M2 |
| ROS | Reactive oxygen species |
| SIRT | Sirtuin |
| SCC | Skin squamous cell carcinoma |
| SUMO | Small ubiquitin-related modifier |
| TNF- α | Tumor necrosis factor α |
| TRF2 | Telomere repeat binding factor 2 |
| UTR | Untranslated region |
| XIAP | X-linked inhibitor of apoptosis protein |
References
- Finkel, T.; Deng, C.X.; Mostoslavsky, R. Recent progress in the biology and physiology of sirtuins. Nature 2009, 460, 587–591. [Google Scholar] [CrossRef] [Green Version]
- Michishita, E.; McCord, R.A.; Berber, E.; Kioi, M.; Padilla-Nash, H.; Damian, M.; Cheung, P.; Kusumoto, R.; Kawahara, T.L.A.; Barrett, J.C.; et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature 2008, 452, 492–496. [Google Scholar] [CrossRef]
- Michishita, E.; McCord, R.A.; Boxer, L.D.; Barber, M.F.; Hong, T.; Gozani, O.; Chua, K.F. Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle 2009, 8, 2664–2666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, B.; Zwaans, B.M.M.; Eckersdorff, M.; Lombard, D.B. The sirtuin SIRT6 deacetylates H3 K56Ac in vivo to promote genomic stability. Cell Cycle 2009, 8, 2662–2663. [Google Scholar] [CrossRef] [Green Version]
- Tasselli, L.; Xi, Y.; Zheng, W.; Tennen, R.I.; Odrowaz, Z.; Simeoni, F.; Li, W.; Chua, K.F. SIRT6 deacetylates H3K18ac at pericentric chromatin to prevent mitotic errors and cellular senescence. Nat. Struct. Mol. Biol. 2016, 23, 434–440. [Google Scholar] [CrossRef] [Green Version]
- Chang, A.R.; Ferrer, C.M.; Mostoslavsky, R. SIRT6, a mammalian deacylase with multitasking abilities. Physiol. Rev. 2020, 100, 145–169. [Google Scholar] [CrossRef] [PubMed]
- Klein, M.A.; Denu, J.M. Biological and catalytic functions of sirtuin 6 as targets for small-molecule modulators. J. Biol. Chem. 2020, 295, 11021–11041. [Google Scholar] [CrossRef]
- Michishita, E.; Park, J.Y.; Burneskis, J.M.; Barrett, J.C.; Horikawa, I. Evolutionarily Conserved and Nonconserved Cellular Localizations and Functions of Human SIRT Proteins. Mol. Biol. Cell 2005, 16, 4623–4635. [Google Scholar] [CrossRef] [Green Version]
- Beauharnois, J.M.; Bolivar, B.E.; Welch, J.T. Sirtuin 6: A review of biological effects and potential therapeutic properties. Mol. Biosyst. 2013, 9, 1789–1806. [Google Scholar] [CrossRef]
- Pan, P.W.; Feldman, J.L.; Devries, M.K.; Dong, A.; Edwards, A.M.; Denu, J.M. Structure and biochemical functions of SIRT6. J. Biol. Chem. 2011, 286, 14575–14587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feldman, J.L.; Baeza, J.; Denu, J.M. Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by Mammalian Sirtuins. J. Biol. Chem. 2013, 288, 31350–31356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, H.; Khan, S.; Wang, Y.; Charron, G.; He, B.; Sebastian, C.; Du, J.; Kim, R.; Ge, E.; Mostoslavsky, R.; et al. SIRT6 regulates TNF-α secretion through hydrolysis of long-chain fatty acyl lysine. Nature 2013, 496, 110–113. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Spiegelman, N.A.; Nelson, O.D.; Jing, H.; Lin, H. SIRT6 regulates Ras-related protein R-Ras2 by lysine defatty-acylation. eLife 2017, 6. [Google Scholar] [CrossRef]
- Toiber, D.; Erdel, F.; Bouazoune, K.; Silberman, D.; Zhong, L.; Mulligan, P.; Sebastian, C.; Cosentino, C.; Martinez-Pastor, B.; Giacosa, S.; et al. SIRT6 recruits SNF2H to DNA break sites, preventing genomic instability through chromatin remodeling. Mol. Cell 2013, 51, 454–468. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.; Liu, N.; Zhang, H.; Zhang, H.; Qiao, J.; Jia, W.; Zhu, S.; Mao, Z.; Kang, J. Sirt6 Promotes DNA End Joining in iPSCs Derived from Old Mice. Cell Rep. 2017, 18, 2880–2892. [Google Scholar] [CrossRef] [Green Version]
- Mao, Z.; Hine, C.; Tian, X.; Van Meter, M.; Au, M.; Vaidya, A.; Seluanov, A.; Gorbunova, V. SIRT6 promotes DNA repair under stress by activating PARP1. Science 2011, 332, 1443–1446. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.; Zhang, L.; Zhang, W.; Meng, D.; Zhang, H.; Jiang, Y.; Xu, X.; Van Meter, M.; Seluanov, A.; Gorbunova, V.; et al. SIRT6 rescues the age related decline in base excision repair in a PARP1-dependent manner. Cell Cycle 2015, 14, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Hwang, B.J.; Jin, J.; Gao, Y.; Shi, G.; Madabushi, A.; Yan, A.; Guan, X.; Zalzman, M.; Nakajima, S.; Lan, L.; et al. SIRT6 protein deacetylase interacts with MYH DNA glycosylase, APE1 endonuclease, and Rad9-Rad1-Hus1 checkpoint clamp. BMC Mol. Biol. 2015, 16. [Google Scholar] [CrossRef] [Green Version]
- Tennen, R.I.; Bua, D.J.; Wright, W.E.; Chua, K.F. SIRT6 is required for maintenance of telomere position effect in human cells. Nat. Commun. 2011, 2. [Google Scholar] [CrossRef] [Green Version]
- Multani, A.S.; Chang, S. WRN at telomeres: Implications for aging and cancer. J. Cell Sci. 2007, 120, 713–721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rizzo, A.; Iachettini, S.; Salvati, E.; Zizza, P.; Maresca, C.; D’Angelo, C.; Benarroch-Popivker, D.; Capolupo, A.; Del Gaudio, F.; Cosconati, S.; et al. SIRT6 interacts with TRF2 and promotes its degradation in response to DNA damage. Nucleic Acids Res. 2017, 45, 1820–1834. [Google Scholar] [CrossRef]
- Van Meter, M.; Kashyap, M.; Rezazadeh, S.; Geneva, A.J.; Morello, T.D.; Seluanov, A.; Gorbunova, V. SIRT6 represses LINE1 retrotransposons by ribosylating KAP1 but this repression fails with stress and age. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef]
- Imai, S.-I.; Guarente, L. It takes two to tango: NAD(+) and sirtuins in aging/longevity control. Npj Aging Mech. Dis. 2016, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mostoslavsky, R.; Chua, K.F.; Lombard, D.B.; Pang, W.W.; Fischer, M.R.; Gellon, L.; Liu, P.; Mostoslavsky, G.; Franco, S.; Murphy, M.M.; et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell 2006, 124, 315–329. [Google Scholar] [CrossRef] [Green Version]
- Kanfi, Y.; Naiman, S.; Amir, G.; Peshti, V.; Zinman, G.; Nahum, L.; Bar-Joseph, Z.; Cohen, H.Y. The sirtuin SIRT6 regulates lifespan in male mice. Nature 2012, 483, 218–221. [Google Scholar] [CrossRef]
- Sebastián, C.; Zwaans, B.M.M.; Silberman, D.M.; Gymrek, M.; Goren, A.; Zhong, L.; Ram, O.; Truelove, J.; Guimaraes, A.R.; Toiber, D.; et al. The histone deacetylase SIRT6 Is a tumor suppressor that controls cancer metabolism. Cell 2012, 151, 1185–1199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bauer, I.; Grozio, A.; Lasiglie, D.; Basile, G.; Sturla, L.; Magnone, M.; Sociali, G.; Soncini, D.; Caffa, I.; Poggi, A.; et al. The NAD+-dependent histone deacetylase SIRT6 promotes cytokine production and migration in pancreatic cancer cells by regulating Ca 2+ responses. J. Biol. Chem. 2012, 287, 40924–40937. [Google Scholar] [CrossRef] [Green Version]
- Kawahara, T.L.A.; Michishita, E.; Adler, A.S.; Damian, M.; Berber, E.; Lin, M.; McCord, R.A.; Ongaigui, K.C.L.; Boxer, L.D.; Chang, H.Y.; et al. SIRT6 Links Histone H3 Lysine 9 Deacetylation to NF-κB-Dependent Gene Expression and Organismal Life Span. Cell 2009, 136, 62–74. [Google Scholar] [CrossRef] [Green Version]
- Zhang, N.; Li, Z.; Mu, W.; Li, L.; Liang, Y.; Lu, M.; Wang, Z.; Qiu, Y.; Wang, Z. Calorie restriction-induced SIRT6 activation delays aging by suppressing NF-κB signaling. Cell Cycle 2016, 15, 1009–1018. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.; Ka, S.O.; Cha, H.N.; Chae, Y.N.; Kim, M.K.; Park, S.Y.; Bae, E.J.; Park, B.H. Myeloid sirtuin 6 deficiency causes insulin resistance in high-fat diet–fed mice by eliciting macrophage polarization toward an M1 phenotype. Diabetes 2017, 66, 2659–2668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ouyang, L.; Shi, Z.; Zhao, S.; Wang, F.T.; Zhou, T.T.; Liu, B.; Bao, J.K. Programmed cell death pathways in cancer: A review of apoptosis, autophagy and programmed necrosis. Cell Prolif. 2012, 45, 487–498. [Google Scholar] [CrossRef]
- Van Meter, M.; Mao, Z.; Gorbunova, V.; Seluanov, A. SIRT6 overexpression induces massive apoptosis in cancer cells but not in normal cells. Cell Cycle 2011, 10, 3153–3158. [Google Scholar] [CrossRef] [Green Version]
- Kugel, S.; Feldman, J.L.; Klein, M.A.; Silberman, D.M.; Sebastián, C.; Mermel, C.; Dobersch, S.; Clark, A.R.; Getz, G.; Denu, J.M.; et al. Identification of and Molecular Basis for SIRT6 Loss-of-Function Point Mutations in Cancer. Cell Rep. 2015, 13, 479–488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qu, N.; Hu, J.Q.; Liu, L.; Zhang, T.T.; Sun, G.H.; Shi, R.L.; Ji, Q.H. SIRT6 is upregulated and associated with cancer aggressiveness in papillary thyroid cancer via BRAF/ERK/Mcl1 pathway. Int. J. Oncol. 2017, 50, 1683–1692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Ceu Teixeira, M.; Sanchez-Lopez, E.; Espina, M.; Garcia, M.L.; Durazzo, A.; Lucarini, M.; Novellino, E.; Souto, S.B.; Santini, A.; Souto, E.B. Sirtuins and SIRT6 in Carcinogenesis and in Diet. Int. J. Mol. Sci. 2019, 20. [Google Scholar] [CrossRef] [Green Version]
- McGlynn, L.M.; Zino, S.; MacDonald, A.I.; Curle, J.; Reilly, J.E.; Mohammed, Z.M.; McMillan, D.C.; Mallon, E.; Payne, A.P.; Edwards, J.; et al. SIRT2: Tumour suppressor or tumour promoter in operable breast cancer? Eur. J. Cancer 2014, 50, 290–301. [Google Scholar] [CrossRef]
- Li, Z.; Xie, Q.R.; Chen, Z.; Lu, S.; Xia, W. Regulation of SIRT2 levels for human non-small cell lung cancer therapy. Lung Cancer 2013, 82, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Karwaciak, I.; Salkowska, A.; Karas, K.; Sobalska-Kwapis, M.; Walczak-Drzewiecka, A.; Pulaski, L.; Strapagiel, D.; Dastych, J.; Ratajewski, M. SIRT2 Contributes to the Resistance of Melanoma Cells to the Multikinase Inhibitor Dasatinib. Cancers 2019, 11. [Google Scholar] [CrossRef] [Green Version]
- Min, L.; Ji, Y.; Bakiri, L.; Qiu, Z.; Cen, J.; Chen, X.; Chen, L.; Scheuch, H.; Zheng, H.; Qin, L.; et al. Liver cancer initiation is controlled by AP-1 through SIRT6-dependent inhibition of survivin. Nat. Cell Biol. 2012, 14, 1203–1211. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Seto, E.; Zhang, J. E2F1 enhances glycolysis through suppressing Sirt6 transcription in cancer cells. Oncotarget 2015, 6, 11252–11263. [Google Scholar] [CrossRef] [Green Version]
- Wencel, P.L.; Lukiw, W.J.; Strosznajder, J.B.; Strosznajder, R.P. Inhibition of Poly(ADP-ribose) Polymerase-1 Enhances Gene Expression of Selected Sirtuins and APP Cleaving Enzymes in Amyloid Beta Cytotoxicity. Mol. Neurobiol. 2018, 55, 4612–4623. [Google Scholar] [CrossRef] [Green Version]
- Dávalos, A.; Goedeke, L.; Smibert, P.; Ramirez, C.M.; Warrier, N.P.; Andreo, U.; Cirera-Salinas, D.; Rayner, K.; Suresh, U.; Pastor-Pareja, J.C.; et al. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc. Natl. Acad. Sci. USA 2011, 108, 9232–9237. [Google Scholar] [CrossRef] [Green Version]
- Elhanati, S.; Kanfi, Y.; Varvak, A.; Roichman, A.; Carmel-Gross, I.; Barth, S.; Gibor, G.; Cohen, H. Multiple regulatory layers of SREBP1/2 by SIRT6. Cell Rep. 2013, 4, 905–912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, M.; Qiao, L.; Li, B.; Wang, J.; Zhang, G.; Shi, W.; Liu, Z.; Gu, N.; Di, Z.; Wang, X.; et al. Suppression of SIRT6 by miR-33a facilitates tumor growth of glioma through apoptosis and oxidative stress resistance. Oncol. Rep. 2017, 38, 1251–1258. [Google Scholar] [CrossRef] [Green Version]
- Lefort, K.; Brooks, Y.; Ostano, P.; Cario-André, M.; Calpini, V.; Guinea-Viniegra, J.; Albinger-Hegyi, A.; Hoetzenecker, W.; Kolfschoten, I.; Wagner, E.F.; et al. A miR-34a-SIRT6 axis in the squamous cell differentiation network. EMBO J. 2013, 32, 2248–2263. [Google Scholar] [CrossRef] [Green Version]
- Baker, J.R.; Vuppusetty, C.; Colley, T.; Papaioannou, A.I.; Fenwick, P.; Donnelly, L.; Ito, K.; Barnes, P.J. Oxidative stress dependent microRNA-34a activation via PI3Kα reduces the expression of sirtuin-1 and sirtuin-6 in epithelial cells. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [Green Version]
- Elhanati, S.; Ben-Hamo, R.; Kanfi, Y.; Varvak, A.; Glazz, R.; Lerrer, B.; Efroni, S.; Cohen, H.Y. Reciprocal Regulation between SIRT6 and miR-122 Controls Liver Metabolism and Predicts Hepatocarcinoma Prognosis. Cell Rep. 2016, 14, 234–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, S.; Yang, Y.; Liu, M.; Liu, B.; Yang, X.; Yu, M.; Qi, H.; Ren, M.; Wang, Z.; Zou, J.; et al. MiR-125b attenuates human hepatocellular carcinoma malignancy through targeting SIRT6. Am. J. Cancer Res. 2018, 8, 993–1007. [Google Scholar] [PubMed]
- Sharma, A.; Diecke, S.; Zhang, W.Y.; Lan, F.; He, C.; Mordwinkin, N.M.; Chua, K.F.; Wu, J.C. The role of SIRT6 protein in aging and reprogramming of human induced pluripotent stem cells. J. Biol. Chem. 2013, 288, 18439–18447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thirumurthi, U.; Shen, J.; Xia, W.; LaBaff, A.M.; Wei, Y.; Li, C.W.; Chang, W.C.; Chen, C.H.; Lin, H.K.; Yu, D.; et al. MDM2-mediated degradation of SIRT6 phosphorylated by AKT1 promotes tumorigenesis and trastuzumab resistance in breast cancer. Sci. Signal. 2014, 7. [Google Scholar] [CrossRef] [Green Version]
- Kim, E.J.; Juhnn, Y.S. Cyclic AMP signaling reduces sirtuin 6 expression in non-small cell lung cancer cells by promoting ubiquitin-proteasomal degradation via inhibition of the Raf-MEK-ERK (raf/mitogen-activated extracellular signal-regulated kinase/ extracellular signal-regulated kinase) pathway. J. Biol. Chem. 2015, 290, 9604–9613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ronnebaum, S.M.; Wu, Y.; McDonough, H.; Patterson, C. The ubiquitin ligase CHIP prevents sirT6 degradation through noncanonical ubiquitination. Mol. Cell. Biol. 2013, 33, 4461–4472. [Google Scholar] [CrossRef] [Green Version]
- Lin, Z.; Yang, H.; Tan, C.; Li, J.; Liu, Z.; Quan, Q.; Kong, S.; Ye, J.; Gao, B.; Fang, D. USP10 Antagonizes c-Myc transcriptional activation through SIRT6 stabilization to suppress tumor formation. Cell Rep. 2013, 5, 1639–1649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, J.; Zuo, Y.; Wang, T.; Cao, Y.; Cai, R.; Chen, F.L.; Cheng, J.; Mu, J. A crucial role of SUMOylation in modulating Sirt6 deacetylation of H3 at lysine 56 and its tumor suppressive activity. Oncogene 2016, 35, 4949–4956. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Yu, Y.; Huang, Q.; Tang, K. SIRT6 regulates the proliferation and apoptosis of hepatocellular carcinoma via the ERK1/2 signaling pathway. Mol. Med. Rep. 2019, 20, 1575–1582. [Google Scholar] [CrossRef] [Green Version]
- The Broad Institute of MIT & Harvard. Cancer Cell Line Encyclopedia. Available online: https://portals.broadinstitute.org/ccle/page?gene=SIRT6 (accessed on 6 March 2021).
- Zhang, J.; Yin, X.J.; Xu, C.J.; Ning, Y.X.; Chen, M.; Zhang, H.; Chen, S.F.; Yao, L.Q. The histone deacetylase SIRT6 inhibits ovarian cancer cell proliferation via down-regulation of Notch 3 expression. Eur. Rev. Med. Pharmacol. Sci. 2015, 19, 818–824. [Google Scholar]
- Jung, S.G.; Kwon, Y.D.; Song, J.A.; Back, M.J.; Lee, S.Y.; Lee, C.; Hwang, Y.Y.; An, H.J. Prognostic significance of Notch 3 gene expression in ovarian serous carcinoma. Cancer Sci. 2010, 101, 1977–1983. [Google Scholar] [CrossRef]
- Chen, X.; Hao, B.; Liu, Y.; Dai, D.; Han, G.; Li, Y.; Wu, X.; Zhou, X.; Yue, Z.; Wang, L.; et al. The histone deacetylase SIRT6 suppresses the expression of the RNA-binding protein PCBP2 in glioma. Biochem. Biophys. Res. Commun. 2014, 446, 364–369. [Google Scholar] [CrossRef] [PubMed]
- Makeyev, A.V.; Liebhaber, S.A. The poly(C)-binding proteins: A multiplicity of functions and a search for mechanisms. RNA 2002, 8. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.-G.; Qin, C.-Y. Sirt6 suppresses hepatocellular carcinoma cell growth via inhibiting the extracellular signal-regulated kinase signaling pathway. Mol. Med. Rep. 2014, 9, 882–888. [Google Scholar] [CrossRef] [Green Version]
- Han, Z.; Liu, L.; Yaxin, L.; Shanqing, L. Sirtuin SIRT6 suppresses cell proliferation through inhibition of Twist1 expression in non-small cell lung cancer. Int. J. Clin. Exp. Pathol. 2014, 7, 4774–4781. [Google Scholar]
- Ouyang, L.; Yi, L.; Li, J.; Yi, S.; Li, S.; Liu, P.; Yang, X. SIRT6 overexpression induces apoptosis of nasopharyngeal carcinoma by inhibiting NF-kappaB signaling. OncoTargets Ther. 2018, 11, 7613–7624. [Google Scholar] [CrossRef] [Green Version]
- Bhardwaj, A.; Das, S. SIRT6 deacetylates PKM2 to suppress its nuclear localization and oncogenic functions. Proc. Natl. Acad. Sci. USA 2016, 113, E538–E547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kugel, S.; Sebastián, C.; Fitamant, J.; Ross, K.N.; Saha, S.K.; Jain, E.; Gladden, A.; Arora, K.S.; Kato, Y.; Rivera, M.N.; et al. SIRT6 suppresses pancreatic cancer through control of Lin28b. Cell 2016, 165, 1401–1415. [Google Scholar] [CrossRef] [Green Version]
- Zhong, L.; D’Urso, A.; Toiber, D.; Sebastian, C.; Henry, R.E.; Vadysirisack, D.D.; Guimaraes, A.; Marinelli, B.; Wikstrom, J.D.; Nir, T.; et al. The Histone Deacetylase Sirt6 Regulates Glucose Homeostasis via Hif1α. Cell 2010, 140, 280–293. [Google Scholar] [CrossRef] [Green Version]
- Warburg, O. On the origin of cancer cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Mao, D.; Cao, Y.; Li, H.; Ren, F.; Li, K. Downregulation of SIRT6 by miR-34c-5p is associated with poor prognosis and promotes colon cancer proliferation through inhibiting apoptosis via the JAK2/STAT3 signaling pathway. Int. J. Oncol. 2018, 52, 1515–1527. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Shen, S.; Verma, I.M. NF-κB, an active player in human cancers. Cancer Immunol. Res. 2014, 2, 823–830. [Google Scholar] [CrossRef] [Green Version]
- Fukuda, T.; Wada-Hiraike, O.; Oda, K.; Tanikawa, M.; Makii, C.; Inaba, K.; Miyasaka, A.; Miyamoto, Y.; Yano, T.; Maeda, D.; et al. Putative tumor suppression function of SIRT6 in endometrial cancer. FEBS Lett. 2015, 589, 2274–2281. [Google Scholar] [CrossRef] [Green Version]
- Dong, Z.; Yang, J.; Li, L.; Tan, L.; Shi, P.; Zhang, J.; Zhong, X.; Ge, L.; Wu, Z.; Cui, H. FOXO3aSIRT6 axis suppresses aerobic glycolysis in melanoma. Int. J. Oncol. 2020, 56, 728–742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Myatt, S.S.; Lam, E.W. The emerging roles of forkhead box (Fox) proteins in cancer. Nat. Rev. Cancer 2007, 7, 847–859. [Google Scholar] [CrossRef] [PubMed]
- Skurk, C.; Maatz, H.; Kim, H.S.; Yang, J.; Abid, M.R.; Aird, W.C.; Walsh, K. The Akt-regulated forkhead transcription factor FOXO3a controls endothelial cell viability through modulation of the caspase-8 inhibitor FLIP. J. Biol. Chem. 2004, 279, 1513–1525. [Google Scholar] [CrossRef] [Green Version]
- Ekoff, M.; Kaufmann, T.; Engstrom, M.; Motoyama, N.; Villunger, A.; Jonsson, J.I.; Strasser, A.; Nilsson, G. The BH3-only protein Puma plays an essential role in cytokine deprivation induced apoptosis of mast cells. Blood 2007, 110, 3209–3217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, H.; Fellows, A.; Foote, K.; Yang, Z.; Figg, N.; Littlewood, T.; Bennett, M. FOXO3a (Forkhead Transcription Factor O Subfamily Member 3a) Links Vascular Smooth Muscle Cell Apoptosis, Matrix Breakdown, Atherosclerosis, and Vascular Remodeling Through a Novel Pathway Involving MMP13 (Matrix Metalloproteinase 13). Arter. Thromb. Vasc. Biol. 2018, 38, 555–565. [Google Scholar] [CrossRef] [Green Version]
- Kops, G.J.P.L.; Dansen, T.B.; Polderman, P.E.; Saarloos, I.; Wirtz, K.W.A.; Coffer, P.J.; Huang, T.T.; Bos, J.L.; Medema, R.H.; Burgering, B.M.T. Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. Nature 2002, 419, 316–321. [Google Scholar] [CrossRef]
- Tao, R.; Xiong, X.; DePinho, R.A.; Deng, C.X.; Dong, X.C. Hepatic SREBP-2 and cholesterol biosynthesis are regulated by FoxO3 and Sirt6. J. Lipid Res. 2013, 54, 2745–2753. [Google Scholar] [CrossRef] [Green Version]
- Tao, R.; Xiong, X.; DePinho, R.A.; Deng, C.X.; Dong, X.C. FoxO3 transcription factor and Sirt6 deacetylase regulate low density lipoprotein (LDL)-cholesterol homeostasis via control of the proprotein convertase subtilisin/kexin type 9 (Pcsk9) gene expression. J. Biol. Chem. 2013, 288, 29252–29259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strub, T.; Ghiraldini, F.G.; Carcamo, S.; Li, M.; Wroblewska, A.; Singh, R.; Goldberg, M.S.; Hasson, D.; Wang, Z.; Gallagher, S.J.; et al. SIRT6 haploinsufficiency induces BRAFV600E melanoma cell resistance to MAPK inhibitors via IGF signalling. Nat. Commun. 2018, 9. [Google Scholar] [CrossRef]
- Lu, C.T.; Hsu, C.M.; Lin, P.M.; Lai, C.C.; Lin, H.C.; Yang, C.H.; Hsiao, H.H.; Liu, Y.C.; Lin, H.Y.H.; Lin, S.F.; et al. The potential of SIRT6 and SIRT7 as circulating markers for head and neck squamous cell carcinoma. Anticancer Res. 2014, 34, 7137–7143. [Google Scholar]
- Han, L.L.; Jia, L.; Wu, F.; Huang, C. Sirtuin6 (SIRT6) Promotes the EMT of Hepatocellular Carcinoma by Stimulating Autophagic Degradation of E-Cadherin. Mol. Cancer Res. 2019, 17, 2267–2280. [Google Scholar] [CrossRef] [Green Version]
- Ran, L.K.; Chen, Y.; Zhang, Z.Z.; Tao, N.N.; Ren, J.H.; Zhou, L.; Tang, H.; Chen, X.; Chen, K.; Li, W.Y.; et al. SIRT6 overexpression potentiates apoptosis evasion in hepatocellular carcinoma via BCL2-associated X protein-dependent apoptotic pathway. Clin. Cancer Res. 2016, 22, 3372–3382. [Google Scholar] [CrossRef] [Green Version]
- Zhou, H.Z.; Zeng, H.Q.; Yuan, D.; Ren, J.H.; Cheng, S.T.; Yu, H.B.; Ren, F.; Wang, Q.; Qin, Y.P.; Huang, A.L.; et al. NQO1 potentiates apoptosis evasion and upregulates XIAP via inhibiting proteasome-mediated degradation SIRT6 in hepatocellular carcinoma. Cell Commun. Signal 2019, 17, 168. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Xie, Q.R.; Wang, B.; Shao, J.; Zhang, T.; Liu, T.; Huang, G.; Xia, W. Inhibition of SIRT6 in prostate cancer reduces cell viability and increases sensitivity to chemotherapeutics. Protein Cell 2013, 4, 702–710. [Google Scholar] [CrossRef] [Green Version]
- Khongkow, M.; Olmos, Y.; Gong, C.; Gomesl, A.R.; Monteiro, L.J.; Yagüe, E.; Cavaco, T.B.; Khongkow, P.; Man, E.P.S.; Laohasinnarong, S.; et al. SIRT6 modulates paclitaxel and epirubicin resistance and survival in breast cancer. Carcinogenesis 2013, 34, 1476–1486. [Google Scholar] [CrossRef] [PubMed]
- Ming, M.; Han, W.; Zhao, B.; Sundaresan, N.R.; Deng, C.X.; Gupta, M.P.; He, Y.Y. SIRT6 promotes COX-2 expression and acts as an oncogene in skin cancer. Cancer Res. 2014, 74, 5925–5933. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Peterson, L.M.; Ndiaye, M.A.; Singh, C.K.; Chhabra, G.; Huang, W.; Ahmad, N. SIRT6 histone deacetylase functions as a potential oncogene in human melanoma. Genes Cancer 2017, 8, 701–712. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Peterson, L.M.; Ndiaye, M.A.; Chhabra, G.; Singh, C.K.; Guzman-Perez, G.; Iczkowski, K.A.; Ahmad, N. CRISPR/Cas9-mediated Knockout of SIRT6 Imparts Remarkable Antiproliferative Response in Human Melanoma Cells in vitro and in vivo. Photochem. Photobiol. 2020, 96, 1314–1320. [Google Scholar] [CrossRef]
- Wang, L.; Guo, W.; Ma, J.; Dai, W.; Liu, L.; Guo, S.; Chen, J.; Wang, H.; Yang, Y.; Yi, X.; et al. Aberrant SIRT6 expression contributes to melanoma growth: Role of the autophagy paradox and IGF-AKT signaling. Autophagy 2018, 14, 518–533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Peterson, L.M.; Guzman-Perez, G.; Krier, C.R.; Ahmad, N. The sirtuin 6: An overture in skin cancer. Exp. Derm. 2020, 29, 124–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Li, Y.; Zhang, Y.; Fang, X.; Chen, N.; Zhou, X.; Wang, X. Sirt6 promotes tumorigenesis and drug resistance of diffuse large B-cell lymphoma by mediating PI3K/Akt signaling. J. Exp. Clin. Cancer Res. 2020, 39, 142. [Google Scholar] [CrossRef] [PubMed]
- Cagnetta, A.; Soncini, D.; Orecchioni, S.; Talarico, G.; Minetto, P.; Guolo, F.; Retali, V.; Colombo, N.; Carminati, E.; Clavio, M.; et al. Depletion of SIRT6 enzymatic activity increases acute myeloid leukemia cells’ vulnerability to DNA-damaging agents. Haematologica 2018, 103, 80–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Speidel, D. Transcription-independent p53 apoptosis: An alternative route to death. Trends Cell Biol. 2010, 20, 14–24. [Google Scholar] [CrossRef]
- Tao, N.N.; Ren, J.H.; Tang, H.; Ran, L.K.; Zhou, H.Z.; Liu, B.; Huang, A.L.; Chen, J. Deacetylation of Ku70 by SIRT6 attenuates Bax-mediated apoptosis in hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 2017, 485, 713–719. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Ha, S.H.; Moon, Y.J.; Hussein, U.K.; Song, Y.; Kim, K.M.; Park, S.H.; Park, H.S.; Park, B.H.; Ahn, A.R.; et al. Inhibition of SIRT6 potentiates the anti-tumor effect of doxorubicin through suppression of the DNA damage repair pathway in osteosarcoma. J. Exp. Clin. Cancer Res. 2020, 39, 247. [Google Scholar] [CrossRef]
- Cea, M.; Cagnetta, A.; Adamia, S.; Acharya, C.; Tai, Y.-T.; Fulciniti, M.; Ohguchi, H.; Munshi, A.; Acharya, P.; Bhasin, M.K.; et al. Evidence for a role of the histone deacetylase SIRT6 in DNA damage response of multiple myeloma cells. Blood 2016, 127, 1138–1150. [Google Scholar] [CrossRef] [Green Version]
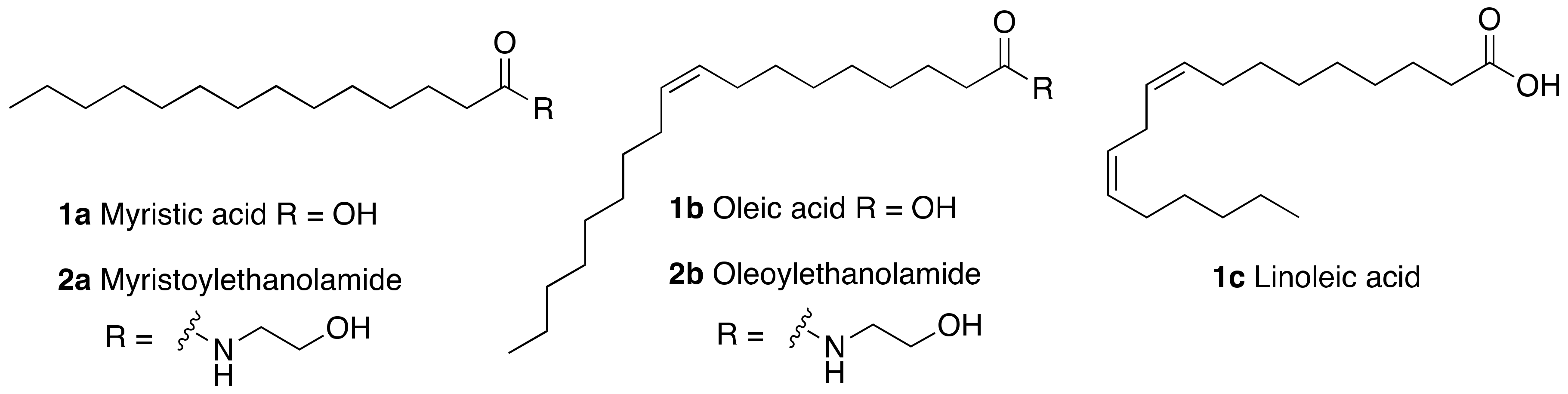
- Rahnasto-Rilla, M.; Kokkola, T.; Jarho, E.; Lahtela-Kakkonen, M.; Moaddel, R. N-Acylethanolamines Bind to SIRT6. ChemBioChem 2016, 17, 77–81. [Google Scholar] [CrossRef] [Green Version]
- Rahnasto-Rilla, M.; Tyni, J.; Huovinen, M.; Jarho, E.; Kulikowicz, T.; Ravichandran, S.; Bohr, V.A.; Ferrucci, L.; Lahtela-Kakkonen, M.; Moaddel, R. Natural polyphenols as sirtuin 6 modulators. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef]
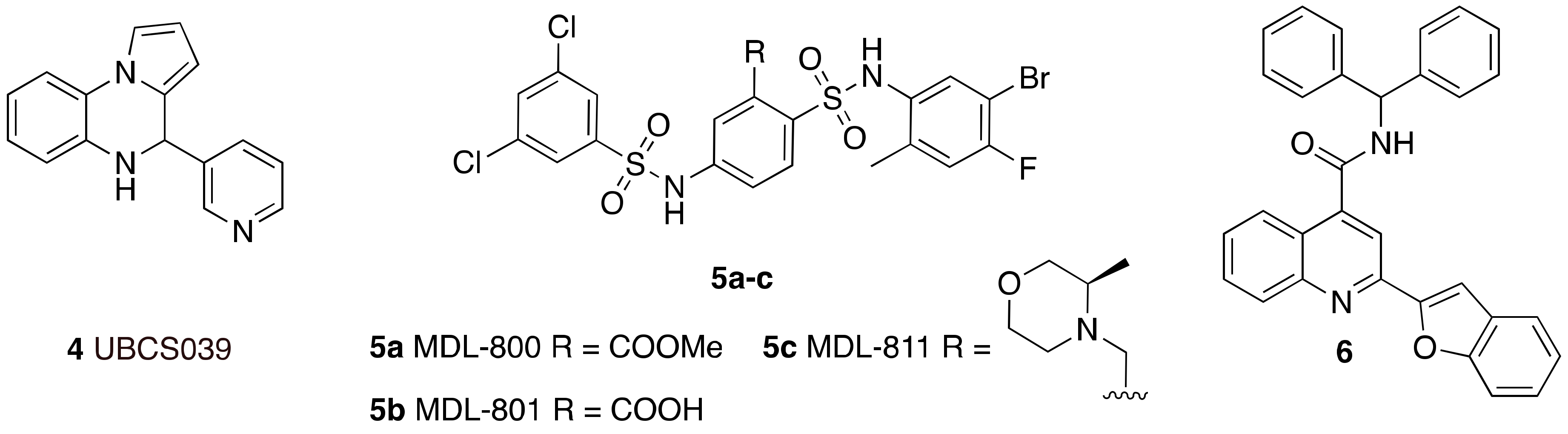
- You, W.; Rotili, D.; Li, T.M.; Kambach, C.; Meleshin, M.; Schutkowski, M.; Chua, K.F.; Mai, A.; Steegborn, C. Structural Basis of Sirtuin 6 Activation by Synthetic Small Molecules. Angew. Chem. Int. Ed. 2017, 56, 1007–1011. [Google Scholar] [CrossRef]
- Iachettini, S.; Trisciuoglio, D.; Rotili, D.; Lucidi, A.; Salvati, E.; Zizza, P.; Di Leo, L.; Del Bufalo, D.; Ciriolo, M.R.; Leonetti, C.; et al. Pharmacological activation of SIRT6 triggers lethal autophagy in human cancer cells. Cell Death Dis. 2018, 9. [Google Scholar] [CrossRef]
- Huang, Z.; Zhao, J.; Deng, W.; Chen, Y.; Shang, J.; Song, K.; Zhang, L.; Wang, C.; Lu, S.; Yang, X.; et al. Identification of a cellularly active SIRT6 allosteric activator. Nat. Chem. Biol. 2018, 14, 1118–1126. [Google Scholar] [CrossRef]
- Shang, J.L.; Ning, S.B.; Chen, Y.Y.; Chen, T.X.; Zhang, J. MDL-800, an allosteric activator of SIRT6, suppresses proliferation and enhances EGFR-TKIs therapy in non-small cell lung cancer. Acta Pharmacologica Sinica 2020. [Google Scholar] [CrossRef]
- Shang, J.; Zhu, Z.; Chen, Y.; Song, J.; Huang, Y.; Song, K.; Zhong, J.; Xu, X.; Wei, J.; Wang, C.; et al. Small-molecule activating SIRT6 elicits therapeutic effects and synergistically promotes anti-tumor activity of vitamin D3 in colorectal cancer. Theranostics 2020, 10, 5845–5864. [Google Scholar] [CrossRef]
- Chen, X.; Sun, W.; Huang, S.; Zhang, H.; Lin, G.; Li, H.; Qiao, J.; Li, L.; Yang, S. Discovery of Potent Small-Molecule SIRT6 Activators: Structure–Activity Relationship and Anti-Pancreatic Ductal Adenocarcinoma Activity. J. Med. Chem. 2020, 63, 10474–10495. [Google Scholar] [CrossRef] [PubMed]
- You, W.; Zheng, W.; Weiss, S.; Chua, K.F.; Steegborn, C. Structural basis for the activation and inhibition of Sirtuin 6 by quercetin and its derivatives. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, D.; Sarikhani, M.; Dasgupta, S.; Maniyadath, B.; Pandit, A.S.; Mishra, S.; Ahamed, F.; Dubey, A.; Fathma, N.; Atreya, H.S.; et al. SIRT6 deacetylase transcriptionally regulates glucose metabolism in heart. J. Cell Physiol. 2018, 233, 5478–5489. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chen, J.; Sun, X.; Yu, J.; Qian, Z.; Wu, L.; Xu, X.; Wan, X.; Jiang, Y.; Zhang, J.; et al. The SIRT6 activator MDL-800 improves genomic stability and pluripotency of old murine-derived iPS cells. Aging Cell 2020, 19, e13185. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Wang, T.T.; Liu, X.Z.; Wang, N.Y.; Sun, L.H.; Zhang, Z.Q.; Chen, H.Z.; Lv, X.; Huang, Y.; Liu, D.P. Sirt6 regulates efficiency of mouse somatic reprogramming and maintenance of pluripotency. Stem Cell Res. 2019, 10, 9. [Google Scholar] [CrossRef]
- Etchegaray, J.P.; Chavez, L.; Huang, Y.; Ross, K.N.; Choi, J.; Martinez-Pastor, B.; Walsh, R.M.; Sommer, C.A.; Lienhard, M.; Gladden, A.; et al. The histone deacetylase SIRT6 controls embryonic stem cell fate via TET-mediated production of 5-hydroxymethylcytosine. Nat. Cell Biol. 2015, 17, 545–557. [Google Scholar] [CrossRef]
- Hobaus, J.; Hummel, D.M.; Thiem, U.; Fetahu, I.S.; Aggarwal, A.; Mullauer, L.; Heller, G.; Egger, G.; Mesteri, I.; Baumgartner-Parzer, S.; et al. Increased copy-number and not DNA hypomethylation causes overexpression of the candidate proto-oncogene CYP24A1 in colorectal cancer. Int. J. Cancer 2013, 133, 1380–1388. [Google Scholar] [CrossRef] [Green Version]
- Sun, H.; Wang, C.; Hao, M.; Sun, R.; Wang, Y.; Liu, T.; Cong, X.; Liu, Y. CYP24A1 is a potential biomarker for the progression and prognosis of human colorectal cancer. Hum. Pathol. 2016, 50, 101–108. [Google Scholar] [CrossRef]
- Pereira, F.; Larriba, M.J.; Muñoz, A. Vitamin D and colon cancer. Endocr. Relat. Cancer 2012, 19, R51–R71. [Google Scholar] [CrossRef] [Green Version]
- Ng, K.; Nimeiri, H.S.; McCleary, N.J.; Abrams, T.A.; Yurgelun, M.B.; Cleary, J.M.; Rubinson, D.A.; Schrag, D.; Miksad, R.; Bullock, A.J.; et al. Effect of High-Dose vs Standard-Dose Vitamin D3 Supplementation on Progression-Free Survival Among Patients With Advanced or Metastatic Colorectal Cancer: The SUNSHINE Randomized Clinical Trial. JAMA 2019, 321, 1370–1379. [Google Scholar] [CrossRef] [Green Version]
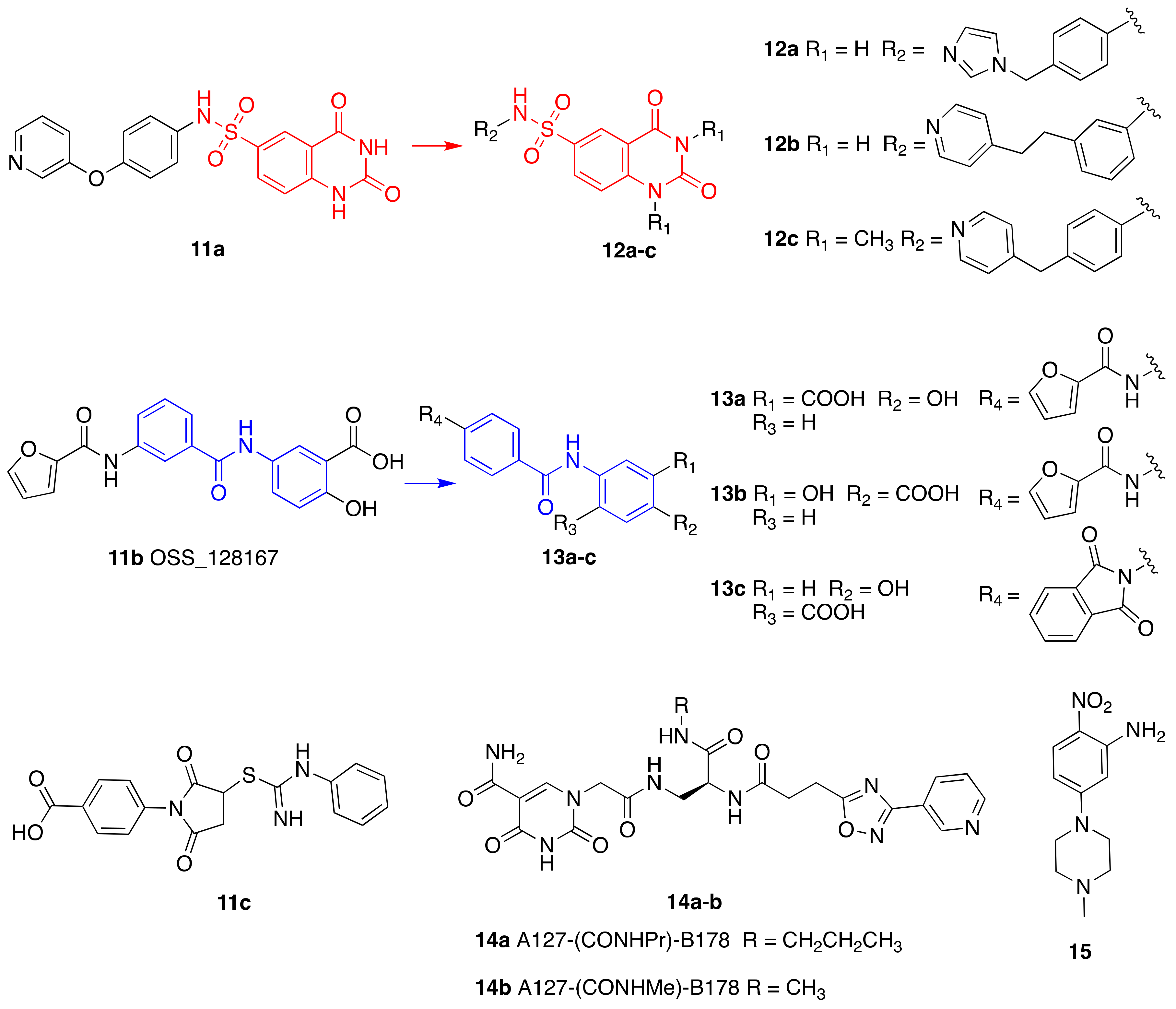
- He, B.; Hu, J.; Zhang, X.; Lin, H. Thiomyristoyl peptides as cell-permeable Sirt6 inhibitors. Org. Biomol. Chem. 2014, 12, 7498–7502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parenti, M.D.; Grozio, A.; Bauer, I.; Galeno, L.; Damonte, P.; Millo, E.; Sociali, G.; Franceschi, C.; Ballestrero, A.; Bruzzone, S.; et al. Discovery of Novel and Selective SIRT6 Inhibitors. J. Med. Chem. 2014, 57, 4796–4804. [Google Scholar] [CrossRef]
- Sociali, G.; Galeno, L.; Parenti, M.D.; Grozio, A.; Bauer, I.; Passalacqua, M.; Boero, S.; Donadini, A.; Millo, E.; Bellotti, M.; et al. Quinazolinedione SIRT6 inhibitors sensitize cancer cells to chemotherapeutics. Eur. J. Med. Chem. 2015, 102, 530–539. [Google Scholar] [CrossRef]
- Damonte, P.; Sociali, G.; Parenti, M.D.; Soncini, D.; Bauer, I.; Boero, S.; Grozio, A.; von Holtey, M.; Piacente, F.; Becherini, P.; et al. SIRT6 inhibitors with salicylate-like structure show immunosuppressive and chemosensitizing effects. Bioorganic Med. Chem. 2017, 25, 5849–5858. [Google Scholar] [CrossRef]
- Cardus, A.; Uryga, A.K.; Walters, G.; Erusalimsky, J.D. SIRT6 protects human endothelial cells from DNA damage, telomere dysfunction, and senescence. Cardiovasc. Res. 2013, 97, 571–579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, W.; Chen, X.; Huang, S.; Li, W.; Tian, C.; Yang, S.; Li, L. Discovery of 5-(4-methylpiperazin-1-yl)-2-nitroaniline derivatives as a new class of SIRT6 inhibitors. Bioorg. Med. Chem. Lett. 2020, 30, 127215. [Google Scholar] [CrossRef] [PubMed]
- Feldman, J.L.; Dittenhafer-Reed, K.E.; Kudo, N.; Thelen, J.N.; Ito, A.; Yoshida, M.; Denu, J.M. Kinetic and structural basis for Acyl-group selectivity and NAD+ dependence in sirtuin-catalyzed deacylation. Biochemistry 2015, 54, 3037–3050. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.; He, B.; Bhargava, S.; Lin, H. A fluorogenic assay for screening Sirt6 modulators. Org. Biomol. Chem. 2013, 11, 5213–5216. [Google Scholar] [CrossRef] [Green Version]
- Bolivar, B.E.; Welch, J.T. Studies of the Binding of Modest Modulators of the Human Enzyme, Sirtuin 6, by STD NMR. ChemBioChem 2017, 18, 931–940. [Google Scholar] [CrossRef]
- Madsen, A.S.; Andersen, C.; Daoud, M.; Anderson, K.A.; Laursen, J.S.; Chakladar, S.; Huynh, F.K.; Colaco, A.R.; Backos, D.S.; Fristrup, P.; et al. Investigating the Sensitivity of NAD(+)-dependent Sirtuin Deacylation Activities to NADH. J. Biol. Chem. 2016, 291, 7128–7141. [Google Scholar] [CrossRef] [Green Version]
- Fatkins, D.G.; Monnot, A.D.; Zheng, W. N-epsilon-thioacetyl-lysine: A multi-facet functional probe for enzymatic protein lysine N-epsilon-deacetylation. Bioorganic Med. Chem. Lett. 2006, 16, 3651–3656. [Google Scholar] [CrossRef] [PubMed]
- Sociali, G.; Liessi, N.; Grozio, A.; Caffa, I.; Parenti, M.D.; Ravera, S.; Tasso, B.; Benzi, A.; Nencioni, A.; Del Rio, A.; et al. Differential modulation of SIRT6 deacetylase and deacylase activities by lysine-based small molecules. Mol. Divers. 2020, 24, 655–671. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Khan, S.; Jiang, H.; Antonyak, M.A.; Chen, X.; Spiegelman, N.A.; Shrimp, J.H.; Cerione, R.A.; Lin, H. Identifying the functional contribution of the defatty-Acylase activity of SIRT6. Nat. Chem. Biol. 2016, 12, 614–620. [Google Scholar] [CrossRef] [Green Version]
- Sociali, G.; Magnone, M.; Ravera, S.; Damonte, P.; Vigliarolo, T.; von Holtey, M.; Vellone, V.G.; Millo, E.; Caffa, I.; Cea, M.; et al. Pharmacological Sirt6 inhibition improves glucose tolerance in a type 2 diabetes mouse model. FASEB J. 2017, 31, 3138–3149. [Google Scholar] [CrossRef] [Green Version]
- Yuen, L.H.; Dana, S.; Liu, Y.; Bloom, S.I.; Thorsell, A.G.; Neri, D.; Donato, A.J.; Kireev, D.; Schuler, H.; Franzini, R.M. A Focused DNA-Encoded Chemical Library for the Discovery of Inhibitors of NAD(+)-Dependent Enzymes. J. Am. Chem. Soc. 2019, 141, 5169–5181. [Google Scholar] [CrossRef]


| Interactor | Modification | Influence on SIRT6 | Reference |
|---|---|---|---|
| AKT1 | Ser338 Phosphorylation | Degradation | [50] |
| CHIP | Lys170 Ubiquitination | Stabilization | [52] |
| USP10 | Deubiquitination | Stabilization | [53] |
| UBC9 | SUMOylation | Increased H3K56 deacetylation | [54] |
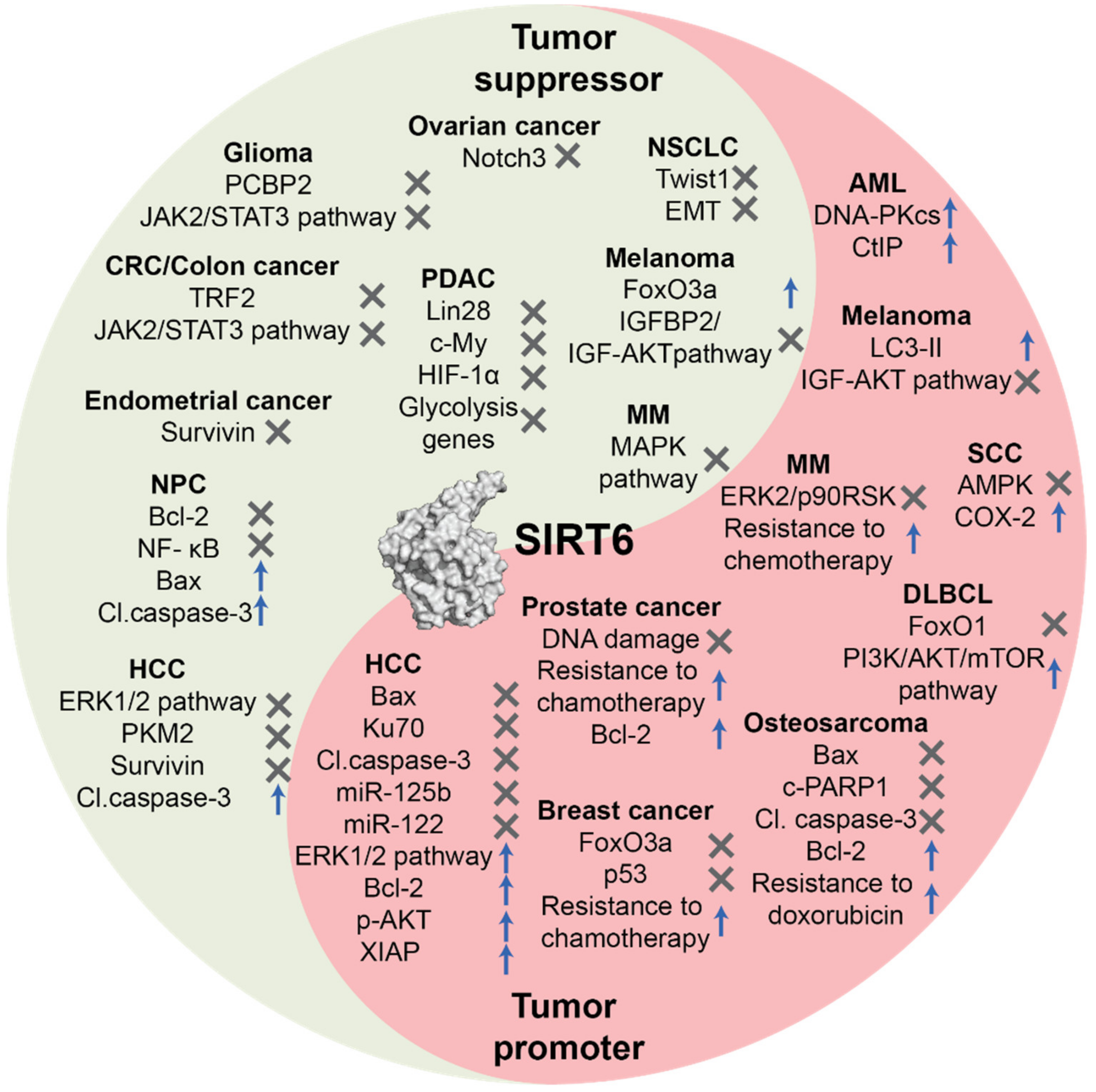
| Cancer Type | SIRT6 Expression | Cell Death Pathways and Regulators | Reference(s) |
|---|---|---|---|
| Ovarian cancer | Downregulated | SIRT6 inhibits Notch3 expression and cell proliferation. | [57] |
| Glioma | Downregulated | SIRT6 suppresses PCBP2 expression and cell proliferation. | [59] |
| SIRT6 inhibits JAK2/STAT3 pathway and reduces cell survival. | [44] | ||
| Non-Small Cell Lung Cancer (NSCLC) | Downregulated | SIRT6 inhibits Twist-1 expression suppressing metastatization and EMT. | [62] |
| Pancreatic Ductal Adenocarcinoma (PDAC) | Downregulated | SIRT6 inhibits Lin28b expression and c-Myc recruitment suppressing cancer progression and metastatization. | [65] |
| Colorectal cancer (CRC)/Colon cancer | Downregulated | SIRT6-mediated deacetylation induces TRF2 ubiquitination, leading to proteasomal degradation | [21] |
| USP10 inhibits SIRT6 degradation and blocks tumor growth via p53 and SIRT6-mediated degradation of c-Myc. | [53] | ||
| miRNA-34c-5p inhibits SIRT6 expression and activates JAK2/STAT3 pathway inhibiting apoptosis. | [68] | ||
| Nasopharyngeal Carcinoma (NPC) | Downregulated | SIRT6 downregulates Bcl-2 and NF-κB and induces apoptosis mediated through upregulation of Bax and cleaved caspase-3. | [63] |
| Endometrial cancer | Downregulated | SIRT6 inhibits survivin expression promoting apoptosis | [70] |
| Melanoma | Downregulated | SIRT6 increases FoxO3a expression levels | [71] |
| SIRT6 downregulates IGFBP2 through, thus impairing the activation of IGF-1R and AKT pathway, responsible of cancer cell survival and drug resistance. | [79] | ||
| SIRT6-mediated suppression of IGF-AKT signaling stimulates autophagy, a tumor protective factor only at early stages. | [89] | ||
| Hepatocellular Carcinoma (HCC) | Downregulated | SIRT6 induces apoptosis via upregulation of cleaved caspase-3 and ERK1/2 pathway inhibition. | [61] |
| SIRT6 deacetylates PKM2 inducing its nuclear export and blocks metastatization. | [64] | ||
| SIRT6 inhibits survivin expression and induces apoptosis. | [39] | ||
| Hepatocellular Carcinoma (HCC) | Upregulated | SIRT6 downregulates Bax and blocks apoptosis. | [82] |
| SIRT6 deacetylates Ku70 and blocks Bax-mediated apoptosis. | [94] | ||
| SIRT6 promotes proliferation and invasion inducing ERK1/2 pathway and Bcl-2 expression and downregulating Bax and cleaved caspase-3. | [55] | ||
| SIRT6 promotes proliferation and cell death evasion increasing p-AKT levels and XIAP expression. | [83] | ||
| SIRT6 inhibits miR-125b expression and apoptosis. | [48] | ||
| Reciprocal regulation between SIRT6 and miR-122 correlates with tumor progression. | [47] | ||
| Prostate cancer | Upregulated | SIRT6 knockdown increases sensitivity to chemotherapy, DNA damage, cell cycle arrest in G1, Bcl-2 downregulation and apoptosis induction. | [84] |
| Breast cancer | Upregulated | SIRT6 increases resistance to epirubicin and paclitaxel and negatively modulates the acetylation status and expression of FoxO3a and p53. | [85] |
| Osteosarcoma | Upregulated | SIRT6 facilitates DNA repair in cancer cells, leading to doxorubicin resistance. SIRT6 also inhibits the expression of Bax, cleaved-PARP1 and cleaved-caspase3 and increases Bcl-2 expression leading to apoptosis evasion. | [95] |
| Squamous Cell Carcinoma (SCC) | Upregulated | Under UVB radiation, AKT induces SIRT6 expression that promotes COX-2 action and AMP-activated protein kinase (AMPK) repression leading to cell survival and proliferation. | [86] |
| SIRT6 is silenced by miR-34a and its downregulation induces cell differentiation and reduces cancer cell proliferation potential | [45] | ||
| Melanoma | Upregulated | SIRT6 activity increases the levels of the phosphatidylethanolamine-conjugated protein LC3-II, a crucial autophagosome initiator. | [87] |
| SIRT6 suppresses IGF-AKT signaling, thus stimulating autophagy which is a protective factor at early stages, but promotes tumor development at later stages. | [89] | ||
| Diffuse Large B-Cell Lymphoma (DLBCL) | Upregulated | SIRT6 activates PI3K/AKT/mTOR pathway, thus promoting cancer proliferation. | [91] |
| SIRT6 knockdown determines upregulation of the oncosuppressor FoxO1. | |||
| Acute Myeloid Leukemia (AML) | Upregulated | SIRT6 increases cell death resistance via DNA-PKcs and CtIP deacetylation. | [92] |
| Multiple Myeloma (MM) | Upregulated | SIRT6 downregulates MAPK pathway genes suppressing cell proliferation. | [96] |
| SIRT6 increases resistance to DNA-damaging agents through inhibition of ERK2/p90RSK signaling. |
| Compound | Structure | Effect on SIRT6 Activity | Cellular and In Vivo Effects | Reference(s) |
|---|---|---|---|---|
| 3c Cyanidin |  | EC50 = 460 μM ×55 max activation (deacetylation) |
| [98] |
| 4 UBCS039 | 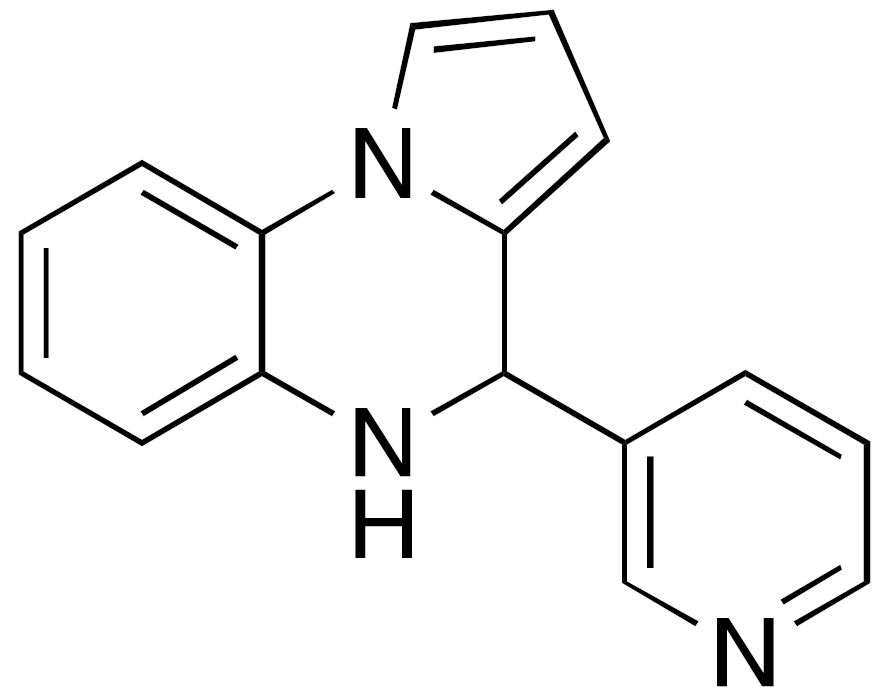 | EC50 = 38 μM ×3.5 max activation (deacetylation) |
| [99,100] |
| 5a MDL-800 | 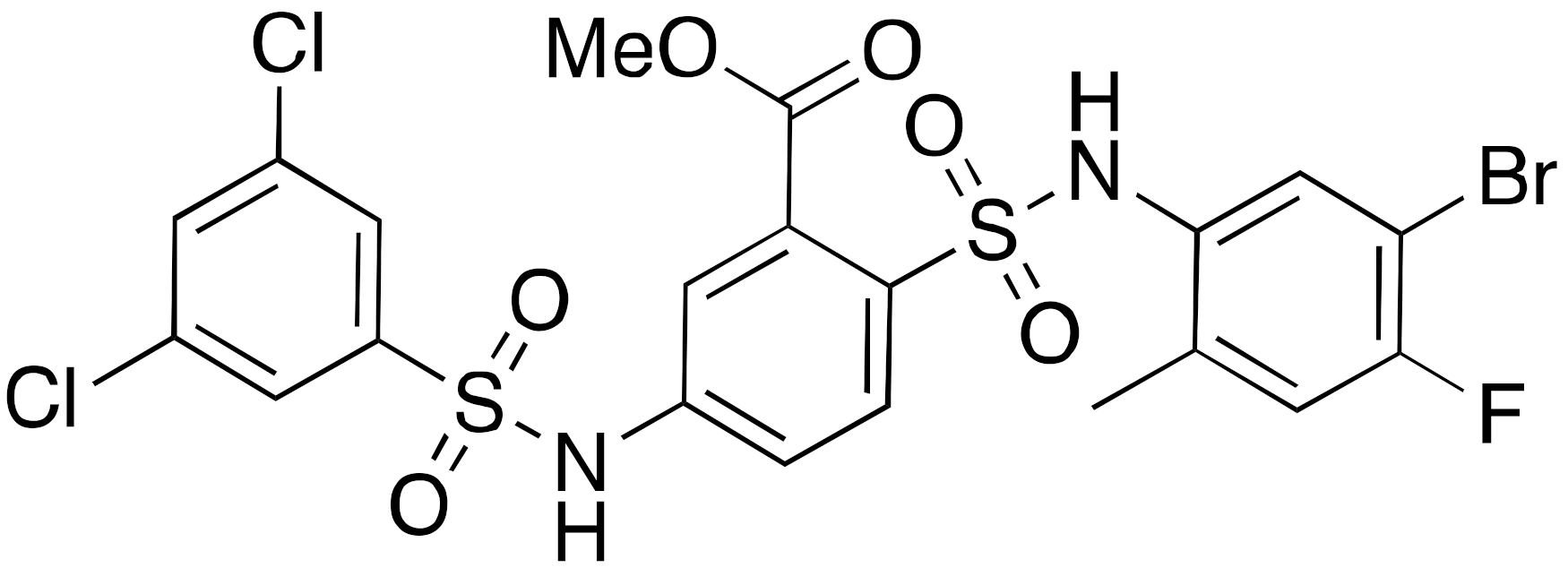 | EC50 = 10.3 μM ×22 max activation (deacetylation) |
| [101,102] |
| 5c MDL-811 | 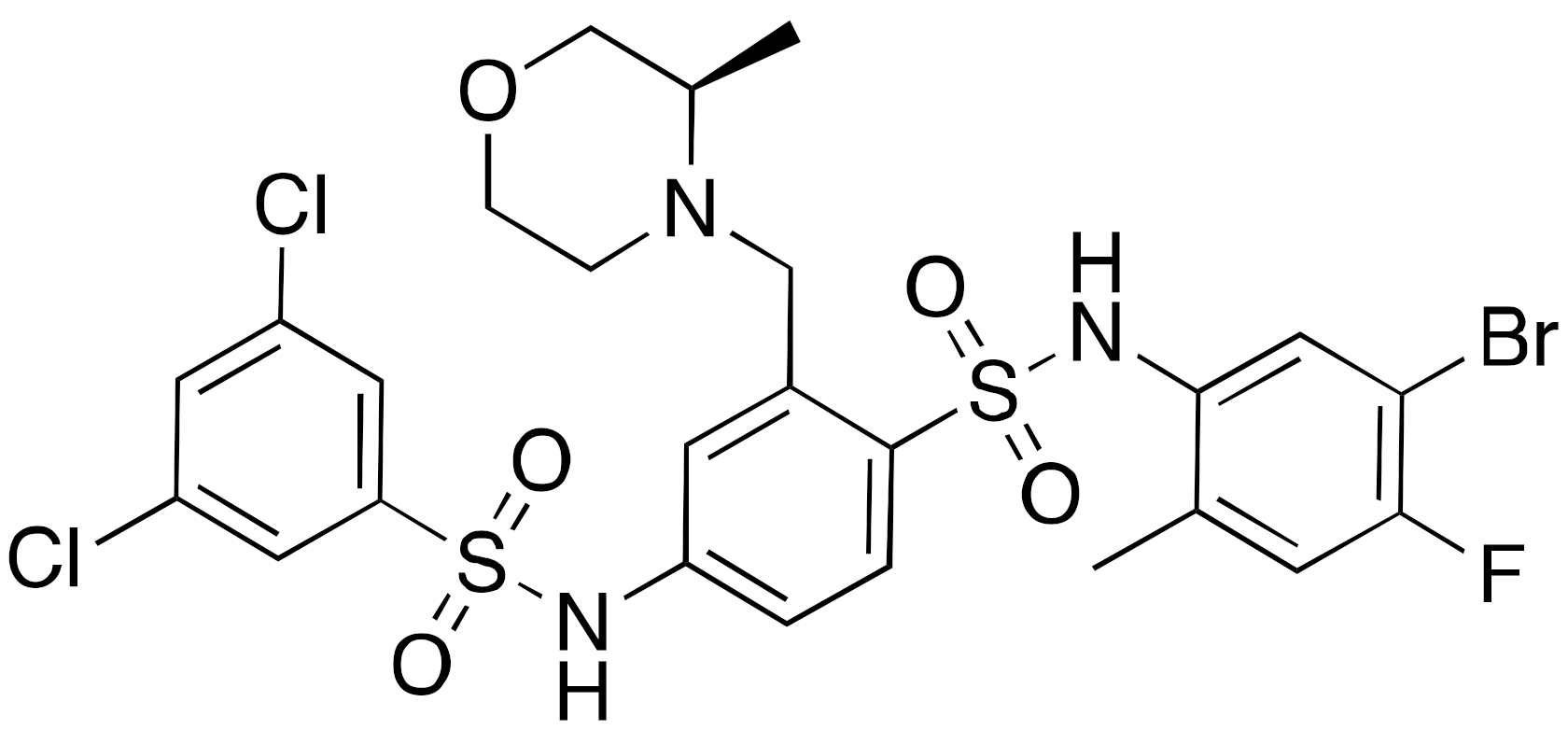 | EC50 = 5.7 μM (deacetylation) |
| [103] |
| 6 | 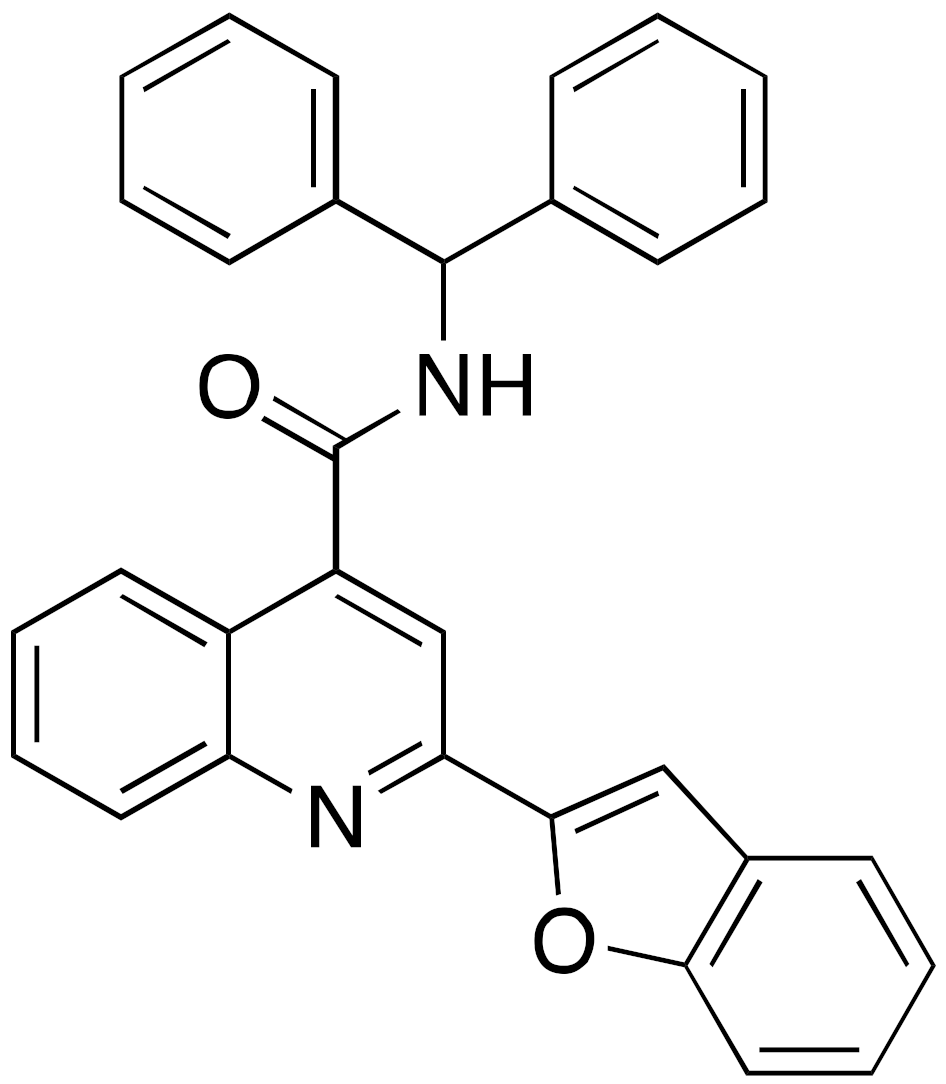 | EC50 = 5.35 μM (deacetylation) EC50 = 8.91 μM (demyristoylation) |
| [104] |
| Compound | Structure | Effect on SIRT6 Activity | Cellular and In Vivo Effects | Reference(s) |
|---|---|---|---|---|
| 9b BHJH-TM3 |  | IC50 = 8.1 μM (demyristoylation) |
| [114] |
| 11b OSS_128167 | 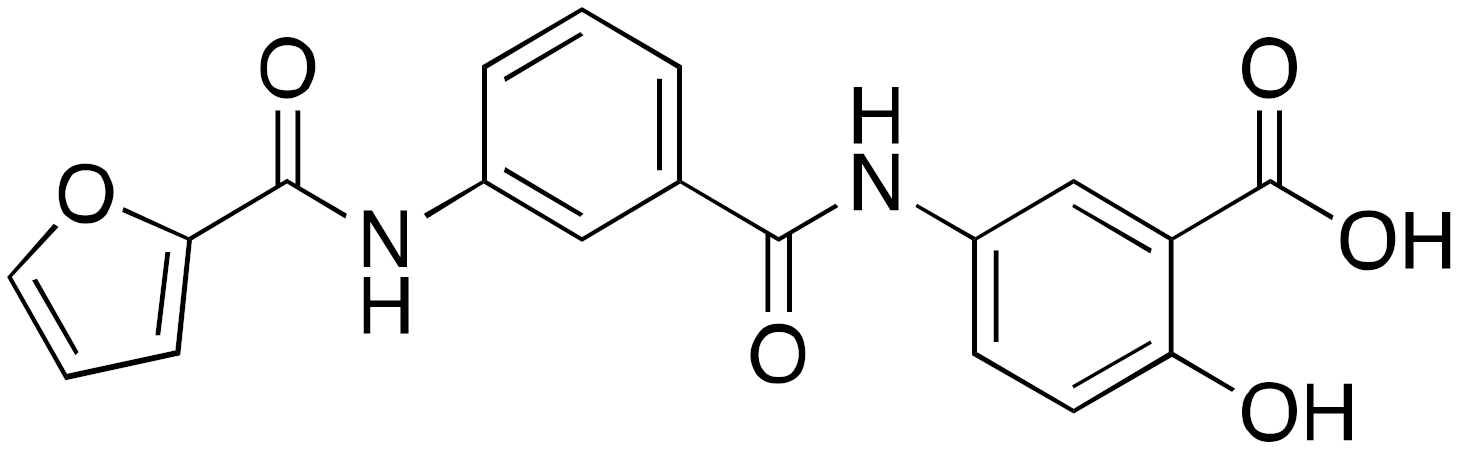 | IC50 = 89 μM (deacetylation) |
| [115] |
| [96] | |||
| [91] | |||
| ||||
| 12b | 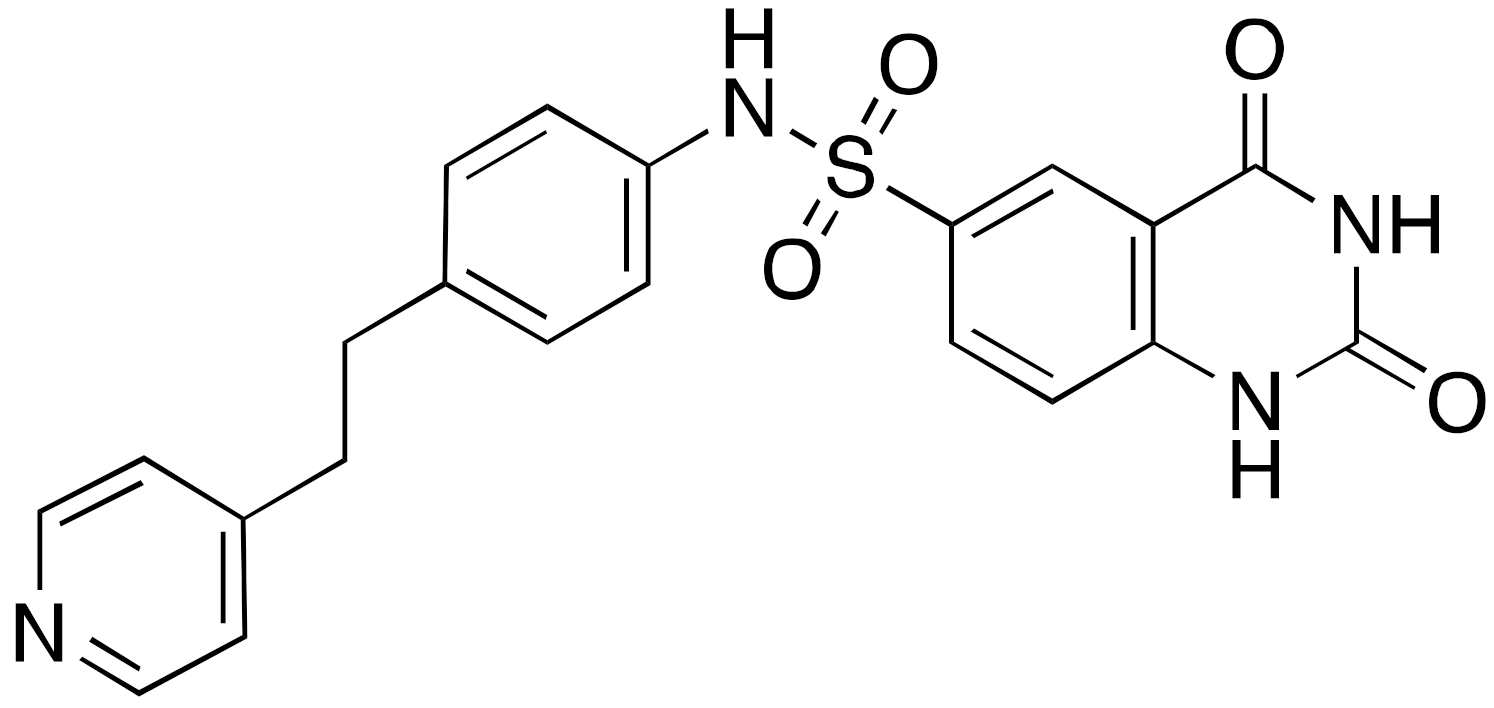 | IC50 = 37 μM (deacetylation) |
| [116] |
| 13b |  | IC50 = 22 μM (deacetylation) |
| [117] |
| 14a A127-(CONHPr)-B178 | 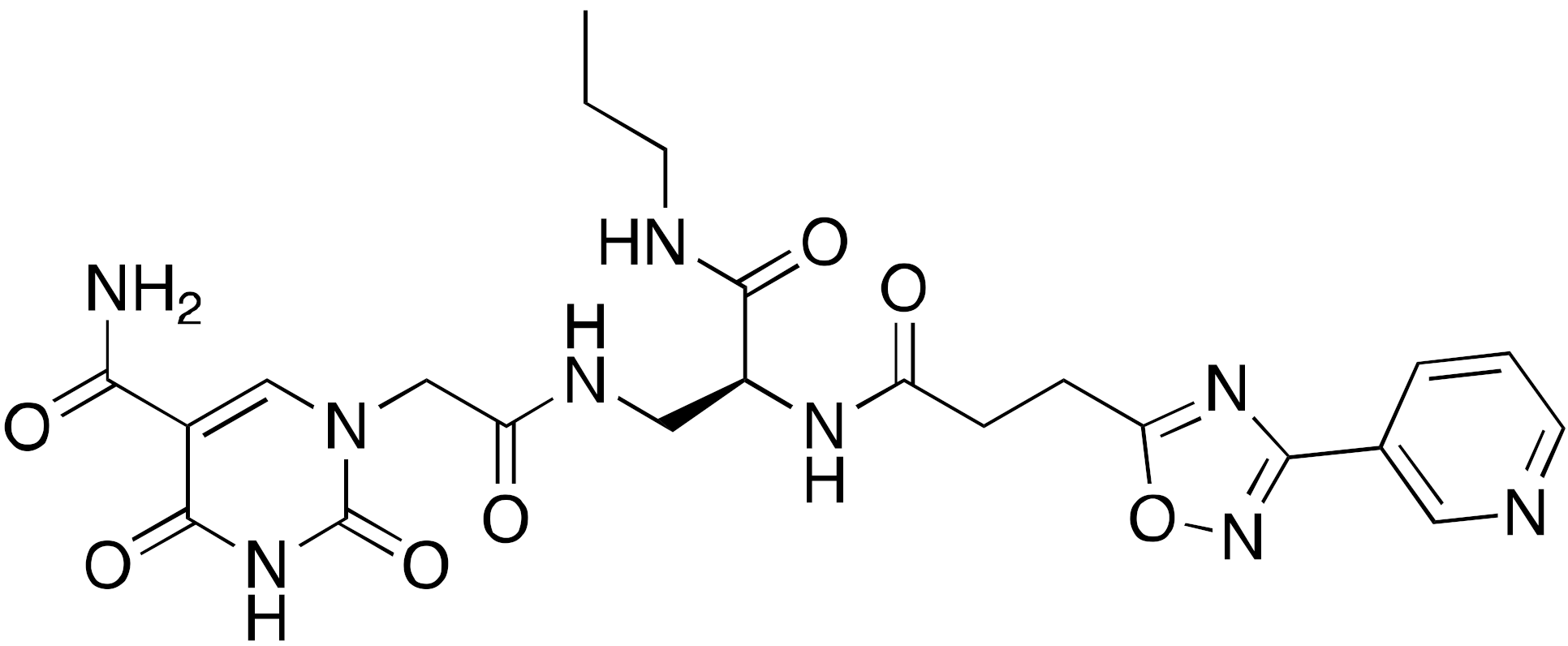 | IC50 = 6.7 μM (demyristoylation) |
| [118] |
| 15 | 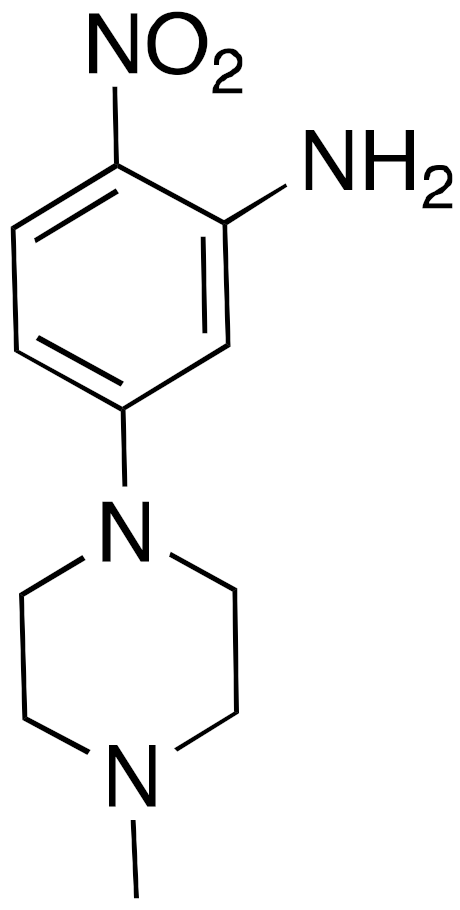 | IC50 = 4.93 μM (deacetylation) |
| [119] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fiorentino, F.; Carafa, V.; Favale, G.; Altucci, L.; Mai, A.; Rotili, D. The Two-Faced Role of SIRT6 in Cancer. Cancers 2021, 13, 1156. https://doi.org/10.3390/cancers13051156
Fiorentino F, Carafa V, Favale G, Altucci L, Mai A, Rotili D. The Two-Faced Role of SIRT6 in Cancer. Cancers. 2021; 13(5):1156. https://doi.org/10.3390/cancers13051156
Chicago/Turabian StyleFiorentino, Francesco, Vincenzo Carafa, Gregorio Favale, Lucia Altucci, Antonello Mai, and Dante Rotili. 2021. "The Two-Faced Role of SIRT6 in Cancer" Cancers 13, no. 5: 1156. https://doi.org/10.3390/cancers13051156
APA StyleFiorentino, F., Carafa, V., Favale, G., Altucci, L., Mai, A., & Rotili, D. (2021). The Two-Faced Role of SIRT6 in Cancer. Cancers, 13(5), 1156. https://doi.org/10.3390/cancers13051156









