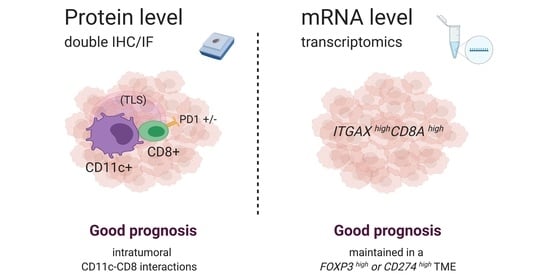
CD11c-CD8 Spatial Cross Presentation: A Novel Approach to Link Immune Surveillance and Patient Survival in Soft Tissue Sarcoma
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Inclusion and Follow Up
2.2. Flow Cytometry and Cell Sorting
2.3. IHC, Histopathological Scoring, and Digital Image Analysis
2.4. OpalTM Multiplexing and Digital Image Analysis
2.5. Statistical Analysis and Transcriptomics
3. Results
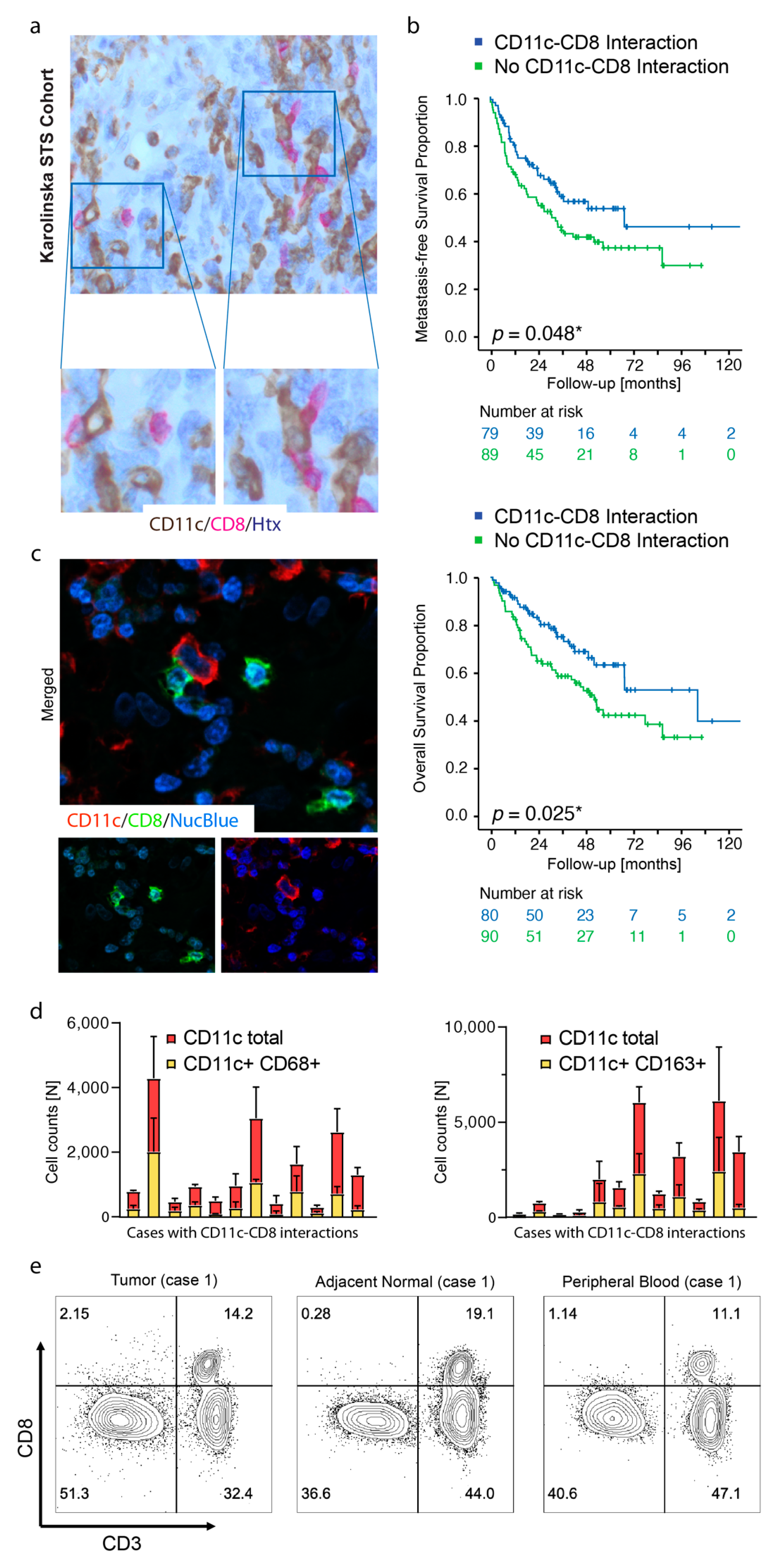
3.1. Spatial Cross Presentation in Human STS Is Prognostic for Patient Survival
3.2. TLSs Are Found in Smaller Tumors and Display High Density of CD11c+ Cells and CD8+ Cells
3.3. Foxp3+ Cells Reside in TA-TLSs
3.4. ITGAX Expression Correlates with HLA Genes, Whereas CD8A Correlates with CD3 Chains and CD8B
3.5. High Gene Expression Levels of ITGAX Together with CD8A Are Prognostic by Transcriptomics, Irrespective of FOXP3 TME Levels, in Human STS
3.6. The Prognostic Value of the ITGAXhighCD8Ahigh Gene Signature Is Maintained in a CD274 (PD-L1)-Rich TME
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- von Mehren, M.; Randall, R.L.; Benjamin, R.S.; Boles, S.; Bui, M.M.; Conrad, E.U., 3rd; Ganjoo, K.N.; George, S.; Gonzalez, R.J.; Heslin, M.J.; et al. Soft Tissue Sarcoma, Version 2.2016, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2016, 14, 758–786. [Google Scholar] [CrossRef] [Green Version]
- ESMO/European Sarcoma Network Working Group. Soft tissue and visceral sarcomas: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2014, 25 (Suppl. 3), iii102–iii112. [Google Scholar] [CrossRef] [PubMed]
- Lahat, G.; Tuvin, D.; Wei, C.; Anaya, D.A.; Bekele, B.N.; Lazar, A.J.; Pisters, P.W.; Lev, D.; Pollock, R.E. New perspectives for staging and prognosis in soft tissue sarcoma. Ann Surg Oncol. 2008, 15, 2739–2748. [Google Scholar] [CrossRef]
- Yoon, S.S. The New American Joint Commission on Cancer Staging System for Soft Tissue Sarcomas: Splitting versus Lumping. Ann. Surg. Oncol. 2018, 25, 1101–1102. [Google Scholar] [CrossRef] [Green Version]
- Guillou, L.; Coindre, J.M.; Bonichon, F.; Nguyen, B.B.; Terrier, P.; Collin, F.; Vilain, M.O.; Mandard, A.M.; Le Doussal, V.; Leroux, A.; et al. Comparative study of the National Cancer Institute and French Federation of Cancer Centers Sarcoma Group grading systems in a population of 410 adult patients with soft tissue sarcoma. J. Clin. Oncol. 1997, 15, 350–362. [Google Scholar] [CrossRef]
- Tanaka, K.; Ozaki, T. New TNM classification (AJCC eighth edition) of bone and soft tissue sarcomas: JCOG Bone and Soft Tissue Tumor Study Group. Jpn. J. Clin. Oncol. 2018, 49, 103–107. [Google Scholar] [CrossRef]
- Ottaiano, A.; De Chiara, A.; Fazioli, F.; Talamanca, A.A.; Mori, S.; Botti, G.; Milano, A.; Apice, G. Biological prognostic factors in adult soft tissue sarcomas. Anticancer Res. 2005, 25, 4519–4526. [Google Scholar] [PubMed]
- Lin, X.; Davion, S.; Bertsch, E.C.; Omar, I.; Nayar, R.; Laskin, W.B. Federation Nationale des Centers de Lutte Contre le Cancer grading of soft tissue sarcomas on needle core biopsies using surrogate markers. Hum. Pathol. 2016, 56, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Missiaglia, E.; Williamson, D.; Chisholm, J.; Wirapati, P.; Pierron, G.; Petel, F.; Concordet, J.P.; Thway, K.; Oberlin, O.; Pritchard-Jones, K.; et al. PAX3/FOXO1 fusion gene status is the key prognostic molecular marker in rhabdomyosarcoma and significantly improves current risk stratification. J. Clin. Oncol. 2012, 30, 1670–1677. [Google Scholar] [CrossRef]
- Rutkowski, P.; Van Glabbeke, M.; Rankin, C.J.; Ruka, W.; Rubin, B.P.; Debiec-Rychter, M.; Lazar, A.; Gelderblom, H.; Sciot, R.; Lopez-Terrada, D.; et al. Imatinib mesylate in advanced dermatofibrosarcoma protuberans: Pooled analysis of two phase II clinical trials. J. Clin. Oncol. 2010, 28, 1772–1779. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.T.; Tan, K.Y. Current research and treatment for gastrointestinal stromal tumors. World J. Gastroenterol. 2017, 23, 4856–4866. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. Cancer Genome Atlas Research, Comprehensive and Integrated Genomic Characterization of Adult Soft Tissue Sarcomas. Cell 2017, 171, 950–965.e28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonneville, R.; Krook, M.A.; Kautto, E.A.; Miya, J.; Wing, M.R.; Chen, H.Z.; Reeser, J.W.; Yu, L.; Roychowdhury, S. Landscape of Microsatellite Instability Across 39 Cancer Types. JCO Precis. Oncol. 2017, 2017. [Google Scholar] [CrossRef] [PubMed]
- Widemann, B.C.; Italiano, A. Biology and Management of Undifferentiated Pleomorphic Sarcoma, Myxofibrosarcoma, and Malignant Peripheral Nerve Sheath Tumors: State of the Art and Perspectives. J. Clin. Oncol. 2018, 36, 160–167. [Google Scholar] [CrossRef] [PubMed]
- Yoshimoto, M.; Yamada, Y.; Ishihara, S.; Kohashi, K.; Toda, Y.; Ito, Y.; Yamamoto, H.; Furue, M.; Nakashima, Y.; Oda, Y. Comparative Study of Myxofibrosarcoma With Undifferentiated Pleomorphic Sarcoma: Histopathologic and Clinicopathologic Review. Am. J. Surg. Pathol. 2020, 44, 87–97. [Google Scholar] [CrossRef]
- Marino-Enriquez, A.; Bovee, J.V. Molecular Pathogenesis and Diagnostic, Prognostic and Predictive Molecular Markers in Sarcoma. Surg. Pathol. Clin. 2016, 9, 457–473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sorbye, S.W.; Kilvaer, T.; Valkov, A.; Donnem, T.; Smeland, E.; Al-Shibli, K.; Bremnes, R.M.; Busund, L.T. High expression of CD20+ lymphocytes in soft tissue sarcomas is a positive prognostic indicator. Oncoimmunology 2012, 1, 75–77. [Google Scholar] [CrossRef] [Green Version]
- Sorbye, S.W.; Kilvaer, T.; Valkov, A.; Donnem, T.; Smeland, E.; Al-Shibli, K.; Bremnes, R.M.; Busund, L.T. Prognostic impact of lymphocytes in soft tissue sarcomas. PLoS ONE 2011, 6, e14611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsagozis, P.; Augsten, M.; Zhang, Y.; Li, T.; Hesla, A.; Bergh, J.; Haglund, F.; Tobin, N.P.; Ehnman, M. An immunosuppressive macrophage profile attenuates the prognostic impact of CD20-positive B cells in human soft tissue sarcoma. Cancer Immunol. Immunother. 2019, 68, 927–936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sorbye, S.W.; Kilvaer, T.K.; Valkov, A.; Donnem, T.; Smeland, E.; Al-Shibli, K.; Bremnes, R.M.; Busund, L.T. Prognostic impact of peritumoral lymphocyte infiltration in soft tissue sarcomas. BMC Clin. Pathol. 2012, 12, 5. [Google Scholar] [CrossRef] [Green Version]
- Petitprez, F.; de Reynies, A.; Keung, E.Z.; Chen, T.W.; Sun, C.M.; Calderaro, J.; Jeng, Y.M.; Hsiao, L.P.; Lacroix, L.; Bougouin, A.; et al. B cells are associated with survival and immunotherapy response in sarcoma. Nature 2020, 577, 556–560. [Google Scholar] [CrossRef]
- Helmink, B.A.; Reddy, S.M.; Gao, J.; Zhang, S.; Basar, R.; Thakur, R.; Yizhak, K.; Sade-Feldman, M.; Blando, J.; Han, G.; et al. B cells and tertiary lymphoid structures promote immunotherapy response. Nature 2020, 577, 549–555. [Google Scholar] [CrossRef]
- Sautes-Fridman, C.; Petitprez, F.; Calderaro, J.; Fridman, W.H. Tertiary lymphoid structures in the era of cancer immunotherapy. Nat. Rev. Cancer 2019, 19, 307–325. [Google Scholar] [CrossRef]
- Joshi, N.S.; Akama-Garren, E.H.; Lu, Y.; Lee, D.Y.; Chang, G.P.; Li, A.; DuPage, M.; Tammela, T.; Kerper, N.R.; Farago, A.F.; et al. Regulatory T Cells in Tumor-Associated Tertiary Lymphoid Structures Suppress Anti-tumor T Cell Responses. Immunity 2015, 43, 579–590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheng, J.; Chen, Q.; Soncin, I.; Ng, S.L.; Karjalainen, K.; Ruedl, C. A Discrete Subset of Monocyte-Derived Cells among Typical Conventional Type 2 Dendritic Cells Can Efficiently Cross-Present. Cell Rep. 2017, 21, 1203–1214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osugi, Y.; Vuckovic, S.; Hart, D.N. Myeloid blood CD11c(+) dendritic cells and monocyte-derived dendritic cells differ in their ability to stimulate T lymphocytes. Blood 2002, 100, 2858–2866. [Google Scholar] [CrossRef] [Green Version]
- Wykes, M.; MacPherson, G. Dendritic cell-B-cell interaction: Dendritic cells provide B cells with CD40-independent proliferation signals and CD40-dependent survival signals. Immunology 2000, 100, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Burgdorf, S.; Scholz, C.; Kautz, A.; Tampe, R.; Kurts, C. Spatial and mechanistic separation of cross-presentation and endogenous antigen presentation. Nat. Immunol. 2008, 9, 558–566. [Google Scholar] [CrossRef]
- Abbas, A.K.; Lichtman, A.H.; Pillai, S. Cellular and Molecular Immunology, 8th ed.; Elsevier Saunders: Philadelphia, PA, USA, 2015; p. viii, 535. [Google Scholar]
- Asano, K.; Nabeyama, A.; Miyake, Y.; Qiu, C.H.; Kurita, A.; Tomura, M.; Kanagawa, O.; Fujii, S.; Tanaka, M. CD169-positive macrophages dominate antitumor immunity by crosspresenting dead cell-associated antigens. Immunity 2011, 34, 85–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanchez-Paulete, A.R.; Teijeira, A.; Cueto, F.J.; Garasa, S.; Perez-Gracia, J.L.; Sanchez-Arraez, A.; Sancho, D.; Melero, I. Antigen cross-presentation and T-cell cross-priming in cancer immunology and immunotherapy. Ann. Oncol. 2017, 28 (Suppl. 12), xii74. [Google Scholar] [CrossRef]
- Gardner, A.; Ruffell, B. Dendritic Cells and Cancer Immunity. Trends Immunol. 2016, 37, 855–865. [Google Scholar] [CrossRef] [Green Version]
- Engelhardt, J.J.; Boldajipour, B.; Beemiller, P.; Pandurangi, P.; Sorensen, C.; Werb, Z.; Egeblad, M.; Krummel, M.F. Marginating dendritic cells of the tumor microenvironment cross-present tumor antigens and stably engage tumor-specific T cells. Cancer Cell. 2012, 21, 402–417. [Google Scholar] [CrossRef] [Green Version]
- Lindsay, R.W.; Darrah, P.A.; Quinn, K.M.; Wille-Reece, U.; Mattei, L.M.; Iwasaki, A.; Kasturi, S.P.; Pulendran, B.; Gall, J.G.; Spies, A.G.; et al. CD8+ T cell responses following replication-defective adenovirus serotype 5 immunization are dependent on CD11c+ dendritic cells but show redundancy in their requirement of TLR and nucleotide-binding oligomerization domain-like receptor signaling. J. Immunol. 2010, 185, 1513–1521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salaun, B.; de Saint-Vis, B.; Clair-Moninot, V.; Pin, J.J.; Barthelemy-Dubois, C.; Kissenpfennig, A.; Peronne, C.; Bates, E.; Mattei, M.G.; Lebecque, S. Cloning and characterization of the mouse homologue of the human dendritic cell maturation marker CD208/DC-LAMP. Eur. J. Immunol. 2003, 33, 2619–2629. [Google Scholar] [CrossRef]
- de Saint-Vis, B.; Vincent, J.; Vandenabeele, S.; Vanbervliet, B.; Pin, J.J.; Ait-Yahia, S.; Patel, S.; Mattei, M.G.; Banchereau, J.; Zurawski, S.; et al. A novel lysosome-associated membrane glycoprotein, DC-LAMP, induced upon DC maturation, is transiently expressed in MHC class II compartment. Immunity 1998, 9, 325–336. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Lichtenberg, T.; Hoadley, K.A.; Poisson, L.M.; Lazar, A.J.; Cherniack, A.D.; Kovatich, A.J.; Benz, C.C.; Levine, D.A.; Lee, A.V.; et al. An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell 2018, 173, 400–416.e11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Neo, S.Y.; Chen, Z.; Cui, W.; Chen, Y.; Guo, M.; Wang, Y.; Xu, H.; Kurzay, A.; Alici, E.; et al. Thioredoxin activity confers resistance against oxidative stress in tumor-infiltrating NK cells. J. Clin. Investig. 2020, 130, 5508–5522. [Google Scholar] [CrossRef] [PubMed]
- Bankhead, P.; Loughrey, M.B.; Fernandez, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.; et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017, 7, 16878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal 2013, 6, pl1. [Google Scholar] [CrossRef] [Green Version]
- van Erp, A.E.M.; Versleijen-Jonkers, Y.M.H.; Hillebrandt-Roeffen, M.H.S.; van Houdt, L.; Gorris, M.A.J.; van Dam, L.S.; Mentzel, T.; Weidema, M.E.; Savci-Heijink, C.D.; Desar, I.M.E.; et al. Expression and clinical association of programmed cell death-1, programmed death-ligand-1 and CD8(+) lymphocytes in primary sarcomas is subtype dependent. Oncotarget 2017, 8, 71371–71384. [Google Scholar] [CrossRef] [Green Version]
- Tawbi, H.A.; Burgess, M.; Bolejack, V.; Van Tine, B.A.; Schuetze, S.M.; Hu, J.; D’Angelo, S.; Attia, S.; Riedel, R.F.; Priebat, D.A.; et al. Pembrolizumab in advanced soft-tissue sarcoma and bone sarcoma (SARC028): A multicentre, two-cohort, single-arm, open-label, phase 2 trial. Lancet Oncol. 2017, 18, 1493–1501. [Google Scholar] [CrossRef]
- Kamada, T.; Togashi, Y.; Tay, C.; Ha, D.; Sasaki, A.; Nakamura, Y.; Sato, E.; Fukuoka, S.; Tada, Y.; Tanaka, A.; et al. PD-1(+) regulatory T cells amplified by PD-1 blockade promote hyperprogression of cancer. Proc. Natl. Acad. Sci. USA 2019, 116, 9999–10008. [Google Scholar] [CrossRef] [Green Version]
- Chan, A.S.; Ng, V.Y.; Snider, J.; Kallen, M.E.; Miller, K.D. Hyperprogression of Liver Metastasis With Neoadjuvant Immunotherapy for Soft Tissue Sarcoma. Cureus 2020, 12, e8575. [Google Scholar]
- Barua, S.; Fang, P.; Sharma, A.; Fujimoto, J.; Wistuba, I.; Rao, A.U.K.; Lin, S.H. Spatial interaction of tumor cells and regulatory T cells correlates with survival in non-small cell lung cancer. Lung. Cancer 2018, 117, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Tseng, W.W.; Malu, S.; Zhang, M.; Chen, J.; Sim, G.C.; Wei, W.; Ingram, D.; Somaiah, N.; Lev, D.C.; Pollock, R.E.; et al. Analysis of the intratumoral adaptive immune response in well differentiated and dedifferentiated retroperitoneal liposarcoma. Sarcoma 2015, 2015, 547460. [Google Scholar] [CrossRef] [PubMed]
- Summers de Luca, L.; Gommerman, J.L. Fine-tuning of dendritic cell biology by the TNF superfamily. Nat. Rev. Immunol. 2012, 12, 339–351. [Google Scholar]
- Joffre, O.P.; Segura, E.; Savina, A.; Amigorena, S. Cross-presentation by dendritic cells. Nat. Rev. Immunol. 2012, 12, 557–569. [Google Scholar] [CrossRef]




| Cohort Characteristics | Undifferentiated Pleomorphic Sarcoma (UPS) | Liposarcoma | Other 1 |
|---|---|---|---|
| Patients | 115 | 48 | 14 |
| Age (years) | |||
| Mean/Median | 70/71 | 68/68 | 68/68 |
| Range (min–max) | 20–94 | 26–86 | 24–89 |
| Sex (N) | |||
| Male | 53 | 29 | 8 |
| Female | 62 | 19 | 6 |
| Grade (FNCLCC) | |||
| G1 | 9 | 1 | |
| G2 | 27 | 14 | 5 |
| G3 | 88 | 25 | 6 |
| GX/n.d. | 2 | ||
| Primary tumor size (cm) | |||
| Mean/Median | 9/8 | 18/19 | 9/7 |
| Range (min–max) | 1–35 | 3–38 | 2–28 |
| Location | |||
| Superficial | 48 | 5 | |
| Deep | 65 | 48 | 9 |
| Missing/n.d. | 2 | ||
| Resection margin | |||
| Intralesional | 19 | 21 * (42 **) | 3 |
| Marginal | 49 | 24 * (6 **) | 7 |
| Wide | 44 | 3 * (0 **) | 4 |
| Radical (amputation) | 1 | ||
| Missing/n.d. | 2 | ||
| Growth pattern | |||
| Infiltrative | 88 | 48 | 10 |
| Pushing border | 25 | 2 | |
| Missing/n.d. | 2 | 2 | |
| Adjuvant therapy | |||
| Chemotherapy | 13 | 2 | 2 |
| Radiation | 14 | 4 | 9 |
| Vascular involvement | |||
| Present | 14 | 1 | 3 |
| Absent | 98 | 47 | 11 |
| Missing/n.d. | 3 | ||
| Overall survival (OS) (%) | |||
| 12 months | 84 | 92 | 93 |
| 36 months | 58 | 82 | 79 |
| 60 months | 45 | 66 | 56 |
| Metastasis-free survival (MFS) (%) | |||
| 12 months | 71 | 81 | 79 |
| 36 months | 54 | 42 | 64 |
| 60 months | 47 | 37 | 54 |
| Clinicopathological Parameter | MFSHR (95% C.I.) | P (Cox) | OSHR (95% C.I.) | P (Cox) |
|---|---|---|---|---|
| Age (continuous, years) | 1.016 | 0.072 | 1.022 | 0.027 * |
| (0.999–1.034) | (1.002–1.042) | |||
| Sex (male) | 1.057 | 0.807 | 1.221 | 0.415 |
| (0.678–1.648) | (0.755–1.974) | |||
| Grade, FNCLCC (G3) | 2.412 | 0.001 ** | 3.339 | 0.000 *** |
| (1.426–4.081) | (1.783–6.255) | |||
| Size (cm, continuous) | 1.042 | 0.001 ** | 1.029 | 0.040 * |
| (1.017–1.067) | (1.001–1.057) | |||
| CD11c–CD8 interactions | 0.561 | 0.012 * | 0.479 | 0.004 ** |
| (present) | (0.357–0.881) | (0.290–0.792) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, Y.; Tsagkozis, P.; Papakonstantinou, A.; Tobin, N.P.; Gultekin, O.; Malmerfelt, A.; Ingelshed, K.; Neo, S.Y.; Lundquist, J.; Chaabane, W.; et al. CD11c-CD8 Spatial Cross Presentation: A Novel Approach to Link Immune Surveillance and Patient Survival in Soft Tissue Sarcoma. Cancers 2021, 13, 1175. https://doi.org/10.3390/cancers13051175
Su Y, Tsagkozis P, Papakonstantinou A, Tobin NP, Gultekin O, Malmerfelt A, Ingelshed K, Neo SY, Lundquist J, Chaabane W, et al. CD11c-CD8 Spatial Cross Presentation: A Novel Approach to Link Immune Surveillance and Patient Survival in Soft Tissue Sarcoma. Cancers. 2021; 13(5):1175. https://doi.org/10.3390/cancers13051175
Chicago/Turabian StyleSu, Yanhong, Panagiotis Tsagkozis, Andri Papakonstantinou, Nicholas P. Tobin, Okan Gultekin, Anna Malmerfelt, Katrine Ingelshed, Shi Yong Neo, Johanna Lundquist, Wiem Chaabane, and et al. 2021. "CD11c-CD8 Spatial Cross Presentation: A Novel Approach to Link Immune Surveillance and Patient Survival in Soft Tissue Sarcoma" Cancers 13, no. 5: 1175. https://doi.org/10.3390/cancers13051175
APA StyleSu, Y., Tsagkozis, P., Papakonstantinou, A., Tobin, N. P., Gultekin, O., Malmerfelt, A., Ingelshed, K., Neo, S. Y., Lundquist, J., Chaabane, W., Nisancioglu, M. H., Leiss, L. W., Östman, A., Bergh, J., Sedimbi, S., Lehti, K., Lundqvist, A., Stragliotto, C. L., Haglund, F., & Ehnman, M. (2021). CD11c-CD8 Spatial Cross Presentation: A Novel Approach to Link Immune Surveillance and Patient Survival in Soft Tissue Sarcoma. Cancers, 13(5), 1175. https://doi.org/10.3390/cancers13051175