Response and Toxicity to Cytarabine Therapy in Leukemia and Lymphoma: From Dose Puzzle to Pharmacogenomic Biomarkers
Simple Summary
Abstract
1. Introduction
2. Spectrum of Clinical Uses and Serious Toxicities of Cytarabine
2.1. Leukemia
2.2. Lymphoma
2.3. Toxicity Profiles
3. Clinical Pharmacology and Cellular Metabolism of Cytarabine
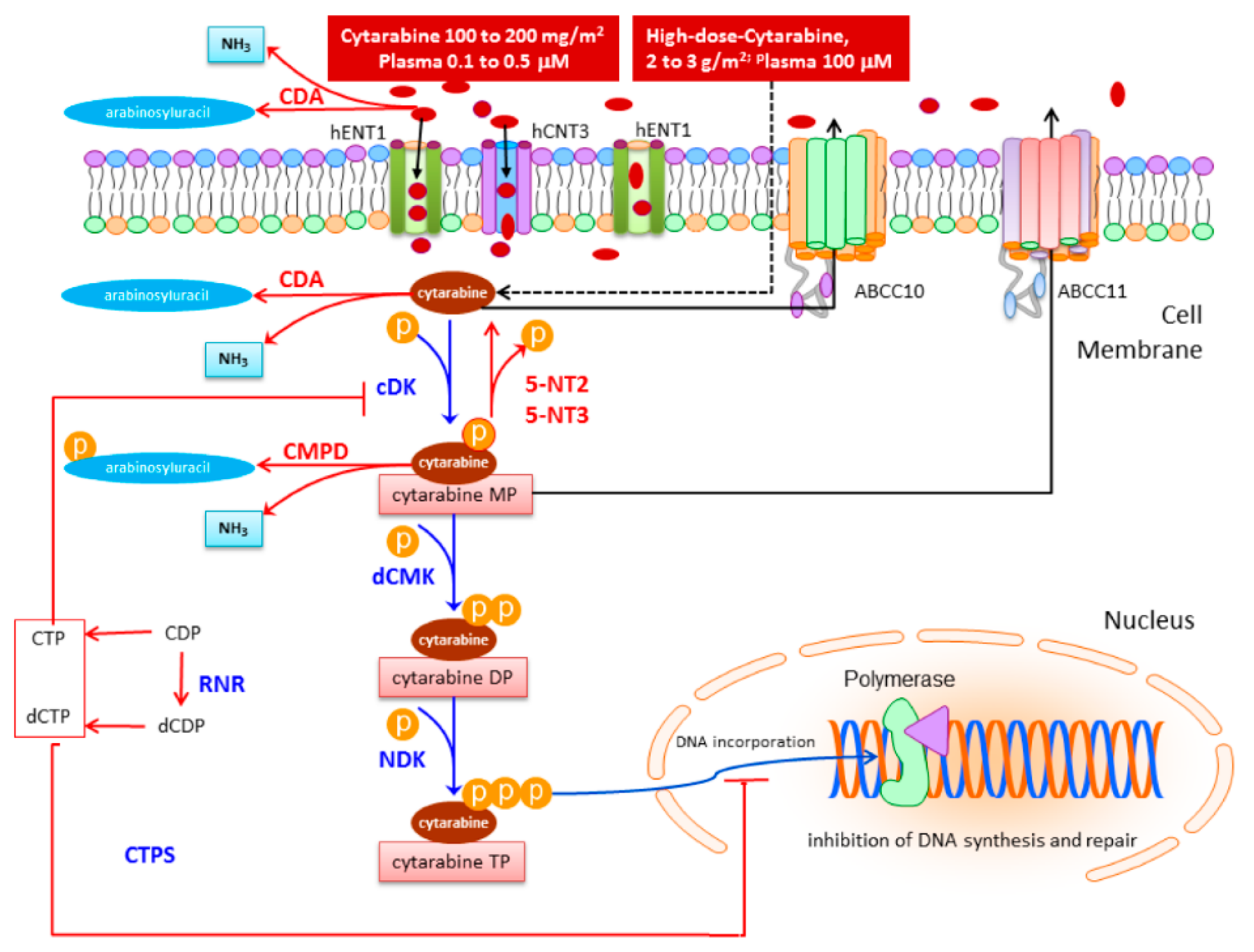
3.1. Drug Uptake
3.2. Drug Activation
3.3. Drug Deactivation
3.4. Mechanisms of Resistance within the Cytarabine Pathway
4. Genetic Determinants of Response and Toxicity to Cytarabine
4.1. Old and Novel Approaches to the Discovery of Pharmacogenomics Markers
4.2. Drug Uptake/Efflux
4.2.1. Genetic Variants of Cytarabine Transporters
4.2.2. Transporters and Response and Toxicity to Cytarabine
5. Drug Activation
5.1. Genetic Variants of Kinases
5.2. Kinases and Response and Toxicity to Cytarabine
5.3. Genetic Variants of Ribonucleotide Reductase and Cytidine 5′-Triphosphate Synthetase
5.4. Ribonucleotide Reductase, Cytidine 5′-Triphosphate Synthetase and Response and Toxicity to Cytarabine
6. Drug Deactivation
6.1. Genetic Variants of Deaminases and 5′-Nucleotidases
6.2. Deaminases, 5′-Nucleotidases and Response and Toxicity to Cytarabine
7. Drug Damage Repair
7.1. Genetic Variants in DNA Repair Genes
7.1.1. DNA Repair Genes and Response and Toxicity to Cytarabine
7.1.2. ‘Out of Pathway’ Genes
8. Consistency and Applicability of Pharmacogenomic Studies on Cytarabine
8.1. New Cytarabine Formulations
8.1.1. Elacytarabine
8.1.2. Sapacitabine
8.1.3. Pronucleotides
8.1.4. Liposomal Formulations
9. Conclusions and Future Outlook
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Burnett, A.; Wetzler, M.; Lowenberg, B. Therapeutic advances in acute myeloid leukemia. J. Clin. Oncol. 2011, 29, 487–494. [Google Scholar] [CrossRef]
- Aldoss, I.T.; Weisenburger, D.D.; Fu, K.; Chan, W.C.; Vose, J.M.; Bierman, P.J.; Bociek, R.G.; Armitage, J.O. Adult Burkitt lymphoma: Advances in diagnosis and treatment. Oncology 2008, 22, 1508–1517. [Google Scholar]
- Friedberg, J.W. Relapsed/refractory diffuse large B-cell lymphoma. Hematol. Am. Soc. Hematol. Educ. Program. 2011, 2011, 498–505. [Google Scholar] [CrossRef] [PubMed]
- Gokbuget, N.; Hoelzer, D. Treatment of adult acute lymphoblastic leukemia. Semin. Hematol. 2009, 46, 64–75. [Google Scholar] [CrossRef]
- Li, L.; Fridley, B.; Kalari, K.; Jenkins, G.; Batzler, A.; Safgren, S.; Hildebrandt, M.; Ames, M.; Schaid, D.; Wang, L. Gemcitabine and cytosine arabinoside cytotoxicity: Association with lymphoblastoid cell expression. Cancer Res. 2008, 68, 7050–7058. [Google Scholar] [CrossRef] [PubMed]
- Ferrara, F.; Pinto, A. Acute myeloid leukemia in the elderly: Current therapeutic results and perspectives for clinical research. Rev. Recent Clin. Trials 2007, 2, 33–41. [Google Scholar] [CrossRef]
- Gisselbrecht, C.; Glass, B.; Mounier, N.; Singh Gill, D.; Linch, D.C.; Trneny, M.; Bosly, A.; Ketterer, N.; Shpilberg, O.; Hagberg, H.; et al. Salvage regimens with autologous transplantation for relapsed large B-cell lymphoma in the rituximab era. J. Clin. Oncol. 2010, 28, 4184–4190. [Google Scholar] [CrossRef] [PubMed]
- Seyfarth, B.; Josting, A.; Dreyling, M.; Schmitz, N. Relapse in common lymphoma subtypes: Salvage treatment options for follicular lymphoma, diffuse large cell lymphoma and Hodgkin disease. Br. J. Haematol. 2006, 133, 3–18. [Google Scholar] [CrossRef]
- Chagnon, K.; Boissel, N.; Raffoux, E.; Dombret, H.; Tazi, A.; Bergeron, A. A new pattern of cytosine-arabinoside-induced lung toxicity. Br. J. Haematol. 2009, 147, 771–774. [Google Scholar] [CrossRef] [PubMed]
- Ciccolini, J.; Evrard, A.; M’Batchi, L.; Pourroy, B.; Mercier, C.; Iliadis, A.; Lacarelle, B.; Verschuur, A.; Ouafik, L.; Andre, N. CDA deficiency as a possible culprit for life-threatening toxicities after cytarabine plus 6-mercaptopurine therapy: Pharmacogenetic investigations. Pharmacogenomics 2012, 13, 393–397. [Google Scholar] [CrossRef] [PubMed]
- Abraham, A.; Varatharajan, S.; Abbas, S.; Zhang, W.; Shaji, R.V.; Ahmed, R.; Abraham, A.; George, B.; Srivastava, A.; Chandy, M.; et al. Cytidine deaminase genetic variants influence RNA expression and cytarabine cytotoxicity in acute myeloid leukemia. Pharmacogenomics 2012, 13, 269–282. [Google Scholar] [CrossRef]
- Eron, J.J.; Benoit, S.L.; Jemsek, J.; MacArthur, R.D.; Santana, J.; Quinn, J.B.; Kuritzkes, D.R.; Fallon, M.A.; Rubin, M. Treatment with lamivudine, zidovudine, or both in HIV-positive patients with 200 to 500 CD4+ cells per cubic millimeter. North American HIV Working Party. N. Engl. J. Med. 1995, 333, 1662–1669. [Google Scholar] [CrossRef] [PubMed]
- Evrard, A.; Lacarelle, B.; Ciccolini, J. Severe or lethal toxicities with nucleosidic analogs: Time for action. Pharmacogenomics 2013, 14, 227–230. [Google Scholar] [CrossRef]
- Baker, W.J.; Royer Jr, G.L.; Weiss, R.B. Cytarabine and neurologic toxicity. J. Clin. Oncol. 1991, 9, 679–693. [Google Scholar] [CrossRef]
- Relling, M.V.; Hancock, M.L.; Rivera, G.K.; Sandlund, J.T.; Ribeiro, R.C.; Krynetski, E.Y.; Pui, C.H.; Evans, W.E. Mercaptopurine therapy intolerance and heterozygosity at the thiopurine S-methyltransferase gene locus. J. Natl. Cancer Inst. 1999, 91, 2001–2008. [Google Scholar] [CrossRef]
- Parmar, S.; Seeringer, A.; Denich, D.; Gartner, F.; Pitterle, K.; Syrovets, T.; Ohmle, B.; Stingl, J.C. Variability in transport and biotransformation of cytarabine is associated with its toxicity in peripheral blood mononuclear cells. Pharmacogenomics 2011, 12, 503–514. [Google Scholar] [CrossRef] [PubMed]
- Lamba, J.K. Genetic factors influencing cytarabine therapy. Pharmacogenomics 2009, 10, 1657–1674. [Google Scholar] [CrossRef] [PubMed]
- Paugh, S.W.; Stocco, G.; McCorkle, J.R.; Diouf, B.; Crews, K.R.; Evans, W.E. Cancer pharmacogenomics. Clin. Pharmacol. Ther. 2011, 90, 461–466. [Google Scholar] [CrossRef]
- Davies, S.M. Pharmacogenetics, pharmacogenomics and personalized medicine: Are we there yet? Hematol. Am. Soc. Hematol. Educ. Program. 2006, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, H.E.; Maitland, M.L.; Dolan, M.E.; Cox, N.J.; Ratain, M.J. Cancer pharmacogenomics: Strategies and challenges. Nat. Rev. Genet. 2013, 14, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Jordheim, L.P.; Durantel, D.; Zoulim, F.; Dumontet, C. Advances in the development of nucleoside and nucleotide analogues for cancer and viral diseases. Nat. Rev. Drug Discov. 2013, 12, 447–464. [Google Scholar] [CrossRef] [PubMed]
- Sampath, D.; Rao, V.A.; Plunkett, W. Mechanisms of apoptosis induction by nucleoside analogs. Oncogene 2003, 22, 9063–9074. [Google Scholar] [CrossRef]
- Hellström-Lindberg, E.; Robèrt, K.H.; Gahrton, G.; Lindberg, G.; Forsblom, A.M.; Kock, Y.; Ost, A. Low-dose ara-C in myelodysplastic syndromes (MDS) and acute leukemia following MDS: Proposal for a predictive model. Leuk. Lymphoma 1994, 12, 343–351. [Google Scholar] [CrossRef]
- Ades, L.; Chevret, S.; Raffoux, E.; de Botton, S.; Guerci, A.; Pigneux, A.; Stoppa, A.M.; Lamy, T.; Rigal-Huguet, F.; Vekhoff, A.; et al. Is cytarabine useful in the treatment of acute promyelocytic leukemia? Results of a randomized trial from the European Acute Promyelocytic Leukemia Group. J. Clin. Oncol. 2006, 24, 5703–5710. [Google Scholar] [CrossRef]
- McClune, B.; Buadi, F.K.; Aslam, N.; Przepiorka, D. Intrathecal liposomal cytarabine for prevention of meningeal disease in patients with acute lymphocytic leukemia and high-grade lymphoma. Leuk. Lymphoma 2007, 48, 1849–1851. [Google Scholar] [CrossRef]
- Gokbuget, N.; Hartog, C.M.; Bassan, R.; Derigs, H.G.; Dombret, H.; Greil, R.; Hernandez-Rivas, J.M.; Huguet, F.; Intermesoli, T.; Jourdan, E.; et al. Liposomal cytarabine is effective and tolerable in the treatment of central nervous system relapse of acute lymphoblastic leukemia and very aggressive lymphoma. Haematologica 2011, 96, 238–244. [Google Scholar] [CrossRef]
- Olmos-Jimenez, R.; Diaz-Carrasco, M.S.; Galera-Minarro, A.; Pascual-Gazquez, J.F.; Espuny-Miro, A. Evaluation of standardized triple intrathecal therapy toxicity in oncohematological pediatric patients. Int. J. Clin. Pharm. 2017, 39, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Tilly, H.; Castaigne, S.; Bordessoule, D.; Casassus, P.; Le Prise, P.Y.; Tertian, G.; Desablens, B.; Henry-Amar, M.; Degos, L. Low-dose cytarabine versus intensive chemotherapy in the treatment of acute nonlymphcytic leukemia in the elderly. J. Clin. Oncol. 1990, 8, 272–279. [Google Scholar] [CrossRef]
- Fenaux, P.; Mufti, G.J.; Hellstrom-Lindberg, E.; Santini, V.; Gattermann, N.; Germing, U.; Sanz, G.; List, A.F.; Gore, S.; Seymour, J.F.; et al. Azacitidine prolongs overall survival compared with conventional care regimens in elderly patients with low bone marrow blast count acute myeloid leukemia. J. Clin. Oncol. 2010, 28, 562–569. [Google Scholar] [CrossRef] [PubMed]
- Burnett, A.K.; Milligan, D.; Prentice, A.G.; Goldstone, A.H.; McMullin, M.F.; Hills, R.K.; Wheatley, K. A comparison of low-dose cytarabine and hydroxyurea with or without all-trans retinoic acid for acute myeloid leukemia and high-risk myelodysplastic syndrome in patients not considered fit for intensive treatment. Cancer 2007, 109, 1114–1124. [Google Scholar] [CrossRef]
- Herzig, R.H.; Lazarus, H.M.; Wolff, S.N.; Phillips, G.L.; Herzig, G.P. High-dose cytosine arabinoside therapy with and without anthracycline antibiotics for remission reinduction of acute nonlymphoblastic leukemia. J. Clin. Oncol. 1985, 3, 992–997. [Google Scholar] [CrossRef] [PubMed]
- Mayer, R.J.; Davis, R.B.; Schiffer, C.A.; Berg, D.T.; Powell, B.L.; Schulman, P.; Omura, G.A.; Moore, J.O.; McIntyre, O.R.; Frei, E., 3rd. Intensive postremission chemotherapy in adults with acute myeloid leukemia. Cancer and Leukemia Group B. N. Engl. J. Med. 1994, 331, 896–903. [Google Scholar] [CrossRef] [PubMed]
- Byrd, J.C.; Ruppert, A.S.; Mrozek, K.; Carroll, A.J.; Edwards, C.G.; Arthur, D.C.; Pettenati, M.J.; Stamberg, J.; Koduru, P.R.; Moore, J.O.; et al. Repetitive cycles of high-dose cytarabine benefit patients with acute myeloid leukemia and inv(16)(p13q22) or t(16;16)(p13;q22): Results from CALGB 8461. J. Clin. Oncol. 2004, 22, 1087–1094. [Google Scholar] [CrossRef]
- Schlenk, R.F. Post-remission therapy for acute myeloid leukemia. Haematologica 2014, 99, 1663–1670. [Google Scholar] [CrossRef] [PubMed]
- Rowe, J.M. Optimal induction and post-remission therapy for AML in first remission. Hematol. Am. Soc. Hematol. Educ. Program. 2009, 2009, 396–405. [Google Scholar] [CrossRef]
- Lowenberg, B. Sense and nonsense of high-dose cytarabine for acute myeloid leukemia. Blood 2013, 121, 26–28. [Google Scholar] [CrossRef]
- Lowenberg, B.; Pabst, T.; Vellenga, E.; van Putten, W.; Schouten, H.C.; Graux, C.; Ferrant, A.; Sonneveld, P.; Biemond, B.J.; Gratwohl, A.; et al. Cytarabine dose for acute myeloid leukemia. N. Engl. J. Med. 2011, 364, 1027–1036. [Google Scholar] [CrossRef] [PubMed]
- Wei, A.H.; Montesinos, P.; Ivanov, V.; DiNardo, C.D.; Novak, J.; Laribi, K.; Kim, I.; Stevens, D.A.; Fiedler, W.; Pagoni, M.; et al. Venetoclax plus LDAC for newly diagnosed AML ineligible for intensive chemotherapy: A phase 3 randomized placebo-controlled trial. Blood 2020, 135, 2137–2145. [Google Scholar] [CrossRef] [PubMed]
- Arlin, Z.; Case, D.C., Jr.; Moore, J.; Wiernik, P.; Feldman, E.; Saletan, S.; Desai, P.; Sia, L.; Cartwright, K. Randomized multicenter trial of cytosine arabinoside with mitoxantrone or daunorubicin in previously untreated adult patients with acute nonlymphocytic leukemia (ANLL). Lederle Cooperative Group. Leukemia 1990, 4, 177–183. [Google Scholar]
- Wiernik, P.H.; Banks, P.L.; Case Jr, D.C.; Arlin, Z.A.; Periman, P.O.; Todd, M.B.; Ritch, P.S.; Enck, R.E.; Weitberg, A.B. Cytarabine plus idarubicin or daunorubicin as induction and consolidation therapy for previously untreated adult patients with acute myeloid leukemia. Blood 1992, 79, 313–319. [Google Scholar] [CrossRef]
- Dillman, R.O.; Davis, R.B.; Green, M.R.; Weiss, R.B.; Gottlieb, A.J.; Caplan, S.; Kopel, S.; Preisler, H.; McIntyre, O.R.; Schiffer, C. A comparative study of two different doses of cytarabine for acute myeloid leukemia: A phase III trial of Cancer and Leukemia Group B. Blood 1991, 78, 2520–2526. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, H.F.; Sun, Z.; Yao, X.; Litzow, M.R.; Luger, S.M.; Paietta, E.M.; Racevskis, J.; Dewald, G.W.; Ketterling, R.P.; Bennett, J.M.; et al. Anthracycline dose intensification in acute myeloid leukemia. N. Eng. J. Med. 2009, 361, 1249–1259. [Google Scholar] [CrossRef]
- Bishop, J.F.; Matthews, J.P.; Young, G.A.; Szer, J.; Gillett, A.; Joshua, D.; Bradstock, K.; Enno, A.; Wolf, M.M.; Fox, R.; et al. A randomized study of high-dose cytarabine in induction in acute myeloid leukemia. Blood 1996, 87, 1710–1717. [Google Scholar] [CrossRef] [PubMed]
- Milligan, D.W.; Wheatley, K.; Littlewood, T.; Craig, J.I.; Burnett, A.K.; Group NHOCS. Fludarabine and cytosine are less effective than standard ADE chemotherapy in high-risk acute myeloid leukemia, and addition of G-CSF and ATRA are not beneficial: Results of the MRC AML-HR randomized trial. Blood 2006, 107, 4614–4622. [Google Scholar] [CrossRef] [PubMed]
- Wrzesien-Kus, A.; Robak, T.; Lech-Maranda, E.; Wierzbowska, A.; Dmoszynska, A.; Kowal, M.; Holowiecki, J.; Kyrcz-Krzemien, S.; Grosicki, S.; Maj, S.; et al. A multicenter, open, non-comparative, phase II study of the combination of cladribine (2-chlorodeoxyadenosine), cytarabine, and G-CSF as induction therapy in refractory acute myeloid leukemia—a report of the Polish Adult Leukemia Group (PALG). Eur. J. Haematol. 2003, 71, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Wierzbowska, A.; Robak, T.; Pluta, A.; Wawrzyniak, E.; Cebula, B.; Holowiecki, J.; Kyrcz-Krzemien, S.; Grosicki, S.; Giebel, S.; Skotnicki, A.B.; et al. Cladribine combined with high doses of arabinoside cytosine, mitoxantrone, and G-CSF (CLAG-M) is a highly effective salvage regimen in patients with refractory and relapsed acute myeloid leukemia of the poor risk: A final report of the Polish Adult Leukemia Group. Eur. J. Haematol. 2008, 80, 115–126. [Google Scholar]
- Montillo, M.; Mirto, S.; Petti, M.C.; Latagliata, R.; Magrin, S.; Pinto, A.; Zagonel, V.; Mele, G.; Tedeschi, A.; Ferrara, F. Fludarabine, cytarabine, and G-CSF (FLAG) for the treatment of poor risk acute myeloid leukemia. Am. J. Hematol. 1998, 58, 105–109. [Google Scholar] [CrossRef]
- Amadori, S.; Arcese, W.; Isacchi, G.; Meloni, G.; Petti, M.C.; Monarca, B.; Testi, A.M.; Mandelli, F. Mitoxantrone, etoposide, and intermediate-dose cytarabine: An effective and tolerable regimen for the treatment of refractory acute myeloid leukemia. J. Clin. Oncol. 1991, 9, 1210–1214. [Google Scholar] [CrossRef]
- Archimbaud, E.; Thomas, X.; Leblond, V.; Michallet, M.; Fenaux, P.; Cordonnier, C.; Dreyfus, F.; Troussard, X.; Jaubert, J.; Travade, P.; et al. Timed sequential chemotherapy for previously treated patients with acute myeloid leukemia: Long-term follow-up of the etoposide, mitoxantrone, and cytarabine-86 trial. J. Clin. Oncol. 1995, 13, 11–18. [Google Scholar] [CrossRef]
- Ades, L.; Sanz, M.A.; Chevret, S.; Montesinos, P.; Chevallier, P.; Raffoux, E.; Vellenga, E.; Guerci, A.; Pigneux, A.; Huguet, F.; et al. Treatment of newly diagnosed acute promyelocytic leukemia (APL): A comparison of French-Belgian-Swiss and PETHEMA results. Blood 2008, 111, 1078–1084. [Google Scholar] [CrossRef]
- Powell, B.L.; Moser, B.; Stock, W.; Gallagher, R.E.; Willman, C.L.; Stone, R.M.; Rowe, J.M.; Coutre, S.; Feusner, J.H.; Gregory, J.; et al. Arsenic trioxide improves event-free and overall survival for adults with acute promyelocytic leukemia: North American Leukemia Intergroup Study C9710. Blood 2010, 116, 3751–3757. [Google Scholar] [CrossRef] [PubMed]
- Rizzieri, D.A.; Johnson, J.L.; Niedzwiecki, D.; Lee, E.J.; Vardiman, J.W.; Powell, B.L.; Barcos, M.; Bloomfield, C.D.; Schiffer, C.A.; Peterson, B.A.; et al. Intensive chemotherapy with and without cranial radiation for Burkitt leukemia and lymphoma: Final results of Cancer and Leukemia Group B Study 9251. Cancer 2004, 100, 1438–1448. [Google Scholar] [CrossRef]
- Mead, G.M.; Barrans, S.L.; Qian, W.; Walewski, J.; Radford, J.A.; Wolf, M.; Clawson, S.M.; Stenning, S.P.; Yule, C.L.; Jack, A.S.; et al. A prospective clinicopathologic study of dose-modified CODOX-M/IVAC in patients with sporadic Burkitt lymphoma defined using cytogenetic and immunophenotypic criteria (MRC/NCRI LY10 trial). Blood 2008, 112, 2248–2260. [Google Scholar] [CrossRef] [PubMed]
- Kantarjian, H.M.; O’Brien, S.; Smith, T.L.; Cortes, J.; Giles, F.J.; Beran, M.; Pierce, S.; Huh, Y.; Andreeff, M.; Koller, C.; et al. Results of treatment with hyper-CVAD, a dose-intensive regimen, in adult acute lymphocytic leukemia. J. Clin. Oncol. 2000, 18, 547–561. [Google Scholar] [CrossRef] [PubMed]
- Stock, W.; Johnson, J.L.; Stone, R.M.; Kolitz, J.E.; Powell, B.L.; Wetzler, M.; Westervelt, P.; Marcucci, G.; DeAngelo, D.J.; Vardiman, J.W.; et al. Dose intensification of daunorubicin and cytarabine during treatment of adult acute lymphoblastic leukemia: Results of Cancer and Leukemia Group B Study 19802. Cancer 2013, 119, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Yang, S.; Chen, F.; Wu, S.; Li, W. The Efficacy and Safety of Cytarabine on Newly Diagnosed Primary Central Nervous System Lymphoma: A Systematic Review and Meta-Analysis. Front. Oncol. 2020, 10, 1213. [Google Scholar] [CrossRef] [PubMed]
- Visco, C.; Finotto, S.; Zambello, R.; Paolini, R.; Menin, A.; Zanotti, R.; Zaja, F.; Semenzato, G.; Pizzolo, G.; D’Amore, E.S.; et al. Combination of rituximab, bendamustine, and cytarabine for patients with mantle-cell non-Hodgkin lymphoma ineligible for intensive regimens or autologous transplantation. J. Clin. Oncol. 2013, 31, 1442–1449. [Google Scholar] [CrossRef]
- Bernstein, S.H.; Epner, E.; Unger, J.M.; Leblanc, M.; Cebula, E.; Burack, R.; Rimsza, L.; Miller, T.P.; Fisher, R.I. A phase II multicenter trial of hyperCVAD MTX/Ara-C and rituximab in patients with previously untreated mantle cell lymphoma; SWOG 0213. Ann. Oncol. 2013, 24, 1587–1593. [Google Scholar] [CrossRef]
- Romero, D. Haematological cancer: Cytarabine—new standard of care for MCL. Nat. Rev. Clin. Oncol. 2016, 13, 464–465. [Google Scholar] [CrossRef] [PubMed]
- Velasquez, W.S.; McLaughlin, P.; Tucker, S.; Hagemeister, F.B.; Swan, F.; Rodriguez, M.A.; Romaguera, J.; Rubenstein, E.; Cabanillas, F. ESHAP--an effective chemotherapy regimen in refractory and relapsing lymphoma: A 4-year follow-up study. J. Clin. Oncol. 1994, 12, 1169–1176. [Google Scholar] [CrossRef]
- Machover, D.; Delmas-Marsalet, B.; Misra, S.C.; Gumus, Y.; Goldschmidt, E.; Schilf, A.; Frenoy, N.; Emile, J.F.; Debuire, B.; Guettier, C.; et al. Dexamethasone, high-dose cytarabine, and oxaliplatin (DHAOx) as salvage treatment for patients with initially refractory or relapsed non-Hodgkin’s lymphoma. Ann. Oncol. 2001, 12, 1439–1443. [Google Scholar] [CrossRef]
- Josting, A.; Rudolph, C.; Reiser, M.; Mapara, M.; Sieber, M.; Kirchner, H.H.; Dorken, B.; Hossfeld, D.K.; Diehl, V.; Engert, A.; et al. Time-intensified dexamethasone/cisplatin/cytarabine: An effective salvage therapy with low toxicity in patients with relapsed and refractory Hodgkin’s disease. Ann. Oncol. 2002, 13, 1628–1635. [Google Scholar] [CrossRef]
- Aparicio, J.; Segura, A.; Garcera, S.; Oltra, A.; Santaballa, A.; Yuste, A.; Pastor, M. ESHAP is an active regimen for relapsing Hodgkin’s disease. Ann. Oncol. 1999, 10, 593–595. [Google Scholar] [CrossRef] [PubMed]
- Chopra, R.; McMillan, A.K.; Linch, D.C.; Yuklea, S.; Taghipour, G.; Pearce, R.; Patterson, K.G.; Goldstone, A.H. The place of high-dose BEAM therapy and autologous bone marrow transplantation in poor-risk Hodgkin’s disease. A single-center eight-year study of 155 patients. Blood 1993, 81, 1137–1145. [Google Scholar] [CrossRef] [PubMed]
- Giebel, S.; Kruzel, T.; Czerw, T.; Sadus-Wojciechowska, M.; Najda, J.; Chmielowska, E.; Grosicki, S.; Jurczyszyn, A.; Pasiarski, M.; Nowara, E.; et al. Intermediate-dose Ara-C plus G-CSF for stem cell mobilization in patients with lymphoid malignancies, including predicted poor mobilizers. Bone Marrow Transpl. 2013, 48, 915–921. [Google Scholar] [CrossRef]
- Ferreri, A.J.; Reni, M.; Foppoli, M.; Martelli, M.; Pangalis, G.A.; Frezzato, M.; Cabras, M.G.; Fabbri, A.; Corazzelli, G.; Ilariucci, F.; et al. High-dose cytarabine plus high-dose methotrexate versus high-dose methotrexate alone in patients with primary CNS lymphoma: A randomised phase 2 trial. Lancet 2009, 374, 1512–1520. [Google Scholar] [CrossRef]
- Larson, R.A.; Dodge, R.K.; Burns, C.P.; Lee, E.J.; Stone, R.M.; Schulman, P.; Duggan, D.; Davey, F.R.; Sobol, R.E.; Frankel, S.R.; et al. A five-drug remission induction regimen with intensive consolidation for adults with acute lymphoblastic leukemia: Cancer and leukemia group B study 8811. Blood 1995, 85, 2025–2037. [Google Scholar] [CrossRef]
- Weiss, M.A.; Aliff, T.B.; Tallman, M.S.; Frankel, S.R.; Kalaycio, M.E.; Maslak, P.G.; Jurcic, J.G.; Scheinberg, D.A.; Roma, T.E. A single, high dose of idarubicin combined with cytarabine as induction therapy for adult patients with recurrent or refractory acute lymphoblastic leukemia. Cancer 2002, 95, 581–587. [Google Scholar] [CrossRef]
- Tsimberidou, A.M.; Wierda, W.G.; Plunkett, W.; Kurzrock, R.; O’Brien, S.; Wen, S.; Ferrajoli, A.; Ravandi-Kashani, F.; Garcia-Manero, G.; Estrov, Z.; et al. Phase I-II study of oxaliplatin, fludarabine, cytarabine, and rituximab combination therapy in patients with Richter’s syndrome or fludarabine-refractory chronic lymphocytic leukemia. J. Clin. Oncol. 2008, 26, 196–203. [Google Scholar] [CrossRef]
- Lee, E.J.; Petroni, G.R.; Schiffer, C.A.; Freter, C.E.; Johnson, J.L.; Barcos, M.; Frizzera, G.; Bloomfield, C.D.; Peterson, B.A. Brief-duration high-intensity chemotherapy for patients with small noncleaved-cell lymphoma or FAB L3 acute lymphocytic leukemia: Results of cancer and leukemia group B study 9251. J. Clin. Oncol. 2001, 19, 4014–4022. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Velasquez, W.S.; Cabanillas, F.; Salvador, P.; McLaughlin, P.; Fridrik, M.; Tucker, S.; Jagannath, S.; Hagemeister, F.B.; Redman, J.R.; Swan, F.; et al. Effective salvage therapy for lymphoma with cisplatin in combination with high-dose Ara-C and dexamethasone (DHAP). Blood 1988, 71, 117–122. [Google Scholar] [CrossRef]
- McGrail, L.H.; Sehn, L.H.; Weiss, R.B.; Robson, M.R.; Antin, J.H.; Byrd, J.C. Pancreatitis during therapy of acute myeloid leukemia: Cytarabine related? Ann. Oncol. 1999, 10, 1373–1376. [Google Scholar] [CrossRef] [PubMed]
- McBride, C.E.; Yavorski, R.T.; Moses, F.M.; Robson, M.E.; Solimando, D.A., Jr.; Byrd, J.C. Acute pancreatitis associated with continuous infusion cytarabine therapy: A case report. Cancer 1996, 77, 2588–2591. [Google Scholar] [CrossRef]
- Powell, B.L.; Zekan, P.J.; Muss, H.B.; Richards, F., 2nd; Lyerly, E.S.; Capizzi, R.L. Ara-C syndrome during low-dose continuous infusion therapy. Med. Pediatr. Oncol. 1986, 14, 310–312. [Google Scholar] [CrossRef] [PubMed]
- Phillips, C.L.; Lane, A.; Gerbing, R.B.; Alonzo, T.A.; Wilkey, A.; Radloff, G.; Lange, B.; Gamazon, E.R.; Dolan, M.E.; Davies, S.M. Genomic Variants of Cytarabine Sensitivity Associated with Treatment-Related Mortality in Pediatric AML: A Report from the Children’s Oncology Group. Clin. Cancer Res. 2020, 26, 2891–2897. [Google Scholar] [CrossRef]
- Moore, J.O.; George, S.L.; Dodge, R.K.; Amrein, P.C.; Powell, B.L.; Kolitz, J.E.; Baer, M.R.; Davey, F.R.; Bloomfield, C.D.; Larson, R.A.; et al. Sequential multiagent chemotherapy is not superior to high-dose cytarabine alone as postremission intensification therapy for acute myeloid leukemia in adults under 60 years of age: Cancer and Leukemia Group B Study 9222. Blood 2005, 105, 3420–3427. [Google Scholar] [CrossRef] [PubMed]
- Herzig, R.H.; Hines, J.D.; Herzig, G.P.; Wolff, S.N.; Cassileth, P.A.; Lazarus, H.M.; Adelstein, D.J.; Brown, R.A.; Coccia, P.F.; Strandjord, S.; et al. Cerebellar toxicity with high-dose cytosine arabinoside. J. Clin. Oncol. 1987, 5, 927–932. [Google Scholar] [CrossRef] [PubMed]
- Salinsky, M.C.; Levine, R.L.; Aubuchon, J.P.; Schutta, H.S. Acute cerebellar dysfunction with high-dose ARA-C therapy. Cancer 1983, 51, 426–429. [Google Scholar] [CrossRef]
- Dunton, S.F.; Nitschke, R.; Spruce, W.E.; Bodensteiner, J.; Krous, H.F. Progressive ascending paralysis following administration of intrathecal and intravenous cytosine arabinoside. A Pediatric Oncology Group study. Cancer 1986, 57, 1083–1088. [Google Scholar] [CrossRef]
- Openshaw, H.; Slatkin, N.E.; Stein, A.S.; Hinton, D.R.; Forman, S.J. Acute polyneuropathy after high dose cytosine arabinoside in patients with leukemia. Cancer 1996, 78, 1899–1905. [Google Scholar] [CrossRef]
- Ventura, G.J.; Keating, M.J.; Castellanos, A.M.; Glass, J.P. Reversible bilateral lateral rectus muscle palsy associated with high-dose cytosine arabinoside and mitoxantrone therapy. Cancer 1986, 58, 1633–1635. [Google Scholar] [CrossRef]
- Damon, L.E.; Mass, R.; Linker, C.A. The association between high-dose cytarabine neurotoxicity and renal insufficiency. J. Clin. Oncol. 1989, 7, 1563–1568. [Google Scholar] [CrossRef]
- Nagahata, Y.; Kondo, T.; Ono, Y.; Hiramoto, N.; Kitano, T.; Hishizawa, M.; Yamashita, K.; Hashimoto, H.; Ishikawa, T.; Takaori-Kondo, A. High-dose cytarabine chemotherapy (>/=4 g/m(2)/day) before allogeneic hematopoietic stem cell transplantation for non-core-binding-factor AML in the first complete remission. Leuk. Lymphoma 2020, 61, 3128–3136. [Google Scholar] [CrossRef]
- Haupt, H.M.; Hutchins, G.M.; Moore, G.W. Ara-C lung: Noncardiogenic pulmonary edema complicating cytosine arabinoside therapy of leukemia. Am. J. Med. 1981, 70, 256–261. [Google Scholar] [CrossRef]
- Deangelis, L.M.; Posner, J.B. Side effects of chemotherapy. In Neurologic Complications of Cancer, 2nd ed.; OUP: New York, NY, USA, 2009; p. 447. [Google Scholar]
- Resar, L.M.; Phillips, P.C.; Kastan, M.B.; Leventhal, B.G.; Bowman, P.W.; Civin, C.I. Acute neurotoxicity after intrathecal cytosine arabinoside in two adolescents with acute lymphoblastic leukemia of B-cell type. Cancer 1993, 71, 117–123. [Google Scholar] [CrossRef]
- Derissen, E.J.B.; Beijnen, J.H. Intracellular Pharmacokinetics of Pyrimidine Analogues used in Oncology and the Correlation with Drug Action. Clin. Pharm. 2020, 59, 1521–1550. [Google Scholar] [CrossRef] [PubMed]
- Chou, T.C.; Hutchinson, D.J.; Schmid, F.A.; Philips, F.S. Metabolism and selective effects of 1-beta-D-arabinofuranosylcytosine in L1210 and Host tissues in vivo. Cancer Res. 1975, 35, 225–236. [Google Scholar] [PubMed]
- Kufe, D.W.; Spriggs, D.R. Biochemical and cellular pharmacology of cytosine arabinoside. Semin. Oncol. 1985, 12 (Suppl. 3), 34–48. [Google Scholar]
- Galmarini, C.M.; Mackey, J.R.; Dumontet, C. Nucleoside analogues: Mechanisms of drug resistance and reversal strategies. Leukemia 2001, 15, 875–890. [Google Scholar] [CrossRef]
- Clarke, M.L.; Mackey, J.R.; Baldwin, S.A.; Young, J.D.; Cass, C.E. The role of membrane transporters in cellular resistance to anticancer nucleoside drugs. Cancer Treat. Res. 2002, 112, 27–47. [Google Scholar] [PubMed]
- Clarke, M.L.; Damaraju, V.L.; Zhang, J.; Mowles, D.; Tackaberry, T.; Lang, T.; Smith, K.M.; Young, J.D.; Tomkinson, B.; Cass, C.E. The role of human nucleoside transporters in cellular uptake of 4′-thio-beta-D-arabinofuranosylcytosine and beta-D-arabinosylcytosine. Mol. Pharmacol. 2006, 70, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Endo, Y.; Obata, T.; Murata, D.; Ito, M.; Sakamoto, K.; Fukushima, M.; Yamasaki, Y.; Yamada, Y.; Natsume, N.; Sasaki, T. Cellular localization and functional characterization of the equilibrative nucleoside transporters of antitumor nucleosides. Cancer Sci. 2007, 98, 1633–1637. [Google Scholar] [CrossRef]
- Galmarini, C.M.; Thomas, X.; Calvo, F.; Rousselot, P.; Rabilloud, M.; El Jaffari, A.; Cros, E.; Dumontet, C. In vivo mechanisms of resistance to cytarabine in acute myeloid leukaemia. Br. J. Haematol. 2002, 117, 860–868. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, M.; Han, T.; Damaraju, V.; Carpenter, P.; Cass, C.E.; Agarwal, R.P. Cytosine arabinoside affects multiple cellular factors and induces drug resistance in human lymphoid cells. Biochem. Pharmacol. 2005, 70, 426–432. [Google Scholar] [CrossRef]
- Yee, S.W.; Mefford, J.A.; Singh, N.; Percival, M.E.; Stecula, A.; Yang, K.; Witte, J.S.; Takahashi, A.; Kubo, M.; Matsuda, K.; et al. Impact of polymorphisms in drug pathway genes on disease-free survival in adults with acute myeloid leukemia. J. Hum. Genet. 2013, 58, 353–361. [Google Scholar] [CrossRef]
- Errasti-Murugarren, E.; Pastor-Anglada, M.; Casado, F.J. Role of CNT3 in the transepithelial flux of nucleosides and nucleoside-derived drugs. J. Physiol 2007, 582 Pt 3, 1249–1260. [Google Scholar] [CrossRef]
- Li, L.; Fridley, B.L.; Kalari, K.; Jenkins, G.; Batzler, A.; Weinshilboum, R.M.; Wang, L. Gemcitabine and arabinosylcytosin pharmacogenomics: Genome-wide association and drug response biomarkers. PLoS ONE 2009, 4, e7765. [Google Scholar] [CrossRef]
- Guo, Y.; Kock, K.; Ritter, C.A.; Chen, Z.S.; Grube, M.; Jedlitschky, G.; Illmer, T.; Ayres, M.; Beck, J.F.; Siegmund, W.; et al. Expression of ABCC-type nucleotide exporters in blasts of adult acute myeloid leukemia: Relation to long-term survival. Clin. Cancer Res. 2009, 15, 1762–1769. [Google Scholar] [CrossRef] [PubMed]
- Hopper-Borge, E.; Xu, X.; Shen, T.; Shi, Z.; Chen, Z.S.; Kruh, G.D. Human multidrug resistance protein 7 (ABCC10) is a resistance factor for nucleoside analogues and epothilone B. Cancer Res. 2009, 69, 178–184. [Google Scholar] [CrossRef]
- Gati, W.P.; Paterson, A.R.; Larratt, L.M.; Turner, A.R.; Belch, A.R. Sensitivity of acute leukemia cells to cytarabine is a correlate of cellular es nucleoside transporter site content measured by flow cytometry with SAENTA-fluorescein. Blood 1997, 90, 346–353. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Visser, F.; King, K.M.; Baldwin, S.A.; Young, J.D.; Cass, C.E. The role of nucleoside transporters in cancer chemotherapy with nucleoside drugs. Cancer Metastasis Rev. 2007, 26, 85–110. [Google Scholar] [CrossRef] [PubMed]
- Eltzschig, H.K.; Abdulla, P.; Hoffman, E.; Hamilton, K.E.; Daniels, D.; Schonfeld, C.; Loffler, M.; Reyes, G.; Duszenko, M.; Karhausen, J.; et al. HIF-1-dependent repression of equilibrative nucleoside transporter (ENT) in hypoxia. J. Exp. Med. 2005, 202, 1493–1505. [Google Scholar] [CrossRef] [PubMed]
- Montero, T.D.; Racordon, D.; Bravo, L.; Owen, G.I.; Bronfman, M.L.; Leisewitz, A.V. PPARalpha and PPARgamma regulate the nucleoside transporter hENT1. Biochem. Biophys. Res. Commun. 2012, 419, 405–411. [Google Scholar] [CrossRef]
- Galmarini, C.M.; Thomas, X.; Calvo, F.; Rousselot, P.; El Jafaari, A.; Cros, E.; Dumontet, C. Potential mechanisms of resistance to cytarabine in AML patients. Leuk. Res. 2002, 26, 621–629. [Google Scholar] [CrossRef]
- Hubeek, I.; Stam, R.W.; Peters, G.J.; Broekhuizen, R.; Meijerink, J.P.; van Wering, E.R.; Gibson, B.E.; Creutzig, U.; Zwaan, C.M.; Cloos, J.; et al. The human equilibrative nucleoside transporter 1 mediates in vitro cytarabine sensitivity in childhood acute myeloid leukaemia. Br. J. Cancer 2005, 93, 1388–1394. [Google Scholar] [CrossRef] [PubMed]
- White, J.C.; Rathmell, J.P.; Capizzi, R.L. Membrane transport influences the rate of accumulation of cytosine arabinoside in human leukemia cells. J. Clin. Investig. 1987, 79, 380–387. [Google Scholar] [CrossRef]
- Kessel, D.; Hall, T.C.; Rosenthal, D. Uptake and phosphorylation of cytosine arabinoside by normal and leukemic human blood cells in vitro. Cancer Res. 1969, 29, 459–463. [Google Scholar]
- Chou, T.C.; Arlin, Z.; Clarkson, B.D.; Phillips, F.S. Metabolism of 1-beta-D-arabinofuranosylcytosine in human leukemic cells. Cancer Res. 1977, 37, 3561–3570. [Google Scholar]
- Heinemann, V.; Hertel, L.W.; Grindey, G.B.; Plunkett, W. Comparison of the cellular pharmacokinetics and toxicity of 2′,2′-difluorodeoxycytidine and 1-beta-D-arabinofuranosylcytosine. Cancer Res. 1988, 48, 4024–4031. [Google Scholar]
- Graham, F.L.; Whitmore, G.F. Studies in mouse L-cells on the incorporation of 1-beta-D-arabinofuranosylcytosine into DNA and on inhibition of DNA polymerase by 1-beta-D-arabinofuranosylcytosine 5′-triphosphate. Cancer Res. 1970, 30, 2636–2644. [Google Scholar] [PubMed]
- Rustum, Y.M.; Raymakers, R.A. 1-Beta-arabinofuranosylcytosine in therapy of leukemia: Preclinical and clinical overview. Pharmacol. Ther. 1992, 56, 307–321. [Google Scholar] [CrossRef]
- Townsend, A.J.; Cheng, Y.C. Sequence-specific effects of ara-5-aza-CTP and ara-CTP on DNA synthesis by purified human DNA polymerases in vitro: Visualization of chain elongation on a defined template. Mol. Pharmacol. 1987, 32, 330–339. [Google Scholar] [PubMed]
- Ross, D.D.; Cuddy, D.P.; Cohen, N.; Hensley, D.R. Mechanistic implications of alterations in HL-60 cell nascent DNA after exposure to 1-beta-D-arabinofuranosylcytosine. Cancer Chemother. Pharmacol. 1992, 31, 61–70. [Google Scholar] [CrossRef]
- Kufe, D.; Spriggs, D.; Egan, E.M.; Munroe, D. Relationships among Ara-CTP pools, formation of (Ara-C)DNA, and cytotoxicity of human leukemic cells. Blood 1984, 64, 54–58. [Google Scholar] [CrossRef]
- Preisler, H.D.; Rustum, Y.; Priore, R.L. Relationship between leukemic cell retention of cytosine arabinoside triphosphate and the duration of remission in patients with acute non-lymphocytic leukemia. Eur. J. Cancer Clin. Oncol. 1985, 21, 23–30. [Google Scholar] [CrossRef]
- Plunkett, W.; Iacoboni, S.; Estey, E.; Danhauser, L.; Liliemark, J.O.; Keating, M.J. Pharmacologically directed ara-C therapy for refractory leukemia. Semin. Oncol. 1985, 12 (Suppl. 3), 20–30. [Google Scholar]
- Plunkett, W.; Liliemark, J.O.; Adams, T.M.; Nowak, B.; Estey, E.; Kantarjian, H.; Keating, M.J. Saturation of 1-beta-D-arabinofuranosylcytosine 5′-triphosphate accumulation in leukemia cells during high-dose 1-beta-D-arabinofuranosylcytosine therapy. Cancer Res. 1987, 47, 3005–3011. [Google Scholar] [PubMed]
- Lauzon, G.J.; Paterson, A.R.; Belch, A.W. Formation of 1-beta-D-arabinofuranosylcytosine diphosphate choline in neoplastic and normal cells. Cancer Res. 1978, 38, 1730–1733. [Google Scholar]
- Sasvari-Szekely, M.; Spasokukotskaja, T.; Staub, M. Deoxyribocytidine is salvaged not only into DNA but also into phospholipid precursors. IV. Exogenous deoxyribocytidine can be used with the same efficacy as (ribo)cytidine for lipid activation. Biochem. Biophys. Res. Commun. 1993, 194, 966–972. [Google Scholar] [CrossRef] [PubMed]
- Chiba, P.; Tihan, T.; Szekeres, T.; Salamon, J.; Kraupp, M.; Eher, R.; Koller, U.; Knapp, W. Concordant changes of pyrimidine metabolism in blasts of two cases of acute myeloid leukemia after repeated treatment with ara-C in vivo. Leukemia 1990, 4, 761–765. [Google Scholar]
- Kawasaki, H.; Kuwabara, H.; Hori, H.; Higashigawa, M.; Ohkubo, T.; Sakurai, M. Intracellular dCTP/ara-CTP ratio and the cytotoxic effect of ara-C. Cancer Investig. 1991, 9, 409–413. [Google Scholar] [CrossRef]
- Tattersall, M.H.; Ganeshaguru, K.; Hoffbrand, A.V. Mechanisms of resistance of human acute leukaemia cells to cytosine arabinoside. Br. J. Haematol. 1974, 27, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Kanno, S.-i.; Hiura, T.; Ohtake, T.; Koiwai, K.; Suzuki, H.; Ujibe, M.; Ishikawa, M. Characterization of resistance to cytosine arabinoside (Ara-C) in NALM-6 human B leukemia cells. Clin. Chim. Acta 2007, 377, 144–149. [Google Scholar] [CrossRef]
- Meuth, M. The molecular basis of mutations induced by deoxyribonucleoside triphosphate pool imbalances in mammalian cells. Exp. Cell Res. 1989, 181, 305–316. [Google Scholar] [CrossRef]
- Verschuur, A.C.; Van Gennip, A.H.; Leen, R.; Voute, P.A.; Brinkman, J.; Van Kuilenburg, A.B. Cyclopentenyl cytosine increases the phosphorylation and incorporation into DNA of 1-beta-D-arabinofuranosyl cytosine in a human T-lymphoblastic cell line. Int. J. Cancer 2002, 98, 616–623. [Google Scholar] [CrossRef]
- Shao, J.; Zhou, B.; Chu, B.; Yen, Y. Ribonucleotide reductase inhibitors and future drug design. Curr. Cancer Drug Targets 2006, 6, 409–431. [Google Scholar] [CrossRef]
- Karp, J.E.; Giles, F.J.; Gojo, I.; Morris, L.; Greer, J.; Johnson, B.; Thein, M.; Sznol, M.; Low, J. A phase I study of the novel ribonucleotide reductase inhibitor 3-aminopyridine-2-carboxaldehyde thiosemicarbazone (3-AP, Triapine) in combination with the nucleoside analog fludarabine for patients with refractory acute leukemias and aggressive myeloproliferative disorders. Leuk. Res. 2008, 32, 71–77. [Google Scholar] [PubMed]
- Gandhi, V.; Estey, E.; Keating, M.J.; Plunkett, W. Biochemical modulation of arabinosylcytosine for therapy of leukemias. Leuk. Lymphoma 1993, 10, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, V.; Estey, E.; Du, M.; Nowak, B.; Keating, M.J.; Plunkett, W. Modulation of the cellular metabolism of cytarabine and fludarabine by granulocyte-colony-stimulating factor during therapy of acute myelogenous leukemia. Clin. Cancer Res. 1995, 1, 169–178. [Google Scholar]
- Gandhi, V.; Kemena, A.; Keating, M.J.; Plunkett, W. Fludarabine infusion potentiates arabinosylcytosine metabolism in lymphocytes of patients with chronic lymphocytic leukemia. Cancer Res. 1992, 52, 897–903. [Google Scholar]
- Dumontet, C.; Fabianowska-Majewska, K.; Mantincic, D.; Callet Bauchu, E.; Tigaud, I.; Gandhi, V.; Lepoivre, M.; Peters, G.J.; Rolland, M.O.; Wyczechowska, D.; et al. Common resistance mechanisms to deoxynucleoside analogues in variants of the human erythroleukaemic line K562. Br. J. Haematol. 1999, 106, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Hunsucker, S.A.; Mitchell, B.S.; Spychala, J. The 5′-nucleotidases as regulators of nucleotide and drug metabolism. Pharmacol. Ther. 2005, 107, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Cacciamani, T.; Vita, A.; Cristalli, G.; Vincenzetti, S.; Natalini, P.; Ruggieri, S.; Amici, A.; Magni, G. Purification of human cytidine deaminase: Molecular and enzymatic characterization and inhibition by synthetic pyrimidine analogs. Arch. Biochem. Biophys. 1991, 290, 285–292. [Google Scholar] [CrossRef]
- Capizzi, R.L.; White, J.C.; Powell, B.L.; Perrino, F. Effect of dose on the pharmacokinetic and pharmacodynamic effects of cytarabine. Semin. Hematol. 1991, 28 (Suppl. 4), 54–69. [Google Scholar]
- Fridland, A.; Verhoef, V. Mechanism for ara-CTP catabolism in human leukemic cells and effect of deaminase inhibitors on this process. Semin. Oncol. 1987, 14 (Suppl. 1), 262–268. [Google Scholar]
- Liliemark, J.O.; Plunkett, W. Regulation of 1-beta-D-arabinofuranosylcytosine 5′-triphosphate accumulation in human leukemia cells by deoxycytidine 5′-triphosphate. Cancer Res. 1986, 46, 1079–1083. [Google Scholar]
- Xu, P.P.; Chen, B.A.; Feng, J.F.; Cheng, L.; Xia, G.H.; Li, Y.F.; Qian, J.; Ding, J.H.; Lu, Z.H.; Wang, X.M.; et al. Association of polymorphisms of cytosine arabinoside-metabolizing enzyme gene with therapeutic efficacy for acute myeloid leukemia. Chin. Med. J. 2012, 125, 2137–2143. [Google Scholar]
- Cao, Y.; Wang, X.; Cao, Z.; Wu, C.; Wu, D.; Cheng, X. Genetic polymorphisms of MBL2 and tuberculosis susceptibility: A meta-analysis of 22 case-control studies. Arch. Med. Sci. 2018, 14, 1212–1232. [Google Scholar] [CrossRef]
- Zhu, K.W.; Chen, P.; Zhang, D.Y.; Yan, H.; Liu, H.; Cen, L.N.; Liu, Y.L.; Cao, S.; Zhou, G.; Zeng, H.; et al. Association of genetic polymorphisms in genes involved in Ara-C and dNTP metabolism pathway with chemosensitivity and prognosis of adult acute myeloid leukemia (AML). J. Transl. Med. 2018, 16, 90. [Google Scholar] [CrossRef]
- McPherson, J.D.; Marra, M.; Hillier, L.; Waterston, R.H.; Chinwalla, A.; Wallis, J.; Sekhon, M.; Wylie, K.; Mardis, E.R.; Wilson, R.K.; et al. A physical map of the human genome. Nature 2001, 409, 934–941. [Google Scholar] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008, 455, 1061–1068. [Google Scholar] [CrossRef]
- Huang, R.S.; Dolan, M.E. Approaches to the discovery of pharmacogenomic markers in oncology: 2000-2010-2020. Pharmacogenomics 2010, 11, 471–474. [Google Scholar] [CrossRef]
- Garner, C. Upward bias in odds ratio estimates from genome-wide association studies. Genet. Epidemiol. 2007, 31, 288–295. [Google Scholar] [CrossRef]
- Myers, S.N.; Goyal, R.K.; Roy, J.D.; Fairfull, L.D.; Wilson, J.W.; Ferrell, R.E. Functional single nucleotide polymorphism haplotypes in the human equilibrative nucleoside transporter 1. Pharm. Genom. 2006, 16, 315–320. [Google Scholar] [CrossRef]
- Joerger, M.; Bosch, T.M.; Doodeman, V.D.; Beijnen, J.H.; Smits, P.H.; Schellens, J.H. Novel deoxycytidine kinase gene polymorphisms: A population screening study in Caucasian healthy volunteers. Eur. J. Clin. Pharmacol. 2006, 62, 681–684. [Google Scholar] [CrossRef]
- Lamba, J.K.; Crews, K.; Pounds, S.; Schuetz, E.G.; Gresham, J.; Gandhi, V.; Plunkett, W.; Rubnitz, J.; Ribeiro, R. Pharmacogenetics of deoxycytidine kinase: Identification and characterization of novel genetic variants. J. Pharmacol. Exp. Ther. 2007, 323, 935–945. [Google Scholar] [CrossRef] [PubMed]
- Braunagel, D.; Schaich, M.; Kramer, M.; Dransfeld, C.L.; Ehninger, G.; Mahlknecht, U. The T_T genotype within the NME1 promoter single nucleotide polymorphism -835 C/T is associated with an increased risk of cytarabine induced neurotoxicity in patients with acute myeloid leukemia. Leuk. Lymphoma 2012, 53, 952–957. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Mitra, A.K.; Pounds, S.; Crews, K.R.; Gandhi, V.; Plunkett, W.; Dolan, M.E.; Hartford, C.; Raimondi, S.; Campana, D.; et al. RRM1 and RRM2 pharmacogenetics: Association with phenotypes in HapMap cell lines and acute myeloid leukemia patients. Pharmacogenomics 2013, 14, 1449–1466. [Google Scholar] [CrossRef] [PubMed]
- Carpi, F.M.; Vincenzetti, S.; Ubaldi, J.; Pucciarelli, S.; Polzonetti, V.; Micozzi, D.; Mignini, F.; Napolioni, V. CDA gene polymorphisms and enzyme activity: Genotype-phenotype relationship in an Italian-Caucasian population. Pharmacogenomics 2013, 14, 769–781. [Google Scholar] [CrossRef]
- Mitra, A.K.; Crews, K.R.; Pounds, S.; Cao, X.; Feldberg, T.; Ghodke, Y.; Gandhi, V.; Plunkett, W.; Dolan, M.E.; Hartford, C.; et al. Genetic variants in cytosolic 5′-nucleotidase II are associated with its expression and cytarabine sensitivity in HapMap cell lines and in patients with acute myeloid leukemia. J. Pharmacol. Exp. Ther. 2011, 339, 9–23. [Google Scholar] [CrossRef]
- Aksoy, P.; Zhu, M.J.; Kalari, K.R.; Moon, I.; Pelleymounter, L.L.; Eckloff, B.W.; Wieben, E.D.; Yee, V.C.; Weinshilboum, R.M.; Wang, L. Cytosolic 5′-nucleotidase III (NT5C3): Gene sequence variation and functional genomics. Pharm. Genom. 2009, 19, 567–576. [Google Scholar] [CrossRef]
- Kuptsova-Clarkson, N.; Ambrosone, C.B.; Weiss, J.; Baer, M.R.; Sucheston, L.E.; Zirpoli, G.; Kopecky, K.J.; Ford, L.; Blanco, J.; Wetzler, M.; et al. XPD DNA nucleotide excision repair gene polymorphisms associated with DNA repair deficiency predict better treatment outcomes in secondary acute myeloid leukemia. Int. J. Mol. Epidemiol. Genet. 2010, 1, 278–294. [Google Scholar] [CrossRef]
- Welsh, M.; Mangravite, L.; Medina, M.W.; Tantisira, K.; Zhang, W.; Huang, R.S.; McLeod, H.; Dolan, M.E. Pharmacogenomic discovery using cell-based models. Pharmacol. Rev. 2009, 61, 413–429. [Google Scholar] [CrossRef]
- Wheeler, H.E.; Dolan, M.E. Lymphoblastoid cell lines in pharmacogenomic discovery and clinical translation. Pharmacogenomics 2012, 13, 55–70. [Google Scholar] [CrossRef] [PubMed]
- Burdick, J.T.; Chen, W.M.; Abecasis, G.R.; Cheung, V.G. In silico method for inferring genotypes in pedigrees. Nat. Genet. 2006, 38, 1002–1004. [Google Scholar] [CrossRef]
- Dausset, J.; Cann, H.; Cohen, D.; Lathrop, M.; Lalouel, J.M.; White, R. Centre d’etude du polymorphisme humain (CEPH): Collaborative genetic mapping of the human genome. Genomics 1990, 6, 575–577. [Google Scholar] [CrossRef]
- Thorisson, G.A.; Smith, A.V.; Krishnan, L.; Stein, L.D. The International HapMap Project Web site. Genome Res. 2005, 15, 1592–1593. [Google Scholar] [CrossRef] [PubMed]
- Wise, J. Consortium hopes to sequence genome of 1000 volunteers. BMJ 2008, 336, 237. [Google Scholar] [CrossRef] [PubMed]
- Thorisson, G.A.; Stein, L.D. The SNP Consortium website: Past, present and future. Nucleic Acids Res. 2003, 31, 124–127. [Google Scholar] [CrossRef]
- Morley, M.; Molony, C.M.; Weber, T.M.; Devlin, J.L.; Ewens, K.G.; Spielman, R.S.; Cheung, V.G. Genetic analysis of genome-wide variation in human gene expression. Nature 2004, 430, 743–747. [Google Scholar] [CrossRef]
- Huang, R.S.; Duan, S.; Shukla, S.J.; Kistner, E.O.; Clark, T.A.; Chen, T.X.; Schweitzer, A.C.; Blume, J.E.; Dolan, M.E. Identification of genetic variants contributing to cisplatin-induced cytotoxicity by use of a genomewide approach. Am. J. Hum. Genet. 2007, 81, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Hartford, C.M.; Duan, S.; Delaney, S.M.; Mi, S.; Kistner, E.O.; Lamba, J.K.; Huang, R.S.; Dolan, M.E. Population-specific genetic variants important in susceptibility to cytarabine arabinoside cytotoxicity. Blood 2009, 113, 2145–2153. [Google Scholar] [CrossRef] [PubMed]
- Gamazon, E.R.; Duan, S.; Zhang, W.; Huang, R.S.; Kistner, E.O.; Dolan, M.E.; Cox, N.J. PACdb: A database for cell-based pharmacogenomics. Pharm. Genom. 2010, 20, 269–273. [Google Scholar] [CrossRef]
- Mackey, J.R.; Baldwin, S.A.; Young, J.D.; Cass, C.E. Nucleoside transport and its significance for anticancer drug resistance. Drug Resist. Updat. 1998, 1, 310–324. [Google Scholar] [CrossRef]
- Leabman, M.K.; Huang, C.C.; DeYoung, J.; Carlson, E.J.; Taylor, T.R.; de la Cruz, M.; Johns, S.J.; Stryke, D.; Kawamoto, M.; Urban, T.J.; et al. Natural variation in human membrane transporter genes reveals evolutionary and functional constraints. Proc. Natl. Acad. Sci. USA 2003, 100, 5896–5901. [Google Scholar] [CrossRef]
- Osato, D.H.; Huang, C.C.; Kawamoto, M.; Johns, S.J.; Stryke, D.; Wang, J.; Ferrin, T.E.; Herskowitz, I.; Giacomini, K.M. Functional characterization in yeast of genetic variants in the human equilibrative nucleoside transporter, ENT1. Pharmacogenetics 2003, 13, 297–301. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.R.; Saito, Y.; Maekawa, K.; Sugiyama, E.; Kaniwa, N.; Ueno, H.; Okusaka, T.; Morizane, C.; Yamamoto, N.; Ikeda, M.; et al. Thirty novel genetic variations in the SLC29A1 gene encoding human equilibrative nucleoside transporter 1 (hENT1). Drug Metab. Pharm. 2006, 21, 248–256. [Google Scholar] [CrossRef]
- Stam, R.W.; den Boer, M.L.; Meijerink, J.P.; Ebus, M.E.; Peters, G.J.; Noordhuis, P.; Janka-Schaub, G.E.; Armstrong, S.A.; Korsmeyer, S.J.; Pieters, R. Differential mRNA expression of Ara-C-metabolizing enzymes explains Ara-C sensitivity in MLL gene-rearranged infant acute lymphoblastic leukemia. Blood 2003, 101, 1270–1276. [Google Scholar] [CrossRef] [PubMed]
- Reiman, T.; Clarke, M.L.; Dabbagh, L.; Vsianska, M.; Coupland, R.W.; Belch, A.R.; Baldwin, S.A.; Young, J.D.; Cass, C.E.; Mackey, J.R. Differential expression of human equilibrative nucleoside transporter 1 (hENT1) protein in the Reed-Sternberg cells of Hodgkin’s disease. Leuk. Lymphoma 2002, 43, 1435–1440. [Google Scholar] [CrossRef]
- Kirschbaum, M. hENT1 and Hodgkin lymphoma: Not just crossing the channel. Leuk. Lymphoma 2008, 49, 1024–1025. [Google Scholar] [CrossRef]
- Lai, R.; Bartlett, N.L.; Mackey, J.R.; Jung, S.H.; Johnson, J.L.; Cook, J.R.; Jones, D.; Cass, C.E.; Young, J.D.; Said, J.; et al. High expression of nucleoside transporter protein hENT1 in Reed-Sternberg cells is associated with treatment failure in relapsed/refractory Hodgkin lymphoma patients treated with gemcitabine, vinorelbine and liposomal doxorubicin—a CALGB 59804 correlative study. Leuk. Lymphoma 2008, 49, 1202–1205. [Google Scholar]
- Chow, L.; Lai, R.; Dabbagh, L.; Belch, A.; Young, J.D.; Cass, C.E.; Mackey, J.R. Analysis of human equilibrative nucleoside transporter 1 (hENT1) protein in non-Hodgkin’s lymphoma by immunohistochemistry. Mod. Pathol. 2005, 18, 558–564. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Guo, Y.; Kotova, E.; Chen, Z.S.; Lee, K.; Hopper-Borge, E.; Belinsky, M.G.; Kruh, G.D. MRP8, ATP-binding cassette C11 (ABCC11), is a cyclic nucleotide efflux pump and a resistance factor for fluoropyrimidines 2′,3′-dideoxycytidine and 9′-(2′-phosphonylmethoxyethyl)adenine. J. Biol. Chem. 2003, 278, 29509–29514. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Gong, S.; Monks, A.; Zaharevitz, D.; Moscow, J.A. Correlation of nucleoside and nucleobase transporter gene expression with antimetabolite drug cytotoxicity. J. Exp. Ther. Oncol. 2002, 2, 200–212. [Google Scholar] [CrossRef]
- Takagaki, K.; Katsuma, S.; Kaminishi, Y.; Horio, T.; Nakagawa, S.; Tanaka, T.; Ohgi, T.; Yano, J. Gene-expression profiling reveals down-regulation of equilibrative nucleoside transporter 1 (ENT1) in Ara-C-resistant CCRF-CEM-derived cells. J. Biochem. 2004, 136, 733–740. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Damaraju, V.L.; Groulx, N.; Mowles, D.; Peng, Y.; Robins, M.J.; Cass, C.E.; Gros, P. Two distinct molecular mechanisms underlying cytarabine resistance in human leukemic cells. Cancer Res. 2008, 68, 2349–2357. [Google Scholar] [CrossRef]
- Zimmerman, E.I.; Huang, M.; Leisewitz, A.V.; Wang, Y.; Yang, J.; Graves, L.M. Identification of a novel point mutation in ENT1 that confers resistance to Ara-C in human T cell leukemia CCRF-CEM cells. FEBS Lett. 2009, 583, 425–429. [Google Scholar] [CrossRef]
- Damaraju, V.L.; Damaraju, S.; Young, J.D.; Baldwin, S.A.; Mackey, J.; Sawyer, M.B.; Cass, C.E. Nucleoside anticancer drugs: The role of nucleoside transporters in resistance to cancer chemotherapy. Oncogene 2003, 22, 7524–7536. [Google Scholar] [CrossRef]
- Naud, J.S.; Ghani, K.; de Campos-Lima, P.O.; Caruso, M. Nilotinib and imatinib inhibit cytarabine cellular uptake: Implications for combination therapy. Leuk. Res. 2012, 36, 1311–1314. [Google Scholar] [CrossRef] [PubMed]
- Toyoda, Y.; Ishikawa, T. Pharmacogenomics of human ABC transporter ABCC11 (MRP8): Potential risk of breast cancer and chemotherapy failure. Anticancer Agents Med. Chem. 2010, 10, 617–624. [Google Scholar] [CrossRef] [PubMed]
- Magdy, T.; Arlanov, R.; Winter, S.; Lang, T.; Klein, K.; Toyoda, Y.; Ishikawa, T.; Schwab, M.; Zanger, U.M. ABCC11/MRP8 polymorphisms affect 5-fluorouracil-induced severe toxicity and hepatic expression. Pharmacogenomics 2013, 14, 1433–1448. [Google Scholar] [CrossRef]
- Gandhi, V.; Plunkett, W. Cell cycle-specific metabolism of arabinosyl nucleosides in K562 human leukemia cells. Cancer Chem. Pharmacol. 1992, 31, 11–17. [Google Scholar] [CrossRef]
- Owens, J.K.; Shewach, D.S.; Ullman, B.; Mitchell, B.S. Resistance to 1-beta-D-arabinofuranosylcytosine in human T-lymphoblasts mediated by mutations within the deoxycytidine kinase gene. Cancer Res. 1992, 52, 2389–2393. [Google Scholar]
- Van der Wilt, C.L.; Kroep, J.R.; Loves, W.J.; Rots, M.G.; Van Groeningen, C.J.; Kaspers, G.J.; Peters, G.J. Expression of deoxycytidine kinase in leukaemic cells compared with solid tumour cell lines, liver metastases and normal liver. Eur. J. Cancer 2003, 39, 691–697. [Google Scholar] [CrossRef]
- Estey, E.; Plunkett, W.; Dixon, D.; Keating, M.; McCredie, K.; Freireich, E.J. Variables predicting response to high dose cytosine arabinoside therapy in patients with refractory acute leukemia. Leukemia 1987, 1, 580–583. [Google Scholar]
- Raza, A.; Gezer, S.; Anderson, J.; Lykins, J.; Bennett, J.; Browman, G.; Goldberg, J.; Larson, R.; Vogler, R.; Preisler, H.D. Relationship of [3H]Ara-C incorporation and response to therapy with high-dose Ara-C in AML patients: A Leukemia Intergroup study. Exp. Hematol. 1992, 20, 1194–1200. [Google Scholar]
- Galmarini, C.M.; Thomas, X.; Graham, K.; El Jafaari, A.; Cros, E.; Jordheim, L.; Mackey, J.R.; Dumontet, C. Deoxycytidine kinase and cN-II nucleotidase expression in blast cells predict survival in acute myeloid leukaemia patients treated with cytarabine. Br. J. Haematol. 2003, 122, 53–60. [Google Scholar] [CrossRef]
- Shi, J.Y.; Shi, Z.Z.; Zhang, S.J.; Zhu, Y.M.; Gu, B.W.; Li, G.; Bai, X.T.; Gao, X.D.; Hu, J.; Jin, W.; et al. Association between single nucleotide polymorphisms in deoxycytidine kinase and treatment response among acute myeloid leukaemia patients. Pharmacogenetics 2004, 14, 759–768. [Google Scholar] [CrossRef]
- Szantai, E.; Ronai, Z.; Sasvari-Szekely, M.; Bonn, G.; Guttman, A. Multicapillary electrophoresis analysis of single-nucleotide sequence variations in the deoxycytidine kinase gene. Clin. Chem. 2006, 52, 1756–1762. [Google Scholar] [CrossRef]
- Mahlknecht, U.; Dransfeld, C.L.; Bulut, N.; Kramer, M.; Thiede, C.; Ehninger, G.; Schaich, M. SNP analyses in cytarabine metabolizing enzymes in AML patients and their impact on treatment response and patient survival: Identification of CDA SNP C-451T as an independent prognostic parameter for survival. Leukemia 2009, 23, 1929–1932. [Google Scholar] [CrossRef][Green Version]
- Baker, J.A.; Wickremsinhe, E.R.; Li, C.H.; Oluyedun, O.A.; Dantzig, A.H.; Hall, S.D.; Qian, Y.W.; Ring, B.J.; Wrighton, S.A.; Guo, Y. Pharmacogenomics of gemcitabine metabolism: Functional analysis of genetic variants in cytidine deaminase and deoxycytidine kinase. Drug Metab. Dispos. 2013, 41, 541–545. [Google Scholar] [CrossRef]
- Verschuur, A.C.; Brinkman, J.; Van Gennip, A.H.; Leen, R.; Vet, R.J.; Evers, L.M.; Voute, P.A.; Van Kuilenburg, A.B. Cyclopentenyl cytosine induces apoptosis and increases cytarabine-induced apoptosis in a T-lymphoblastic leukemic cell-line. Leuk. Res. 2001, 25, 891–900. [Google Scholar] [CrossRef]
- Verschuur, A.C.; van Gennip, A.H.; Leen, R.; Voute, P.A.; van Kuilenburg, A.B. Cyclopentenyl cytosine increases the phosphorylation and incorporation into dna of arabinofu-ranosyl cytosine in a myeloid leukemic cell-line. Adv. Exp. Med. Biol. 2000, 486, 311–317. [Google Scholar] [PubMed]
- Rudd, S.G.; Tsesmetzis, N.; Sanjiv, K.; Paulin, C.B.; Sandhow, L.; Kutzner, J.; Hed Myrberg, I.; Bunten, S.S.; Axelsson, H.; Zhang, S.M.; et al. Ribonucleotide reductase inhibitors suppress SAMHD1 ara-CTPase activity enhancing cytarabine efficacy. EMBO Mol. Med. 2020, 12, e10419. [Google Scholar] [CrossRef]
- Avramis, V.I.; Nandy, P.; Kwock, R.; Solorzano, M.M.; Mukherjee, S.K.; Danenberg, P.; Cohen, L.J. Increased p21/WAF-1 and p53 protein levels following sequential three drug combination regimen of fludarabine, cytarabine and docetaxel induces apoptosis in human leukemia cells. Anticancer Res. 1998, 18, 2327–2338. [Google Scholar]
- Gandhi, V.; Estey, E.; Du, M.; Keating, M.J.; Plunkett, W. Minimum dose of fludarabine for the maximal modulation of 1-beta-D-arabinofuranosylcytosine triphosphate in human leukemia blasts during therapy. Clin. Cancer Res. 1997, 3, 1539–1545. [Google Scholar]
- Kim, S.O.; Jeong, J.Y.; Kim, M.R.; Cho, H.J.; Ju, J.Y.; Kwon, Y.S.; Oh, I.J.; Kim, K.S.; Kim, Y.I.; Lim, S.C.; et al. Efficacy of gemcitabine in patients with non-small cell lung cancer according to promoter polymorphisms of the ribonucleotide reductase M1 gene. Clin. Cancer Res. 2008, 14, 3083–3088. [Google Scholar] [CrossRef] [PubMed]
- Rha, S.Y.; Jeung, H.C.; Choi, Y.H.; Yang, W.I.; Yoo, J.H.; Kim, B.S.; Roh, J.K.; Chung, H.C. An association between RRM1 haplotype and gemcitabine-induced neutropenia in breast cancer patients. Oncologist 2007, 12, 622–630. [Google Scholar] [CrossRef]
- Whelan, J.; Smith, T.; Phear, G.; Rohatiner, A.; Lister, A.; Meuth, M. Resistance to cytosine arabinoside in acute leukemia: The significance of mutations in CTP synthetase. Leukemia 1994, 8, 264–265. [Google Scholar]
- Whelan, J.; Phear, G.; Yamauchi, M.; Meuth, M. Clustered base substitutions in CTP synthetase conferring drug resistance in Chinese hamster ovary cells. Nat. Genet. 1993, 3, 317–322. [Google Scholar] [CrossRef]
- Fitzgerald, S.M.; Goyal, R.K.; Osborne, W.R.; Roy, J.D.; Wilson, J.W.; Ferrell, R.E. Identification of functional single nucleotide polymorphism haplotypes in the cytidine deaminase promoter. Hum. Genet. 2006, 119, 276–283. [Google Scholar] [CrossRef] [PubMed]
- Schroder, J.K.; Kirch, C.; Seeber, S.; Schutte, J. Structural and functional analysis of the cytidine deaminase gene in patients with acute myeloid leukaemia. Br. J. Haematol. 1998, 103, 1096–1103. [Google Scholar] [CrossRef] [PubMed]
- Yue, L.; Saikawa, Y.; Ota, K.; Tanaka, M.; Nishimura, R.; Uehara, T.; Maeba, H.; Ito, T.; Sasaki, T.; Koizumi, S. A functional single-nucleotide polymorphism in the human cytidine deaminase gene contributing to ara-C sensitivity. Pharmacogenetics 2003, 13, 29–38. [Google Scholar] [CrossRef]
- Gilbert, J.A.; Salavaggione, O.E.; Ji, Y.; Pelleymounter, L.L.; Eckloff, B.W.; Wieben, E.D.; Ames, M.M.; Weinshilboum, R.M. Gemcitabine pharmacogenomics: Cytidine deaminase and deoxycytidylate deaminase gene resequencing and functional genomics. Clin. Cancer Res. 2006, 12, 1794–1803. [Google Scholar] [CrossRef]
- Micozzi, D.; Carpi, F.M.; Pucciarelli, S.; Polzonetti, V.; Polidori, P.; Vilar, S.; Williams, B.; Costanzi, S.; Vincenzetti, S. Human cytidine deaminase: A biochemical characterization of its naturally occurring variants. Int. J. Biol. Macromol. 2013, 63C, 64–74. [Google Scholar] [CrossRef]
- Iyer, S.N.; Ankala, A.; Singhal, R.S.; Hegde, M.R. Determination of common genetic variants in cytidine deaminase (CDA) gene in Indian ethnic population. Gene 2013, 524, 35–39. [Google Scholar] [CrossRef]
- Jahns-Streubel, G.; Reuter, C.; Auf der Landwehr, U.; Unterhalt, M.; Schleyer, E.; Wormann, B.; Buchner, T.; Hiddemann, W. Activity of thymidine kinase and of polymerase alpha as well as activity and gene expression of deoxycytidine deaminase in leukemic blasts are correlated with clinical response in the setting of granulocyte-macrophage colony-stimulating factor-based priming before and during TAD-9 induction therapy in acute myeloid leukemia. Blood 1997, 90, 1968–1976. [Google Scholar]
- Bhatla, D.; Gerbing, R.B.; Alonzo, T.A.; Conner, H.; Ross, J.A.; Meshinchi, S.; Zhai, X.; Zamzow, T.; Mehta, P.A.; Geiger, H.; et al. Cytidine deaminase genotype and toxicity of cytosine arabinoside therapy in children with acute myeloid leukemia. Br. J. Haematol. 2009, 144, 388–394. [Google Scholar] [CrossRef]
- Galmarini, C.M.; Cros, E.; Thomas, X.; Jordheim, L.; Dumontet, C. The prognostic value of cN-II and cN-III enzymes in adult acute myeloid leukemia. Haematologica 2005, 90, 1699–1701. [Google Scholar]
- Galmarini, C.M.; Graham, K.; Thomas, X.; Calvo, F.; Rousselot, P.; El Jafaari, A.; Cros, E.; Mackey, J.R.; Dumontet, C. Expression of high Km 5′-nucleotidase in leukemic blasts is an independent prognostic factor in adults with acute myeloid leukemia. Blood 2001, 98, 1922–1926. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, T.; Negoro, E.; Kishi, S.; Takagi, K.; Yoshida, A.; Urasaki, Y.; Iwasaki, H.; Ueda, T. Intracellular cytarabine triphosphate production correlates to deoxycytidine kinase/cytosolic 5′-nucleotidase II expression ratio in primary acute myeloid leukemia cells. Biochem. Pharmacol. 2009, 77, 1780–1786. [Google Scholar] [CrossRef]
- Jordheim, L.P.; Marton, Z.; Rhimi, M.; Cros-Perrial, E.; Lionne, C.; Peyrottes, S.; Dumontet, C.; Aghajari, N.; Chaloin, L. Identification and characterization of inhibitors of cytoplasmic 5′-nucleotidase cN-II issued from virtual screening. Biochem. Pharmacol. 2013, 85, 497–506. [Google Scholar] [CrossRef]
- Helleday, T.; Petermann, E.; Lundin, C.; Hodgson, B.; Sharma, R.A. DNA repair pathways as targets for cancer therapy. Nat. Rev. Cancer 2008, 8, 193–204. [Google Scholar] [CrossRef]
- Liu, X.; Wang, Y.; Benaissa, S.; Matsuda, A.; Kantarjian, H.; Estrov, Z.; Plunkett, W. Homologous recombination as a resistance mechanism to replication-induced double-strand breaks caused by the antileukemia agent CNDAC. Blood 2010, 116, 1737–1746. [Google Scholar] [CrossRef] [PubMed]
- Giachino, D.F.; Ghio, P.; Regazzoni, S.; Mandrile, G.; Novello, S.; Selvaggi, G.; Gregori, D.; DeMarchi, M.; Scagliotti, G.V. Prospective assessment of XPD Lys751Gln and XRCC1 Arg399Gln single nucleotide polymorphisms in lung cancer. Clin. Cancer Res. 2007, 13, 2876–2881. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.Y.; Ren, Z.H.; Jiao, B.; Xiao, R.; Yun, H.Y.; Chen, B.; Zhao, W.L.; Zhu, Q.; Chen, Z.; Chen, S.J. Genetic variations of DNA repair genes and their prognostic significance in patients with acute myeloid leukemia. Int. J. Cancer 2011, 128, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Yin, G.; Zheng, Q.; Yan, C.; Berk, B.C. GIT1 is a scaffold for ERK1/2 activation in focal adhesions. J. Biol. Chem. 2005, 280, 27705–27712. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Xu, S. ERK1/2 MAP kinases in cell survival and apoptosis. IUBMB Life 2006, 58, 621–631. [Google Scholar] [CrossRef]
- Shaw, G.C.; Cope, J.J.; Li, L.; Corson, K.; Hersey, C.; Ackermann, G.E.; Gwynn, B.; Lambert, A.J.; Wingert, R.A.; Traver, D.; et al. Mitoferrin is essential for erythroid iron assimilation. Nature 2006, 440, 96–100. [Google Scholar] [CrossRef]
- Leardi, A.; Caraglia, M.; Selleri, C.; Pepe, S.; Pizzi, C.; Notaro, R.; Fabbrocini, A.; De Lorenzo, S.; Musico, M.; Abbruzzese, A.; et al. Desferioxamine increases iron depletion and apoptosis induced by ara-C of human myeloid leukaemic cells. Br. J. Haematol. 1998, 102, 746–752. [Google Scholar] [CrossRef]
- Mitra, A.K.; Crews, K.; Pounds, S.; Cao, X.; Downing, J.R.; Raimondi, S.; Campana, D.; Ribeiro, R.C.; Rubnitz, J.E.; Lamba, J.K. Impact of genetic variation in FKBP5 on clinical response in pediatric acute myeloid leukemia patients: A pilot study. Leukemia 2011, 25, 1354–1356. [Google Scholar] [CrossRef]
- Xie, C.; Drenberg, C.; Edwards, H.; Caldwell, J.T.; Chen, W.; Inaba, H.; Xu, X.; Buck, S.A.; Taub, J.W.; Baker, S.D.; et al. Panobinostat enhances cytarabine and daunorubicin sensitivities in AML cells through suppressing the expression of BRCA1, CHK1, and Rad51. PLoS ONE 2013, 8, e79106. [Google Scholar] [CrossRef] [PubMed]
- Children’s Oncology Group; Aplenc, R.; Alonzo, T.A.; Gerbing, R.B.; Smith, F.O.; Meshinchi, S.; Ross, J.A.; Perentesis, J.; Woods, W.G.; Lange, B.J.; et al. Ethnicity and survival in childhood acute myeloid leukemia: A report from the Children’s Oncology Group. Blood 2006, 108, 74–80. [Google Scholar] [CrossRef]
- Sekeres, M.A.; Peterson, B.; Dodge, R.K.; Mayer, R.J.; Moore, J.O.; Lee, E.J.; Kolitz, J.; Baer, M.R.; Schiffer, C.A.; Carroll, A.J.; et al. Differences in prognostic factors and outcomes in African Americans and whites with acute myeloid leukemia. Blood 2004, 103, 4036–4042. [Google Scholar] [CrossRef] [PubMed]
- Rubnitz, J.E.; Lensing, S.; Razzouk, B.I.; Pounds, S.; Pui, C.H.; Ribeiro, R.C. Effect of race on outcome of white and black children with acute myeloid leukemia: The St. Jude experience. Pediatr. Blood Cancer 2007, 48, 10–15. [Google Scholar] [CrossRef]
- Pearson, T.A.; Manolio, T.A. How to interpret a genome-wide association study. JAMA 2008, 299, 1335–1344. [Google Scholar] [CrossRef] [PubMed]
- Gamazon, E.R.; Lamba, J.K.; Pounds, S.; Stark, A.L.; Wheeler, H.E.; Cao, X.; Im, H.K.; Mitra, A.K.; Rubnitz, J.E.; Ribeiro, R.C.; et al. Comprehensive genetic analysis of cytarabine sensitivity in a cell-based model identifies polymorphisms associated with outcome in AML patients. Blood 2013, 121, 4366–4376. [Google Scholar] [CrossRef]
- Zeggini, E.; Ioannidis, J.P. Meta-analysis in genome-wide association studies. Pharmacogenomics 2009, 10, 191–201. [Google Scholar] [CrossRef]
- Higgins, J.P.; Thompson, S.G.; Deeks, J.J.; Altman, D.G. Measuring inconsistency in meta-analyses. BMJ 2003, 327, 557–560. [Google Scholar] [CrossRef]
- Salanti, G.; Higgins, J.P.; Trikalinos, T.A.; Ioannidis, J.P. Bayesian meta-analysis and meta-regression for gene-disease associations and deviations from Hardy-Weinberg equilibrium. Stat. Med. 2007, 26, 553–567. [Google Scholar] [CrossRef]
- Fridley, B.L.; Serie, D.; Jenkins, G.; White, K.; Bamlet, W.; Potter, J.D.; Goode, E.L. Bayesian mixture models for the incorporation of prior knowledge to inform genetic association studies. Genet. Epidemiol. 2010, 34, 418–426. [Google Scholar] [CrossRef] [PubMed]
- Visscher, P.M.; Brown, M.A.; McCarthy, M.I.; Yang, J. Five years of GWAS discovery. Am. J. Hum. Genet. 2012, 90, 7–24. [Google Scholar] [CrossRef]
- Tirelli, U.; Berretta, M.; Bearz, A.; Carbone, A. Grouping of molecularly targeted anti-cancer agents based on cost-effectiveness analysis. Eur. Rev. Med. Pharmacol. Sci. 2011, 15, 1355–1356. [Google Scholar] [PubMed]
- Payne, K.; Shabaruddin, F.H. Cost-effectiveness analysis in pharmacogenomics. Pharmacogenomics 2010, 11, 643–646. [Google Scholar] [CrossRef]
- Dhalla, I.A.; Garner, S.; Chalkidou, K.; Littlejohns, P. Perspectives on the National Institute for Health and Clinical Excellence’s recommendations to use health technologies only in research. Int. J. Technol. Assess. Health Care 2009, 25, 272–280. [Google Scholar] [CrossRef] [PubMed]
- Biondi, A.; Cazzaniga, G. Novel clinical trials for pediatric leukemias: Lessons learned from genomic analyses. Hematology Am. Soc. Hematol. Educ. Program. 2013, 2013, 612–619. [Google Scholar] [CrossRef][Green Version]
- Adema, A.D.; Smid, K.; Losekoot, N.; Honeywell, R.J.; Verheul, H.M.; Myhren, F.; Sandvold, M.L.; Peters, G.J. Metabolism and accumulation of the lipophilic deoxynucleoside analogs elacytarabine and CP-4126. Investig. New Drugs 2012, 30, 1908–1916. [Google Scholar] [CrossRef] [PubMed]
- Giles, F.J.; Vey, N.; Rizzieri, D.; Ravandi, F.; Prebet, T.; Borthakur, G.; Jacobsen, T.F.; Hagen, S.; Nilsson, B.; O’Brien, S. Phase I and pharmacokinetic study of elacytarabine, a novel 5′-elaidic acid derivative of cytarabine, in adults with refractory hematological malignancies. Leukemia 2012, 26, 1686–1689. [Google Scholar] [CrossRef][Green Version]
- Robinson, G.M.; Anderson, E.; Salisbury, V.C.; Mehta, P.A.; Sandvold, M.L.; Reynolds, D.M. Detection of intracellular ara-C/ara-CTP following treatment with the new anti-leukemic drug elacytarabine using a bioluminescent bacterial biosensor. In Proceedings of the AACR Annual Meeting, Washington, DC, USA, 6–10 April 2013. [Google Scholar]
- Roboz, G.J.; Rosenblat, T.; Arellano, M.; Gobbi, M.; Altman, J.K.; Montesinos, P.; O’Connell, C.; Solomon, S.R.; Pigneux, A.; Vey, N.; et al. International randomized phase III study of elacytarabine versus investigator choice in patients with relapsed/refractory acute myeloid leukemia. J. Clin. Oncol. 2014, 32, 1919–1926. [Google Scholar] [CrossRef]
- Kantarjian, H.; Faderl, S.; Garcia-Manero, G.; Luger, S.; Venugopal, P.; Maness, L.; Wetzler, M.; Coutre, S.; Stock, W.; Claxton, D.; et al. Oral sapacitabine for the treatment of acute myeloid leukaemia in elderly patients: A randomised phase 2 study. Lancet Oncol. 2012, 13, 1096–1104. [Google Scholar] [CrossRef]
- Hanaoka, K.; Suzuki, M.; Kobayashi, T.; Tanzawa, F.; Tanaka, K.; Shibayama, T.; Miura, S.; Ikeda, T.; Iwabuchi, H.; Nakagawa, A.; et al. Antitumor activity and novel DNA-self-strand-breaking mechanism of CNDAC (1-(2-C-cyano-2-deoxy-beta-D-arabino-pentofuranosyl) cytosine) and its N4-palmitoyl derivative (CS-682). Int. J. Cancer 1999, 82, 226–236. [Google Scholar] [CrossRef]
- Ewald, B.; Sampath, D.; Plunkett, W. Nucleoside analogs: Molecular mechanisms signaling cell death. Oncogene 2008, 27, 6522–6537. [Google Scholar] [CrossRef]
- Liu, X.; Guo, Y.; Li, Y.; Jiang, Y.; Chubb, S.; Azuma, A.; Huang, P.; Matsuda, A.; Hittelman, W.; Plunkett, W. Molecular basis for G2 arrest induced by 2′-C-cyano-2′-deoxy-1-beta-D-arabino-pentofuranosylcytosine and consequences of checkpoint abrogation. Cancer Res. 2005, 65, 6874–6881. [Google Scholar] [CrossRef]
- Jordheim, L.P.; Cros, E.; Gouy, M.H.; Galmarini, C.M.; Peyrottes, S.; Mackey, J.; Perigaud, C.; Dumontet, C. Characterization of a gemcitabine-resistant murine leukemic cell line: Reversion of in vitro resistance by a mononucleotide prodrug. Clin. Cancer Res. 2004, 10, 5614–5621. [Google Scholar] [CrossRef] [PubMed]
- Tobias, S.C.; Borch, R.F. Synthesis and biological evaluation of a cytarabine phosphoramidate prodrug. Mol. Pharm. 2004, 1, 112–116. [Google Scholar] [CrossRef]
- Gouy, M.H.; Jordheim, L.P.; Lefebvre, I.; Cros, E.; Dumontet, C.; Peyrottes, S.; Perigaud, C. Special feature of mixed phosphotriester derivatives of cytarabine. Bioorg. Med. Chem. 2009, 17, 6340–6347. [Google Scholar] [CrossRef] [PubMed]
- Senanayake, T.H.; Warren, G.; Wei, X.; Vinogradov, S.V. Application of activated nucleoside analogs for the treatment of drug-resistant tumors by oral delivery of nanogel-drug conjugates. J. Control. Release 2013, 167, 200–209. [Google Scholar] [CrossRef] [PubMed]
- Chamberlain, M.C. Neurotoxicity of intra-CSF liposomal cytarabine (DepoCyt) administered for the treatment of leptomeningeal metastases: A retrospective case series. J. Neurooncol. 2012, 109, 143–148. [Google Scholar] [CrossRef]
- Hilgendorf, I.; Wolff, D.; Junghanss, C.; Kahl, C.; Leithaeuser, M.; Steiner, B.; Casper, J.; Freund, M. Neurological complications after intrathecal liposomal cytarabine application in patients after allogeneic haematopoietic stem cell transplantation. Ann. Hematol. 2008, 87, 1009–1012. [Google Scholar] [CrossRef] [PubMed]
- Jabbour, E.; O’Brien, S.; Kantarjian, H.; Garcia-Manero, G.; Ferrajoli, A.; Ravandi, F.; Cabanillas, M.; Thomas, D.A. Neurologic complications associated with intrathecal liposomal cytarabine given prophylactically in combination with high-dose methotrexate and cytarabine to patients with acute lymphocytic leukemia. Blood 2007, 109, 3214–3218. [Google Scholar] [CrossRef]
- Ostermann, K.; Pels, H.; Kowoll, A.; Kuhnhenn, J.; Schlegel, U. Neurologic complications after intrathecal liposomal cytarabine in combination with systemic polychemotherapy in primary CNS lymphoma. J. Neurooncol. 2011, 103, 635–640. [Google Scholar] [CrossRef] [PubMed]
- Corazzelli, G.; Frigeri, F.; Russo, F.; Frairia, C.; Arcamone, M.; Esposito, G.; De Chiara, A.; Morelli, E.; Capobianco, G.; Becchimanzi, C.; et al. RD-CODOX-M/IVAC with rituximab and intrathecal liposomal cytarabine in adult Burkitt lymphoma and ‘unclassifiable’ highly aggressive B-cell lymphoma. Br. J. Haematol. 2012, 156, 234–244. [Google Scholar] [CrossRef]
- Spina, M.; Chimienti, E.; Martellotta, F.; Vaccher, E.; Berretta, M.; Zanet, E.; Lleshi, A.; Canzonieri, V.; Bulian, P.; Tirelli, U. Phase 2 study of intrathecal, long-acting liposomal cytarabine in the prophylaxis of lymphomatous meningitis in human immunodeficiency virus-related non-Hodgkin lymphoma. Cancer 2010, 116, 1495–1501. [Google Scholar] [CrossRef] [PubMed]
- Federico, C.; Morittu, V.M.; Britti, D.; Trapasso, E.; Cosco, D. Gemcitabine-loaded liposomes: Rationale, potentialities and future perspectives. Int. J. Nanomed. 2012, 7, 5423–5436. [Google Scholar]
- Feldman, E.J.; Lancet, J.E.; Kolitz, J.E.; Ritchie, E.K.; Roboz, G.J.; List, A.F.; Allen, S.L.; Asatiani, E.; Mayer, L.D.; Swenson, C.; et al. First-in-man study of CPX-351: A liposomal carrier containing cytarabine and daunorubicin in a fixed 5:1 molar ratio for the treatment of relapsed and refractory acute myeloid leukemia. J. Clin. Oncol. 2011, 29, 979–985. [Google Scholar] [CrossRef]
- International HapMap Consortium. The International HapMap Project. Nature 2003, 426, 789–796. [Google Scholar] [CrossRef] [PubMed]
- International HapMap Consortium. A haplotype map of the human genome. Nature 2005, 437, 1299–1320. [Google Scholar] [CrossRef]
- Genomes Project, C.; Abecasis, G.R.; Auton, A.; Brooks, L.D.; DePristo, M.A.; Durbin, R.M.; Handsaker, R.E.; Kang, H.M.; Marth, G.T.; McVean, G.A. An integrated map of genetic variation from 1,092 human genomes. Nature 2012, 491, 56–65. [Google Scholar]
- 1000 Genomes Project Consortium; Abecasis, G.R.; Altshuler, D.; Auton, A.; Brooks, L.D.; Durbin, R.M.; Gibbs, R.A.; Hurles, M.E.; McVean, G.A. A map of human genome variation from population-scale sequencing. Nature 2010, 467, 1061–1073. [Google Scholar] [CrossRef]
| Disease | Phase | Regimens | Cytarabine Dosing and Additional Drugs | References |
|---|---|---|---|---|
| Acute myeloid leukemia | induction | 7 + 3 | 100 mg/m2/day continuous infusion for 7 days (in combination with daunorubicin or idarubicin or mitoxantrone) or (Adults < 60 years) 200 mg/m2/day continuous infusion for 7 days (in combination with daunorubicin) | [39,40,41,42] |
| Low-Dose SubQ | Adults ≥ 65 years: SubQ: 20 mg/m2/day for 14 days out of every 28-day cycle for at least 4 cycles or 10 mg/m2 every 12 h for 21 days, or 10 mg/m2 every 12 h for 21 days; may repeat after 15 days | [28,29] | ||
| consolidation | 5 + 2 | 100 mg/m2/day continuous infusion for 5 days (in combination with daunorubicin or idarubicin or mitoxantrone) | [39,40] | |
| 5 + 2 + 5 | 100 mg/m2/day continuous infusion for 5 days (in combination with daunorubicin and etoposide) | [43] | ||
| BCL2 inhibitor/LDAC 4 + 4 | Venetoclax once daily, began at 100 mg on day 1 and increased stepwise over 4 days to reach the target dose of 600 mg (100, 200, 400, and 600 mg); dosing was continued at 600 mg per day from day 4 through day 28 in combination with 20 mg/m2 of Ara-C | [38] | ||
| High-Dose single-agent | Adults ≤ 60 years: 3000 mg/m2 over 3 h every 12 h on days 1, 3, and 5 (total of 6 doses); repeat every 28–35 days for 4 courses | [32] | ||
| Intermediate-dose | cycle I: cytarabine 200 mg/m2 per continuous infusion on days 1–7; included idarubicin at 12 mg/m2 (3-h infusion on days 5, 6 and 7); cycle II: cytarabine 1000 mg/m2 intravenously for 3 h twice daily on days 1–6; included amsacrine 120 mg/m2 per 1-h infusion on days 3, 5 and 7 | [37] | ||
| salvage | ADE | 100 mg/m2 I.V push every 12 h for 10 days (in combination with daunorubicin and etoposide) | [44] | |
| CLAG | 2000 mg/m2/day over 4 h for 5 days (in combination with cladribine and G-CSF) | [45] | ||
| CLAG-M | 2000 mg/m2/day over 4 h for 5 days (in combination with cladribine, G-CSF, and mitoxantrone) | [46] | ||
| FLAG | 2000 mg/m2/day over 4 h for 5 days (in combination with fludarabine and G-CSF) | [47] | ||
| High-Dose | 3000 mg/m2 over 1 h every 12 h for 12 doses (± an anthracycline) | [31] | ||
| MEC | 1000 mg/m2/day over 6 h for 6 days (in combination with mitoxantrone and etoposide)or Adults < 60 years: 500 mg/m2/day continuous infusion days 1, 2, and 3 and days 8, 9, and 10 (in combination with mitoxantrone and etoposide) | [48,49] | ||
| Acute promyelocytic leukemia | induction | APL2000 C9710 | 200 mg/m2/day continuous infusion for 7 days beginning on day 3 of treatment (in combination with tretinoin and daunorubicin) | [50,51] |
| Disease | Phase | Regimens | Cytarabine Dosing and Additional Drugs | References |
|---|---|---|---|---|
| Acute lymphocytic leukemia | induction | Hyper-CVAD | Dose-intensive regimen: I.V.: 3000 mg/m2 over 2 h every 12 h days 2 and 3 (4 doses/cycle) of even numbered cycles (in combination with methotrexate; alternates with Hyper-CVAD) | [54] |
| Larson regime SubQ | Early intensification phase: 75 mg/m2/dose days 1 to 4 and 8 to 11 (4-week cycle; repeat once) Late intensification phase: 75 mg/m2/dose days 29 to 32 and 36 to 39 | [67] | ||
| induction, relapse or progression | High-Dose | 3000 mg/m2 over 3 h daily for 5 days (in combination with idarubicin [day 3]) | [68] | |
| Chronic lymphocytic leukemia | refractory or Richter’s syndrome | OFAR | I.V.: 1000 mg/m2/dose over 2 h days 2 and 3 every 4 weeks for up to 6 cycles (in combination with oxaliplatin, fludarabine, and rituximab) | [69] |
| Burkitt and Burkitt-like lymphoma | induction | CALGB 9251 | Cycles 2, 4, and 6: 150 mg/m2/day continuous infusion days 4 and 5 | [52,70] |
| CODOX-M/IVAC | Adults ≤ 65 years: Cycles 2 and 4 (IVAC): 2000 mg/m2 over 3 h every 12 h days 1 and 2 (total of 4 doses/cycle) (1000 mg/m2 if age > 65) (IVAC is combination with ifosfamide, mesna, and etoposide; IVAC alternates with CODOX-M) | [53] | ||
| Mantle cell lymphoma | induction | R-BAC | cytarabine 800 mg/m2 IV on days 2 to 4) every 28 days for four to six cycles (in combination with rituximab and bendamustine) | [57] |
| CNS lymphoma, primary | induction | I.V.: 2000 mg/m2 over 1 h every 12 h days 2 and 3 (total of 4 doses) every 3 weeks (in combination with methotrexate and followed by whole brain irradiation) for a total of 4 courses | [66] | |
| Hodgkin and non-Hodgkin lymphoma | relapse or progression | DHAP | 2000 mg/m2 over 3 h every 12 h day 2 (total of 2 doses/cycle) for 2 cycles (in combination with dexamethasone and cisplatin) | [62,71] |
| ESHAP | 2000 mg/m2 day 5 (in combination with etoposide, methylprednisolone, and cisplatin) every 3 to 4 weeks for 3 or 6 cycles | [60,63] | ||
| DHAOX | 2000 mg/m2 every 12 h day 2 (in combination with dexamethasone, and oxaliplatin) every 3 weeks | [61] | ||
| BEAM | transplant preparative regimen: 200 mg/m2 twice daily for 4 days beginning 5 days prior to transplant (in combination with carmustine, etoposide, and melphalan) | [64] |
| Factor | Result |
|---|---|
| Mechanism of action | The active form, cytarabine triphosphate, induces miscoding after incorporation into DNA, terminates DNA chain elongation and inhibits DNA polymerase |
| Cellular influx and transporters | Free diffusion into the cell if plasma concentration >10 μM which is achieved with high-dose cytarabine Nucleoside transporters required if plasma concentration <1 μM which is achieved with 100–200 mg/m2 daily (hENT1 responsible for up to 80% influx; hENT2, hCNT3, ABCC10 and ABCC11 transporters also involved) |
| Metabolism enzymes | Stepwise phosphorylation of cytarabine in tumor cells at the 5′position of arabinoside up to triphosphate form by Deoxycytidine kinase (→cytarabine monophosphate) Deoxycytidine momophosphate kinase (→cytarabine diphosphate) Nucleoside diphosphate kinase (→cytarabine triphosphate) Irreversible deamination of cytarabine and its monophosphate intermediateinto inactive derivatives by Cytidinedeaminase (cytarabine→uracilarabinoside) Deoxycytidine momophosphate deaminase (cytarabine monophosphate →arabinosyluracil monophosphate) Catalytic dephosphorilation of cytarabine monophosphate by Cytosolic5′-nucleotidase II(cytarabinemonophosphate→cytarabine) |
| Pharmacokinetics | Plasma t ½α 7–20 min, t ½β 1–3 h; CSFt ½ 2–6 h |
| Elimination | Dose rate ranging from 86% to 96% is deaminated to inactive uracil arabinoside Deamination in liver, plasma, and peripheral tissue Intrathecal administration results in little conversion to uracil arabinoside due to the low level of deaminase in the cerebral spinal fluid Escretion: urine (~80%; 90% as metabolite uracil arabinoside) within 24 h |
| Drug interactions | Methotrexate and fludarabine increase cytarabine triphosphate formation Cytarabine blocks DNA repair and enhances activity of alkylating agents |
| Myelosuppression | Neutropenia (onset: 1–7 days; nadir [biphasic]: 7–9 days and at 15–24 days; recovery [biphasic]: 9–12 days and at 24–34 days); thrombocytopenia (onset: 5 days; nadir: 12–15 days; recovery 15–25 days); megaloblastosis |
| Severe adverse events of high dose regimen | Neurologic: cerebellar toxicity, coma, personality changes, cognitive dysfunctions, neurotoxicity (up to 55% in patients with renal impairment), motor and sensor peripheral neuropathy Gastrointestinal: esophageal and intestinalulceration, bowel necrosis, hepatic sinusoidal obstruction syndrome, pancreatitis, pneumatosis cystoides intestinalis Cutaneous: skin eruptions and ulceration, exanthematous pustulosis Ocular: vision loss, keratitis, hemorrhagic conjunctivitis Cardiopulmonary: non-cardiogenic pulmonary edema, syndrome of sudden respiratory distress, interstitial pneumonitis, cardiomiopathy (in combination with cyclophosphamide) Sepsis |
| 1. Deficiency of cytarabine cellular uptake and retention from |
|
|
|
| 2. Overexpression of enzymes inactivating cytarabine, primarily CDA, CMPD and NT5C2 |
| 3. Increased cellular dCTP pools following overexpression of RNR or CTPS and followed in turn by |
|
|
| 4. Altered DNA polymerase and increased DNA repairgenes i.e., XRCC and ERCC groups. |
| GENE (Protein) | DNA Variants | Minor Allele Frequency * | Phenotype | Activities Related SNP | Ref. | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs # | Nucleotide | Location (Codon) | Afr | Eur | Asn | Clinical | Cell Model | ||||
| Transport | SLC29A1 (hENT1) | rs747199 | −706G>C | 5′UTR | (C) 0 | (C) 0.21 | (C) 0.23 | increase in hENT1 mRNA expression in PBMCs and cytarabine uptake | [145] | ||
| SLC28A3 (hCNT3) | rs11140500 | T>C | 5′UTR | (T) 0 | (T) 0.01 | (T) 0.29 | [96] | ||||
| Activation | DCK (DCK) | NA/ rs2306744 | −360C>G/−201C>T | 5′UTR | NA | NA | NA | mRNA expression −360GG/−201TT haplotype shown good clinical response in AML | altered enzyme kinetics, increased cytarabine triphosphate intracellular concentrations | [146] | |
| rs66878317 | 70A>G | Exon 1 (Ile24Val) | (G) 0 | (G) 0 | (G) 0.03 | Altered substrate kinetics | [147] | ||||
| rs67437265 | 364C>T | Exon 3 (Pro122Ser) | (T) 0.06 | (T) 0.01 | (T) 0.04 | variant Pro122 shown lower enzyme activity | [147] | ||||
| NDK-NME1 (NDK) | rs2302254 | 835C>T | 5′UTR | (T) 0.37 | (T) 0.18 | (T) 0.25 | −835 T/T increased risk of neurotoxicity. | [148] | |||
| RRM1 (RRM1) | rs1561876 | −2993A>G | 3′UTR | (G) 0.72 | (G) 0.12 | (G) 0.21 | cytarabine triphosphate levels Increased, response and survival improved | [149] | |||
| Deactivation | CDA (CDA) | rs532545 | −451C>T | 5′UTR | (T) 0.07 | (T) 0.32 | (T) 0.13 | Decreased expression of CDA | PBMC from healthy donors, report that among individuals carrying two *2A alleles (seeTable 1), cytarabine-induced toxicity was approximately 53% higher whencompared with carriers of no *2A alleles and nearly 74% higher compared with carries of two wild-type *1A alleles. | Low mRNA level. High level of cytarabine triphosphate resulting in bone marrow depression | [16] |
| rs602950 | −92A>G | 5′UTR | (G) 0.07 | (G) 0.32 | (G) 0.13 | ||||||
| rs3215400 | −31Del C | 5′UTR | (C) 0.33 | (C) 0.43 | (C) 0.44 | del/del shown 1.37fold increased expression than ins C/insC Parmar | |||||
| rs2072671 | 79A>C | Exon 1 (Lys27Gln) | (C) 0.08 | (C) 0.33 | (C) 0.13 | Low enzyme activity in Lys27Lys than Gln27Gln | Cytidine is the 2.4 fold lower Km for Lys27Lys than Gln27Gln in European | [150] | |||
| rs1048977 | 435T>C | Exon 4 (Thr145Thr) | (T) 0.37 | (T) 0.33 | (T) 0.25 | Haplotype CDA *2A(435A) display higher CDA activity compared with others | |||||
| NT5C2 (NT5C2) | rs11598702 | 175+1178A>G | 5′UTR | (C) 0.19 | (C) 0.35 | (C) 0.22 | Low mRNA expression and cytarabine sensitivity in cells derived from AML patients | Low mRNA DFS 17.5 months vs 11 months in AML | Low enzyme activity | [151] | |
| NT5C3 (NT5C3) | rs3750117 | 276T>C | Tyr92Tyr | (T) 0.17 | (T) 0.29 | (T) 0.5 | In PBMCs haplotypes T276/H283 and T276/C306 decreased enzyme activity | [152] | |||
| DNA repair | XRCC1 (XR_) | rs25487 | 28152G>A | Exon (Arg399Gln) | (T) 0.12 | (T) 0.35 | (T) 0.25 | Mutations are correlated to prediction of better treatment outcomes in patients with AML | Low activity in Base excision DNA repair | [153] | |
| ERCC2 (XDP) | rs13181 | 35931A>C | Exon (Lys751Gln) | (G) 0.21 | (G) 0.38 | (G) 0.09 | Low activity in Nucleotide excision DNA repair | [153] | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Francia, R.; Crisci, S.; De Monaco, A.; Cafiero, C.; Re, A.; Iaccarino, G.; De Filippi, R.; Frigeri, F.; Corazzelli, G.; Micera, A.; et al. Response and Toxicity to Cytarabine Therapy in Leukemia and Lymphoma: From Dose Puzzle to Pharmacogenomic Biomarkers. Cancers 2021, 13, 966. https://doi.org/10.3390/cancers13050966
Di Francia R, Crisci S, De Monaco A, Cafiero C, Re A, Iaccarino G, De Filippi R, Frigeri F, Corazzelli G, Micera A, et al. Response and Toxicity to Cytarabine Therapy in Leukemia and Lymphoma: From Dose Puzzle to Pharmacogenomic Biomarkers. Cancers. 2021; 13(5):966. https://doi.org/10.3390/cancers13050966
Chicago/Turabian StyleDi Francia, Raffaele, Stefania Crisci, Angela De Monaco, Concetta Cafiero, Agnese Re, Giancarla Iaccarino, Rosaria De Filippi, Ferdinando Frigeri, Gaetano Corazzelli, Alessandra Micera, and et al. 2021. "Response and Toxicity to Cytarabine Therapy in Leukemia and Lymphoma: From Dose Puzzle to Pharmacogenomic Biomarkers" Cancers 13, no. 5: 966. https://doi.org/10.3390/cancers13050966
APA StyleDi Francia, R., Crisci, S., De Monaco, A., Cafiero, C., Re, A., Iaccarino, G., De Filippi, R., Frigeri, F., Corazzelli, G., Micera, A., & Pinto, A. (2021). Response and Toxicity to Cytarabine Therapy in Leukemia and Lymphoma: From Dose Puzzle to Pharmacogenomic Biomarkers. Cancers, 13(5), 966. https://doi.org/10.3390/cancers13050966







