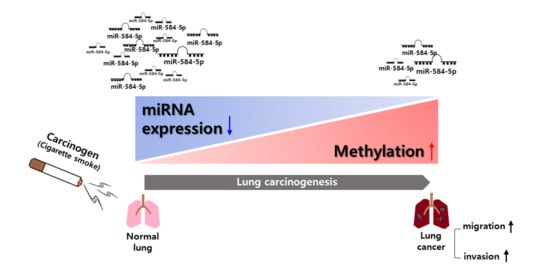
Tumor Suppressor miR-584-5p Inhibits Migration and Invasion in Smoking Related Non-Small Cell Lung Cancer Cells by Targeting YKT6
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Transfection
2.2. Treatment with Demethylation Agent
2.3. Microarray Analysis
2.4. Pyrosequencing Analysis
2.5. Gene Expression Omnibus (GEO) Database Analysis
2.6. RNA Isolation and Real-Time RT-PCR
2.7. Dual-Luciferase Reporter Assay
2.8. Wound-Healing Assay
2.9. Trans-Well Assays
2.10. Western Blot Analysis
2.11. Animal Studies
2.11.1. In Vivo Tumorigenicity Assays
2.11.2. In Vivo Metastasis Assays
2.12. Immunohistochemistry
2.13. TUNEL Assay
2.14. Kaplan–Meier Plot Analysis
2.15. Statistical Analysis
3. Results
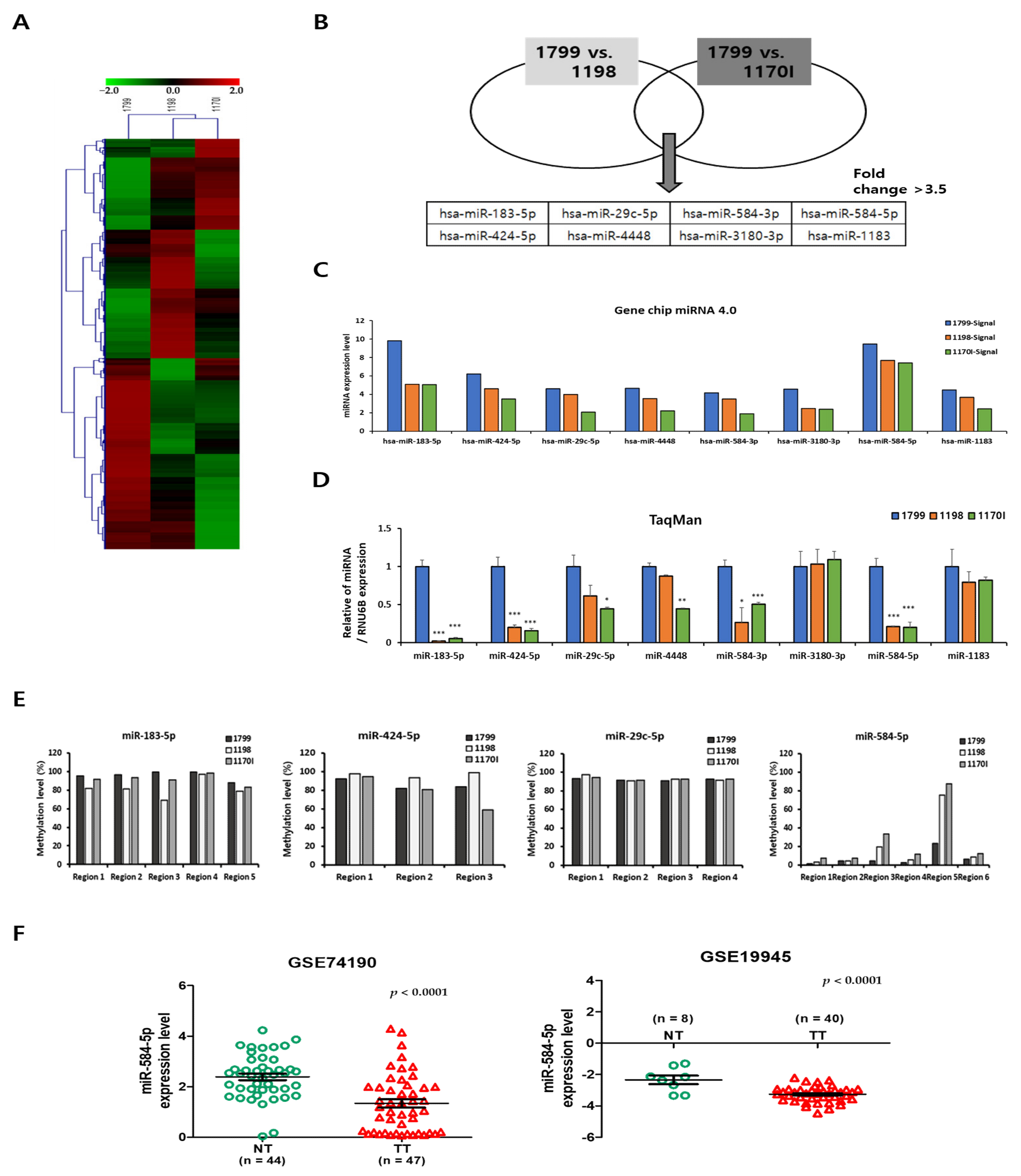
3.1. miR-584-5p Expression Is Downregulated in Lung Carcinogenesis Model Cell Lines
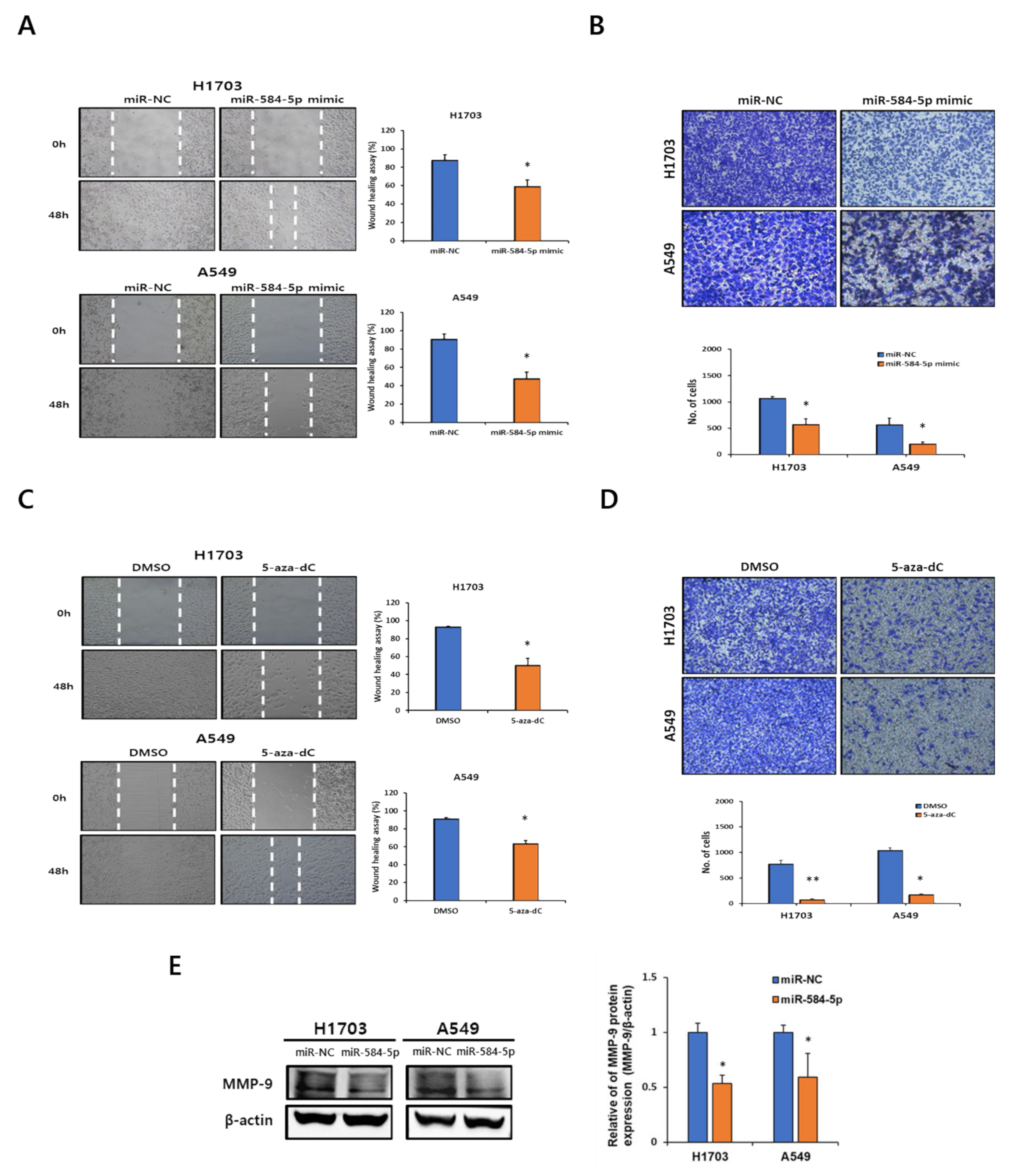
3.2. miR-584-5p Regulates Migration and Invasion in Lung Carcinogenesis Model Cell Lines
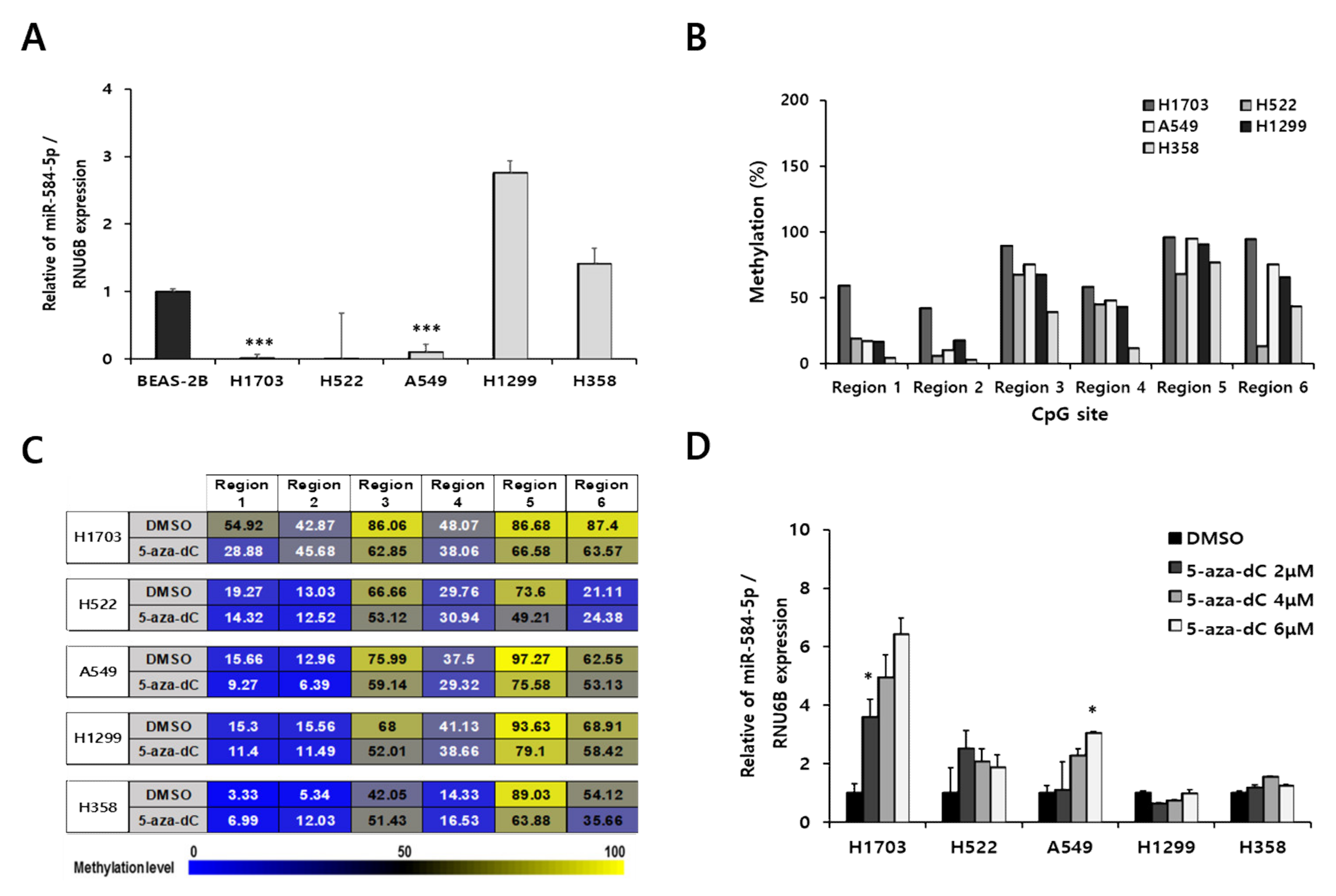
3.3. Downregulation of miR-584-5p Expression Is Associated with Hypermethylation of Promoter CpG Island in Smoking-Related NSCLC Cells
3.4. miR-584-5p Suppresses Migration and Invasion in Smoking-Related NSCLC Cell Lines
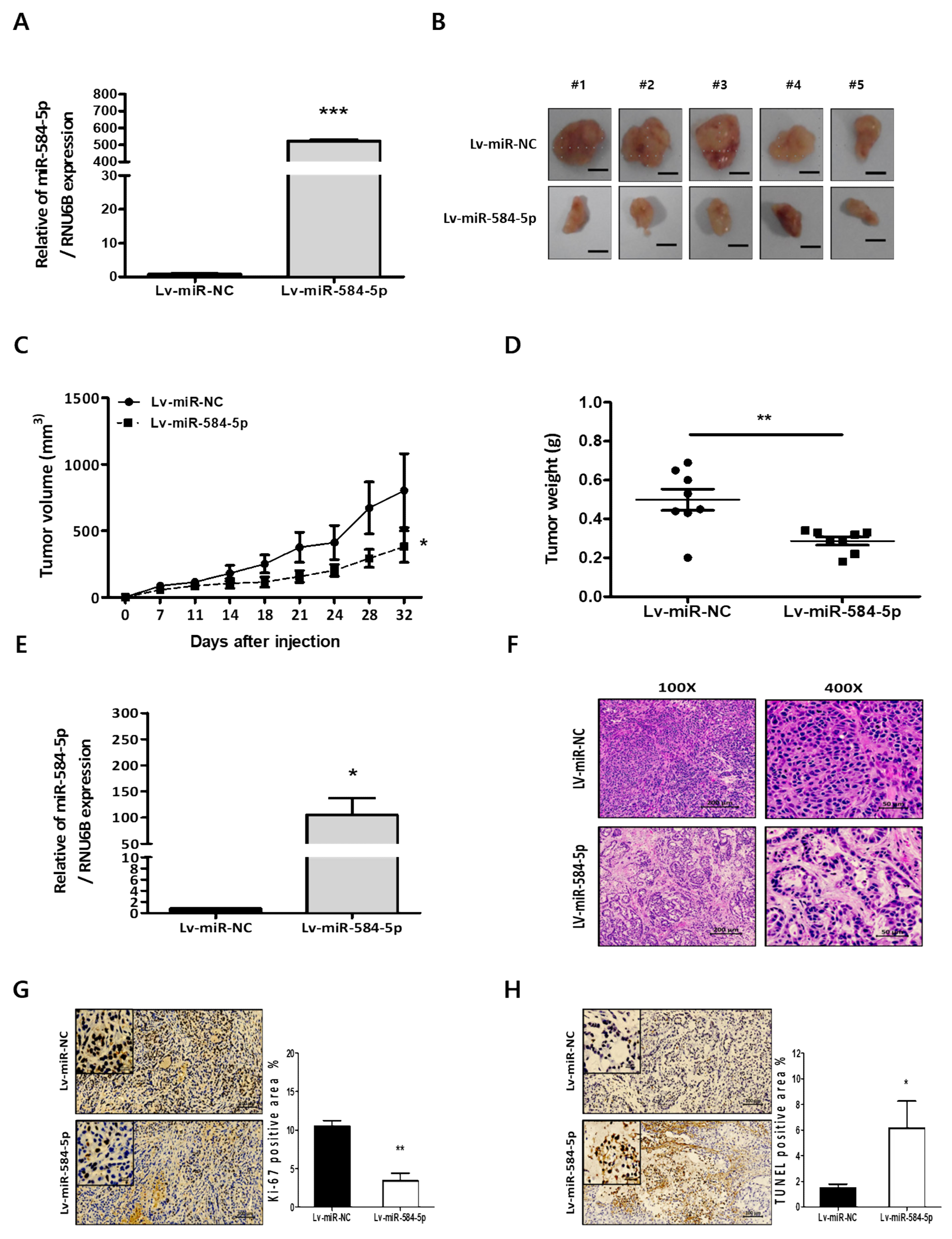
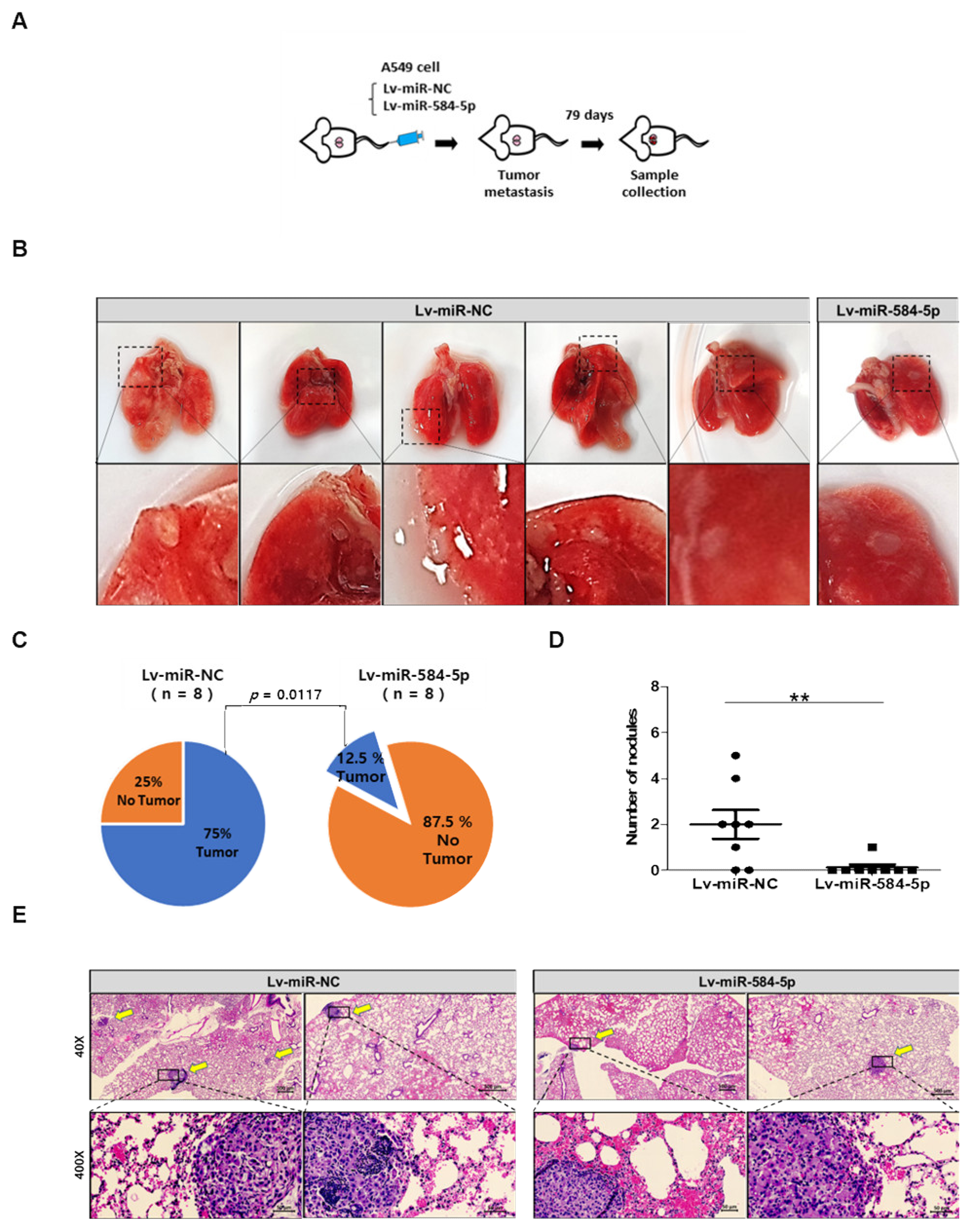
3.5. miR-584-5p Inhibits Tumor Growth and Lung Metastasis Abilities of NSCLC Cells In Vivo
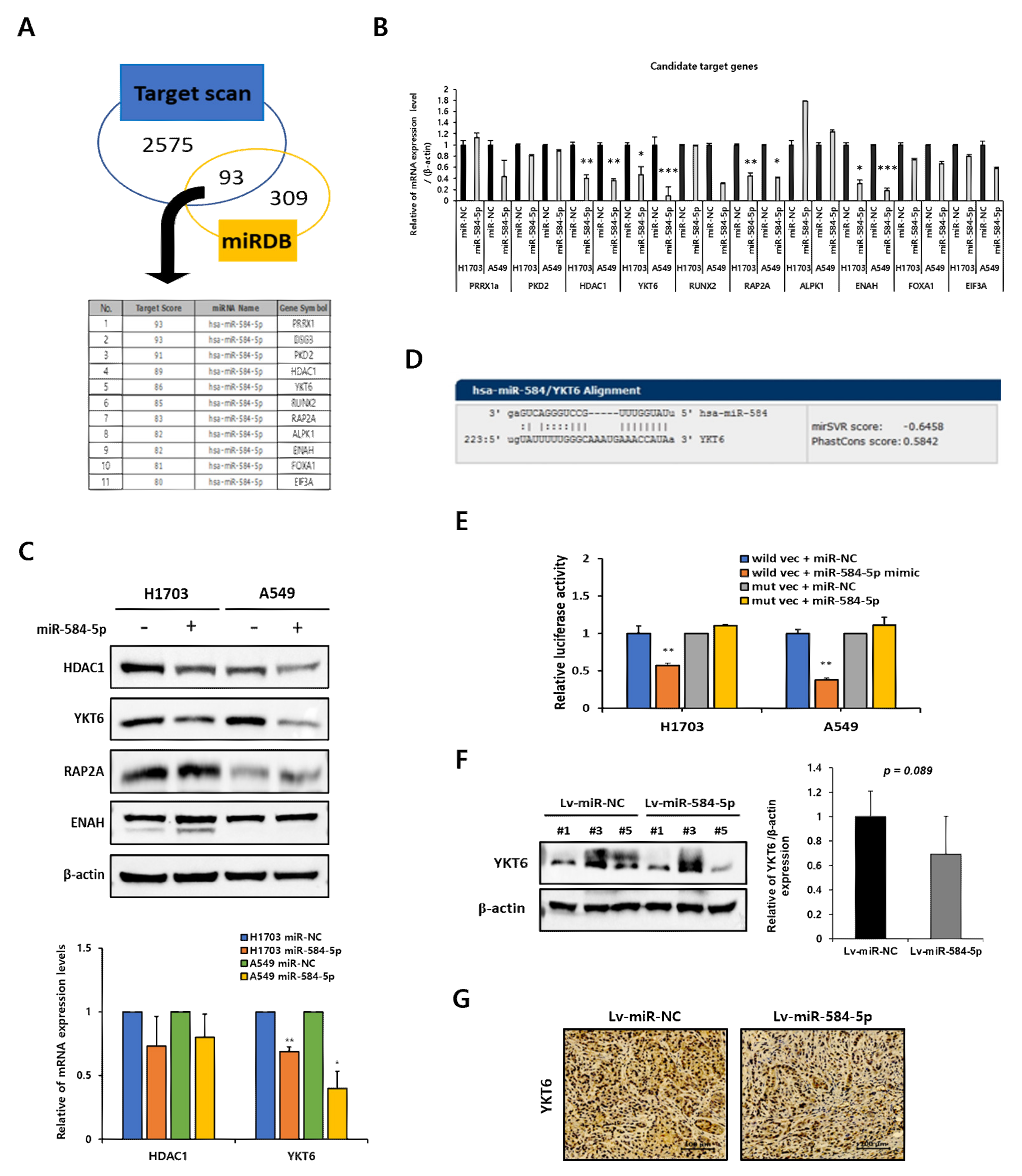
3.6. YKT6 Is a Direct Target of miR-584-5p
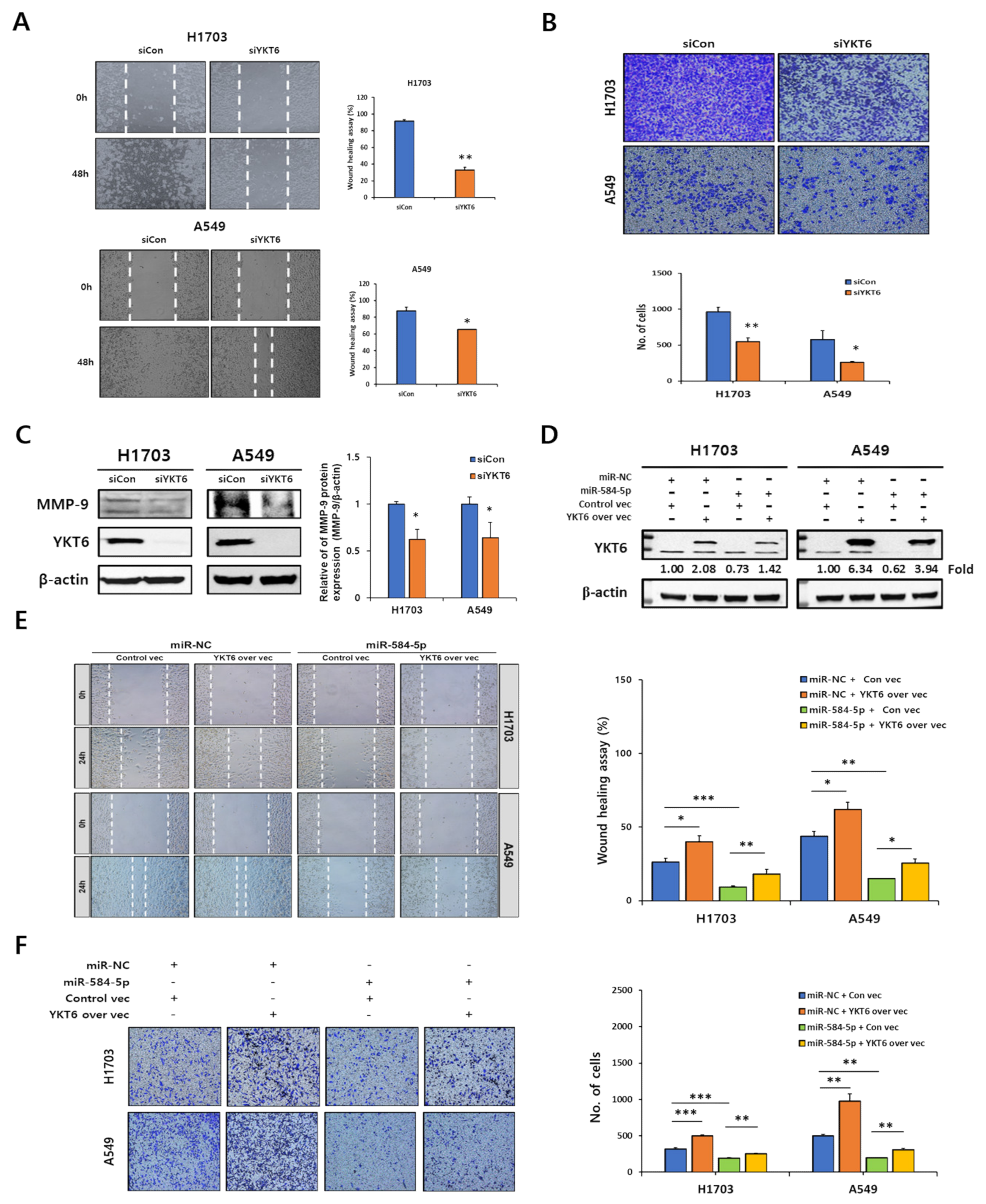
3.7. YKT6 Regulates Migration and Invasion in Smoking-Related NSCLC Cell Lines
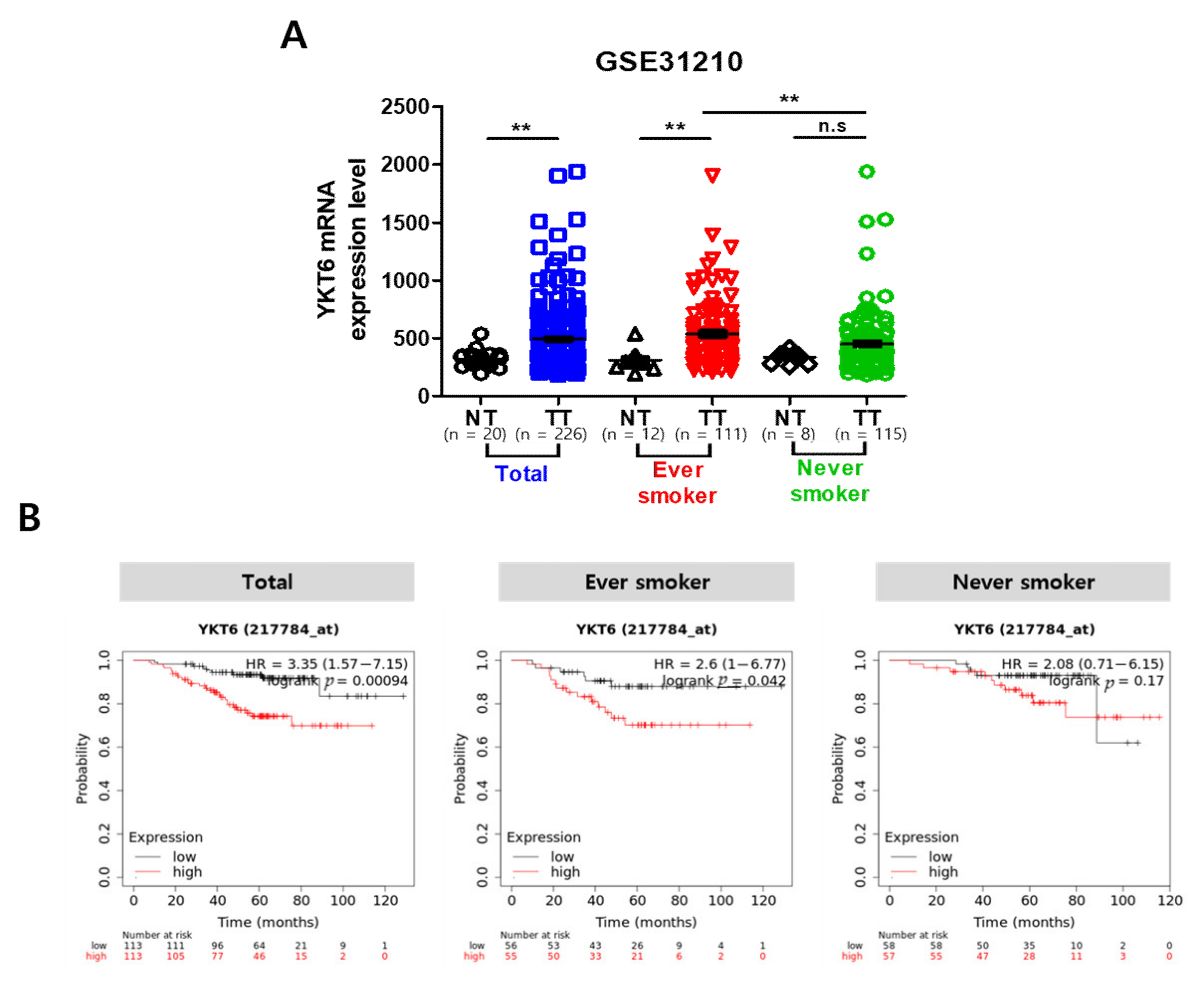
3.8. YKT6 Expression Level Is Associated with Survival Rate in Smoking-Related NSCLC Patients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Khuder, S.A. Effect of cigarette smoking on major histological types of lung cancer: A meta-analysis. Lung Cancer 2001, 31, 139–148. [Google Scholar] [CrossRef]
- Nasim, F.; Sabath, B.F.; Eapen, G.A. Lung Cancer. Med. Clin. N. Am. 2019, 103, 463–473. [Google Scholar] [CrossRef]
- Pikor, L.A.; Ramnarine, V.R.; Lam, S.; Lam, W.L. Genetic alterations defining NSCLC subtypes and their therapeutic implications. Lung Cancer 2013, 82, 179–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inamura, K. Lung Cancer: Understanding Its Molecular Pathology and the 2015 WHO Classification. Front. Oncol. 2017, 7, 193. [Google Scholar] [CrossRef] [Green Version]
- Bakulski, K.M.; Dou, J.; Lin, N.; London, S.J.; Colacino, J.A. DNA methylation signature of smoking in lung cancer is enriched for exposure signatures in newborn and adult blood. Sci. Rep. 2019, 9, 4576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeon, S.Y.; Go, R.E.; Heo, J.R.; Kim, C.W.; Hwang, K.A.; Choi, K.C. Effects of cigarette smoke extracts on the progression and metastasis of human ovarian cancer cells via regulating epithelial-mesenchymal transition. Reprod. Toxicol. 2016, 65, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.K.; Hong, Y.; Kim, S.Y.; London, S.J.; Kim, W.J. DNA methylation and smoking in Korean adults: Epigenome-wide association study. Clin. Epigenetics 2016, 8, 103. [Google Scholar] [CrossRef] [Green Version]
- Peluso, M.E.; Munnia, A.; Bollati, V.; Srivatanakul, P.; Jedpiyawongse, A.; Sangrajrang, S.; Ceppi, M.; Giese, R.W.; Boffetta, P.; Baccarelli, A.A. Aberrant methylation of hypermethylated-in-cancer-1 and exocyclic DNA adducts in tobacco smokers. Toxicol. Sci. 2014, 137, 47–54. [Google Scholar] [CrossRef] [Green Version]
- Heller, G.; Altenberger, C.; Steiner, I.; Topakian, T.; Ziegler, B.; Tomasich, E.; Lang, G.; End-Pfützenreuter, A.; Zehetmayer, S.; Döme, B.; et al. DNA methylation of microRNA-coding genes in non-small-cell lung cancer patients. J. Pathol. 2018, 245, 387–398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kulis, M.; Esteller, M. DNA methylation and cancer. Adv. Genet. 2010, 70, 27–56. [Google Scholar] [CrossRef] [PubMed]
- Shui, I.M.; Wong, C.J.; Zhao, S.; Kolb, S.; Ebot, E.M.; Geybels, M.S.; Rubicz, R.; Wright, J.L.; Lin, D.W.; Klotzle, B.; et al. Prostate tumor DNA methylation is associated with cigarette smoking and adverse prostate cancer outcomes. Cancer 2016, 122, 2168–2177. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Qi, W.; Sun, L.; Zhou, H.; Zhou, B.; Hu, Y. DNA methylation analysis of selected genes for the detection of early-stage lung cancer using circulating cell-free DNA. Adv. Clin. Exp. Med. 2019, 28, 355–360. [Google Scholar] [CrossRef] [Green Version]
- Pan, Y.; Liu, G.; Zhou, F.; Su, B.; Li, Y. DNA methylation profiles in cancer diagnosis and therapeutics. Clin. Exp. Med. 2018, 18, 1–14. [Google Scholar] [CrossRef]
- Yan, F.; Shen, N.; Pang, J.; Xie, D.; Deng, B.; Molina, J.R.; Yang, P.; Liu, S. Restoration of miR-101 suppresses lung tumorigenesis through inhibition of DNMT3a-dependent DNA methylation. Cell Death Dis. 2014, 5, e1413. [Google Scholar] [CrossRef] [Green Version]
- Iqbal, M.A.; Arora, S.; Prakasam, G.; Calin, G.A.; Syed, M.A. MicroRNA in lung cancer: Role, mechanisms, pathways and therapeutic relevance. Mol. Asp. Med. 2019, 70, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.L.; Tsai, Y.M.; Lien, C.T.; Kuo, P.L.; Hung, A.J. The Roles of MicroRNA in Lung Cancer. Int. J. Mol. Sci. 2019, 20, 1611. [Google Scholar] [CrossRef] [Green Version]
- Petrovic, N.; Ergün, S.; Isenovic, E.R. Levels of MicroRNA Heterogeneity in Cancer Biology. Mol. Diagn. 2017, 21, 511–523. [Google Scholar] [CrossRef]
- Lacroix, L.; Feng, G.; Lotan, R. Identification of genes expressed differentially in an in vitro human lung carcinogenesis model. Cancer Biol. 2006, 5, 665–673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klein-Szanto, A.J.; Iizasa, T.; Momiki, S.; Garcia-Palazzo, I.; Caamano, J.; Metcalf, R.; Welsh, J.; Harris, C.C. A tobacco-specific N-nitrosamine or cigarette smoke condensate causes neoplastic transformation of xenotransplanted human bronchial epithelial cells. Proc. Natl. Acad. Sci. USA 1992, 89, 6693–6697. [Google Scholar] [CrossRef] [Green Version]
- Reddel, R.R.; Ke, Y.; Gerwin, B.I.; McMenamin, M.G.; Lechner, J.F.; Su, R.T.; Brash, D.E.; Park, J.B.; Rhim, J.S.; Harris, C.C. Transformation of human bronchial epithelial cells by infection with SV40 or adenovirus-12 SV40 hybrid virus, or transfection via strontium phosphate coprecipitation with a plasmid containing SV40 early region genes. Cancer Res. 1988, 48, 1904–1909. [Google Scholar] [PubMed]
- Whang, Y.M.; Jo, U.; Sung, J.S.; Ju, H.J.; Kim, H.K.; Park, K.H.; Lee, J.W.; Koh, I.S.; Kim, Y.H. Wnt5a is associated with cigarette smoke-related lung carcinogenesis via protein kinase C. PLoS ONE 2013, 8, e53012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, P.; Peng, R.; Peng, H.; Yao, L.; Sun, Y.; Wen, L.; Wu, T.; Zhou, J.; Zhang, Z. MiR-451 suppresses cell proliferation and metastasis in A549 lung cancer cells. Mol. Biotechnol. 2015, 57, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Rajasinghe, L.D.; Pindiprolu, R.H.; Gupta, S.V. Delta-tocotrienol inhibits non-small-cell lung cancer cell invasion via the inhibition of NF-κB, uPA activator, and MMP-9. Onco Targets Ther. 2018, 11, 4301–4314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Wang, H.; Wang, C. Persistence of smoking induced non-small cell lung carcinogenesis by decreasing ERBB pathway-related microRNA expression. Thorac. Cancer 2019, 10, 890–897. [Google Scholar] [CrossRef] [Green Version]
- Tessema, M.; Yingling, C.M.; Picchi, M.A.; Wu, G.; Ryba, T.; Lin, Y.; Bungum, A.O.; Edell, E.S.; Spira, A.; Belinsky, S.A. ANK1 Methylation regulates expression of MicroRNA-486-5p and discriminates lung tumors by histology and smoking status. Cancer Lett. 2017, 410, 191–200. [Google Scholar] [CrossRef]
- Wang, B.; Liu, Y.; Luo, F.; Xu, Y.; Qin, Y.; Lu, X.; Xu, W.; Shi, L.; Liu, Q.; Xiang, Q. Epigenetic silencing of microRNA-218 via EZH2-mediated H3K27 trimethylation is involved in malignant transformation of HBE cells induced by cigarette smoke extract. Arch. Toxicol. 2016, 90, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Fujii, T.; Shimada, K.; Nakai, T.; Ohbayashi, C. MicroRNAs in Smoking-Related Carcinogenesis: Biomarkers, Functions, and Therapy. J. Clin. Med. 2018, 7, 98. [Google Scholar] [CrossRef] [Green Version]
- Peng, H.Y.; Hsiao, J.R.; Chou, S.T.; Hsu, Y.M.; Wu, G.H.; Shieh, Y.S.; Shiah, S.G. MiR-944/CISH mediated inflammation via STAT3 is involved in oral cancer malignance by cigarette smoking. Neoplasia 2020, 22, 554–565. [Google Scholar] [CrossRef]
- Dino, P.; D’Anna, C.; Sangiorgi, C.; Di Sano, C.; Di Vincenzo, S.; Ferraro, M.; Pace, E. Cigarette smoke extract modulates E-Cadherin, Claudin-1 and miR-21 and promotes cancer invasiveness in human colorectal adenocarcinoma cells. Toxicol. Lett. 2019, 317, 102–109. [Google Scholar] [CrossRef]
- Zhang, Y.; Pan, T.; Zhong, X.; Cheng, C. Nicotine upregulates microRNA-21 and promotes TGF-β-dependent epithelial-mesenchymal transition of esophageal cancer cells. Tumour Biol. 2014, 35, 7063–7072. [Google Scholar] [CrossRef]
- Ganji, S.M.; Saidijam, M.; Amini, R.; Mousavi-Bahar, S.H.; Shabab, N.; Seyedabadi, S.; Mahdavinezhad, A. Evaluation of MicroRNA-99a and MicroRNA-205 Expression Levels in Bladder Cancer. Int. J. Mol. Cell. Med. 2017, 6, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Wu, J.; Li, Y.; Li, X.; Yang, T.; Yang, Q.; Jiang, Y. Deregulation of serum microRNA expression is associated with cigarette smoking and lung cancer. Biomed Res. Int. 2014, 2014, 364316. [Google Scholar] [CrossRef]
- Xi, S.; Xu, H.; Shan, J.; Tao, Y.; Hong, J.A.; Inchauste, S.; Zhang, M.; Kunst, T.F.; Mercedes, L.; Schrump, D.S. Cigarette smoke mediates epigenetic repression of miR-487b during pulmonary carcinogenesis. J. Clin. Investig. 2013, 123, 1241–1261. [Google Scholar] [CrossRef]
- Dalle-Donne, I.; Colombo, G.; Gornati, R.; Garavaglia, M.L.; Portinaro, N.; Giustarini, D.; Bernardini, G.; Rossi, R.; Milzani, A. Protein Carbonylation in Human Smokers and Mammalian Models of Exposure to Cigarette Smoke: Focus on Redox Proteomic Studies. Antioxid. Redox Signal. 2017, 26, 406–426. [Google Scholar] [CrossRef]
- Kim, Y.H.; An, Y.J.; Jo, S.; Lee, S.H.; Lee, S.J.; Choi, S.J.; Lee, K. Comparison of volatile organic compounds between cigarette smoke condensate (CSC) and extract (CSE) samples. Environ. Health Toxicol. 2018, 33, e2018010–e2018012. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Wang, J.; Xu, Z.; Lu, Y.; Wu, X.; Zhuo, C.; Tan, C.; Tang, Q.; Pu, J. miR-584-5p regulates hepatocellular carcinoma cell migration and invasion through targeting KCNE2. Mol. Genet. Genom. Med. 2019, 7, e702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdelfattah, N.; Rajamanickam, S.; Panneerdoss, S.; Timilsina, S.; Yadav, P.; Onyeagucha, B.C.; Garcia, M.; Vadlamudi, R.; Chen, Y.; Brenner, A.; et al. MiR-584-5p potentiates vincristine and radiation response by inducing spindle defects and DNA damage in medulloblastoma. Nat. Commun. 2018, 9, 4541. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Li, Z.; Wei, S.; Wang, W.; Chen, Z.; Zhang, L.; Chen, L.; Li, B.; Sun, G.; Xu, J.; et al. Overexpression of miR-584-5p inhibits proliferation and induces apoptosis by targeting WW domain-containing E3 ubiquitin protein ligase 1 in gastric cancer. J. Exp. Clin. Cancer Res. 2017, 36, 59. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Wen, W.; Shan, X.; Zhu, W.; Xu, J.; Guo, R.; Cheng, W.; Wang, F.; Qi, L.W.; Chen, Y.; et al. A six-microRNA panel in plasma was identified as a potential biomarker for lung adenocarcinoma diagnosis. Oncotarget 2017, 8, 6513–6525. [Google Scholar] [CrossRef] [PubMed]
- Guo, T.; Zheng, C.; Wang, Z.; Zheng, X. miR-584-5p regulates migration and invasion in non-small cell lung cancer cell lines through regulation of MMP-14. Mol. Med. Rep. 2019, 19, 1747–1752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poli, E.; Zhang, J.; Nwachukwu, C.; Zheng, Y.; Adedokun, B.; Olopade, O.I.; Han, Y.J. Molecular Subtype-Specific Expression of MicroRNA-29c in Breast Cancer Is Associated with CpG Dinucleotide Methylation of the Promoter. PLoS ONE 2015, 10, e0142224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, P.; Wu, Q.; Yu, J.; Rao, Y.; Kou, Z.; Fang, G.; Shi, X.; Liu, W.; Han, H. A Systematic Way to Infer the Regulation Relations of miRNAs on Target Genes and Critical miRNAs in Cancers. Front. Genet. 2020, 11, 278. [Google Scholar] [CrossRef] [PubMed]
- Naydenov, N.G.; Joshi, S.; Feygin, A.; Saini, S.; Litovchick, L.; Ivanov, A.I. A membrane fusion protein, Ykt6, regulates epithelial cell migration via microRNA-mediated suppression of Junctional Adhesion Molecule A. Cell Cycle 2018, 17, 1812–1831. [Google Scholar] [CrossRef] [PubMed]
- Ooe, A.; Kato, K.; Noguchi, S. Possible involvement of CCT5, RGS3, and YKT6 genes up-regulated in p53-mutated tumors in resistance to docetaxel in human breast cancers. Breast Cancer Res. Treat. 2007, 101, 305–315. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Martinez, M.; Navarro, A.; Marrades, R.M.; Viñolas, N.; Santasusagna, S.; Muñoz, C.; Ramírez, J.; Molins, L.; Monzo, M. YKT6 expression, exosome release, and survival in non-small cell lung cancer. Oncotarget 2016, 7, 51515–51524. [Google Scholar] [CrossRef] [PubMed]
- Fukasawa, M.; Varlamov, O.; Eng, W.S.; Söllner, T.H.; Rothman, J.E. Localization and activity of the SNARE Ykt6 determined by its regulatory domain and palmitoylation. Proc. Natl. Acad. Sci. USA 2004, 101, 4815–4820. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Cell Lines | Histological Stage a | CSC Exposure | Tumorigenicity b | Histology |
|---|---|---|---|---|
| 1799 | Immortalized | - | - | Adenocarcinoma |
| 1198 | Transformed | + | - | Adenocarcinoma |
| 1170I | Tumorigenic | + | + | Adenocarcinoma |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.B.; Park, Y.S.; Sung, J.S.; Lee, J.W.; Kim, B.; Kim, Y.H. Tumor Suppressor miR-584-5p Inhibits Migration and Invasion in Smoking Related Non-Small Cell Lung Cancer Cells by Targeting YKT6. Cancers 2021, 13, 1159. https://doi.org/10.3390/cancers13051159
Lee SB, Park YS, Sung JS, Lee JW, Kim B, Kim YH. Tumor Suppressor miR-584-5p Inhibits Migration and Invasion in Smoking Related Non-Small Cell Lung Cancer Cells by Targeting YKT6. Cancers. 2021; 13(5):1159. https://doi.org/10.3390/cancers13051159
Chicago/Turabian StyleLee, Saet Byeol, Young Soo Park, Jae Sook Sung, Jong Won Lee, Boyeon Kim, and Yeul Hong Kim. 2021. "Tumor Suppressor miR-584-5p Inhibits Migration and Invasion in Smoking Related Non-Small Cell Lung Cancer Cells by Targeting YKT6" Cancers 13, no. 5: 1159. https://doi.org/10.3390/cancers13051159