The Potential of Artificial Intelligence to Detect Lymphovascular Invasion in Testicular Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
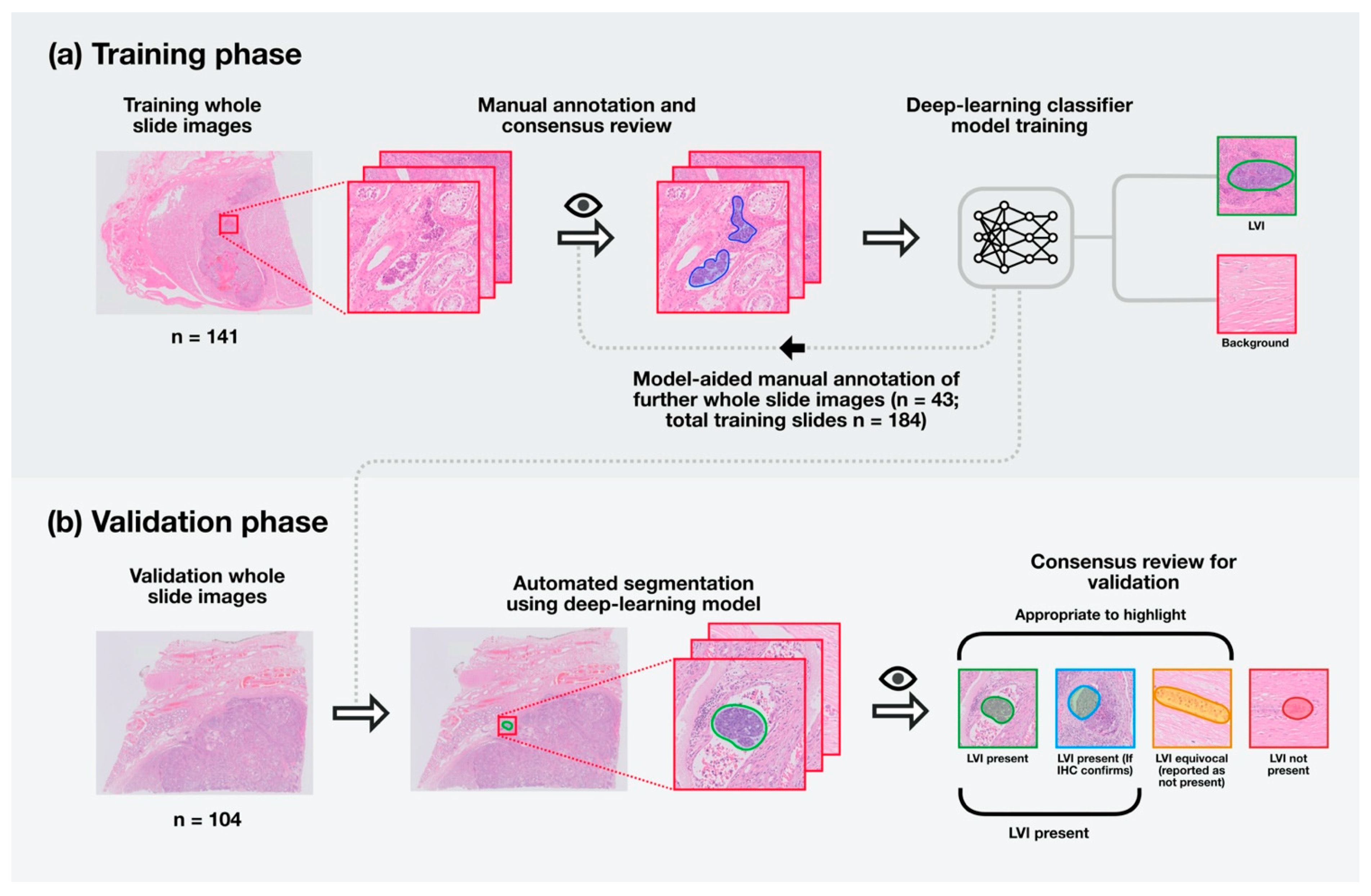
2. Materials and Methods
2.1. Patients
2.2. Digitisation of Slides
2.3. Training the Model
2.4. Assessment of Model Performance
2.5. Statistical Analysis
3. Results
3.1. Classifier Precision
3.2. Interobserver Variability
3.3. Ranked Retrieval Results
3.4. Metastatic Disease
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AI | Artificial intelligence |
| AIDA | Annotation of Image Data by Assignments |
| CNN | Convolutional neural networks |
| DP | Digital pathology |
| GCNIS | Germ cell neoplasia in-situ |
| H&E | Haematoxylin and eosin |
| IHC | Immunohistochemistry |
| ICCR | International Collaboration on Cancer Reporting |
| LVI | Lymphovascular invasion |
| NSGCT | Non-seminomatous germ cell tumour |
| ORB | Oxford Radcliffe Biobank |
| TGCT | Testicular germ cell tumour |
| TIL | Tumour infiltrating lymphocyte |
| WHO | World Health Organisation |
References
- Cheng, L.; Albers, P.; Berney, D.M.; Feldman, D.R.; Daugaard, G.; Gilligan, T.; Looijenga, L.H.J. Testicular Cancer. Nat. Rev. Dis. Primer 2018, 4, 29. [Google Scholar] [CrossRef] [PubMed]
- Fukawa, T.; Kanayama, H. Current Knowledge of Risk Factors for Testicular Germ Cell Tumors. Int. J. Urol. 2018, 25, 337–344. [Google Scholar] [CrossRef] [Green Version]
- Barrett, M.T.; Lenkiewicz, E.; Malasi, S.; Stanton, M.; Slack, J.; Andrews, P.; Pagliaro, L.; Bryce, A.H. Clonal Analyses of Refractory Testicular Germ Cell Tumors. PLoS ONE 2019, 14, e0213815. [Google Scholar] [CrossRef] [Green Version]
- Moch, H.; Humphrey, P.; Ulbright, T.M.; Reuter, V. WHO Classification of Tumors of the Urinary System and Male Genital Organs; France International Agency for Research on Cancer (IARC): Lyon, France, 2016. [Google Scholar]
- International Germ Cell Consensus Classification: A Prognostic Factor-Based Staging System for Metastatic Germ Cell Cancers. International Germ Cell Cancer Collaborative Group. J. Clin. Oncol. 1997, 15, 594–603. [CrossRef]
- Berney, D.M.; Comperat, E.; Feldman, D.R.; Hamilton, R.J.; Idrees, M.T.; Samaratunga, H.; Tickoo, S.K.; Yilmaz, A.; Srigley, J.R. Datasets for the Reporting of Neoplasia of the Testis: Recommendations from the International Collaboration on Cancer Reporting. Histopathology 2019, 74, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Berney, D.M.; Verrill, C. Royal College of Pathologists’ Standards and Datasets for Reporting Cancers: Dataset for the Histological Reporting of Testicular Neoplasms, 4th ed.; The Royal College of Pathologists: London, UK, 2020. [Google Scholar]
- Mortensen, M.S.; Lauritsen, J.; Gundgaard, M.G.; Agerbæk, M.; Holm, N.V.; Christensen, I.J.; von der Maase, H.; Daugaard, G. A Nationwide Cohort Study of Stage I Seminoma Patients Followed on a Surveillance Program. Eur. Urol. 2014, 66, 1172–1178. [Google Scholar] [CrossRef]
- Chung, P.; Daugaard, G.; Tyldesley, S.; Atenafu, E.G.; Panzarella, T.; Kollmannsberger, C.; Warde, P. Evaluation of a Prognostic Model for Risk of Relapse in Stage I Seminoma Surveillance. Cancer Med. 2015, 4, 155–160. [Google Scholar] [CrossRef] [PubMed]
- Kamba, T.; Kamoto, T.; Okubo, K.; Teramukai, S.; Kakehi, Y.; Matsuda, T.; Ogawa, O. Outcome of Different Postorchiectomy Management for Stage I Seminoma: Japanese Multiinstitutional Study Including 425 Patients. Int. J. Urol. 2010, 17, 980–987. [Google Scholar] [CrossRef] [Green Version]
- Scandura, G.; Wagner, T.; Beltran, L.; Alifrangis, C.; Shamash, J.; Berney, D.M. Pathological Predictors of Metastatic Disease in Testicular Non-Seminomatous Germ Cell Tumors: Which Tumor-Node-Metastasis Staging System? Mod. Pathol. 2020. [Google Scholar] [CrossRef]
- Aparicio, J.; Maroto, P.; García del Muro, X.; Sánchez-Muñoz, A.; Gumà, J.; Margelí, M.; Sáenz, A.; Sagastibelza, N.; Castellano, D.; Arranz, J.A.; et al. Prognostic Factors for Relapse in Stage I Seminoma: A New Nomogram Derived from Three Consecutive, Risk-Adapted Studies from the Spanish Germ Cell Cancer Group (SGCCG). Ann. Oncol. 2014, 25, 2173–2178. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, A.; Cheng, T.; Zhang, J.; Trpkov, K. Testicular Hilum and Vascular Invasion Predict Advanced Clinical Stage in Nonseminomatous Germ Cell Tumors. Mod. Pathol. 2013, 26, 579–586. [Google Scholar] [CrossRef]
- Feldman, D.R. Treatment Options for Stage I Nonseminoma. J. Clin. Oncol. 2014, 32, 3797–3800. [Google Scholar] [CrossRef]
- Blok, J.M.; Pluim, I.; Daugaard, G.; Wagner, T.; Jóźwiak, K.; Wilthagen, E.A.; Looijenga, L.H.J.; Meijer, R.P.; Bosch, J.L.H.R.; Horenblas, S. Lymphovascular Invasion and Presence of Embryonal Carcinoma as Risk Factors for Occult Metastatic Disease in Clinical Stage I Nonseminomatous Germ Cell Tumour: A Systematic Review and Meta-Analysis: Prognostic Value of LVI and EC in CS I NSGCT. BJU Int. 2020, 125, 355–368. [Google Scholar] [CrossRef]
- Verrill, C.; Yilmaz, A.; Srigley, J.R.; Amin, M.B.; Compérat, E.; Egevad, L.; Ulbright, T.M.; Tickoo, S.K.; Berney, D.M.; Epstein, J.I. Reporting and Staging of Testicular Germ Cell Tumors: The International Society of Urological Pathology (ISUP) Testicular Cancer Consultation Conference Recommendations. Am. J. Surg. Pathol. 2017, 41, e22–e32. [Google Scholar] [CrossRef]
- French, B.L.; Zynger, D.L. Do Histopathologic Variables Affect the Reporting of Lymphovascular Invasion in Testicular Germ Cell Tumors? Am. J. Clin. Pathol. 2016, 145, 341–349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lobo, J.; Stoop, H.; Gillis, A.J.M.; Looijenga, L.H.J.; Oosterhuis, W. Interobserver Agreement in Vascular Invasion Scoring and the Added Value of Immunohistochemistry for Vascular Markers to Predict Disease Relapse in Stage I Testicular Nonseminomas. Am. J. Surg. Pathol. 2019, 43, 1711–1719. [Google Scholar] [CrossRef]
- Nicolai, N.; Colecchia, M.; Biasoni, D.; Catanzaro, M.; Stagni, S.; Torelli, T.; Necchi, A.; Piva, L.; Milani, A.; Salvioni, R. Concordance and Prediction Ability of Original and Reviewed Vascular Invasion and Other Prognostic Parameters of Clinical Stage I Nonseminomatous Germ Cell Testicular Tumors After Retroperitoneal Lymph Node Dissection. J. Urol. 2011, 186, 1298–1302. [Google Scholar] [CrossRef]
- Brimo, F.; Srigley, J.; Ryan, C. Chapter 59: Testis. In AJCC Cancer Staging Manual, 8th ed.; Amin, M.B., Edge, S., Greene, F., Eds.; American Joint Commission on Cancer, Springer International Publishing: New York, NY, USA, 2016. [Google Scholar]
- Sobin, L.; Gospodarowicz, M.; Brierley, J. UICC International Union against Cancer. In TNM Classification of Malignant Tumors, 7th ed.; John Wiley and Sons: Hoboken, NJ, USA, 2010. [Google Scholar]
- NHS Long Term Plan 2019. Available online: https://www.longtermplan.nhs.uk/online-version/overview-and-summary/ (accessed on 1 May 2020).
- Williams, B.J.; Bottoms, D.; Treanor, D. Future-Proofing Pathology: The Case for Clinical Adoption of Digital Pathology. J. Clin. Pathol. 2017, 70, 1010–1018. [Google Scholar] [CrossRef] [Green Version]
- UK Government, Office for Life Sciences. Life Sciences: Industrial Strategy. A Report to Government from the Life Sciences Sector. Office for Life Sciences 30th August 2017. Available online: https://www.gov.uk/government/publications/life-sciences-industrial-strategy (accessed on 19 May 2020).
- Niazi, M.K.K.; Parwani, A.V.; Gurcan, M.N. Digital Pathology and Artificial Intelligence. Lancet Oncol. 2019, 20, e253–e261. [Google Scholar] [CrossRef]
- Cancer Research UK (CRUK). Testing Times to Come? An Evaluation of Pathology Capacity across the UK. Available online: https://www.cancerresearchuk.org/sites/default/files/testing_times_to_come_nov_16_cruk.pdf (accessed on 1 May 2020).
- Pantanowitz, L.; Quiroga-Garza, G.M.; Bien, L.; Heled, R.; Laifenfeld, D.; Linhart, C.; Sandbank, J.; Albrecht Shach, A.; Shalev, V.; Vecsler, M.; et al. An Artificial Intelligence Algorithm for Prostate Cancer Diagnosis in Whole Slide Images of Core Needle Biopsies: A Blinded Clinical Validation and Deployment Study. Lancet Digit. Health 2020, 2, e407–e416. [Google Scholar] [CrossRef]
- Campanella, G.; Hanna, M.G.; Geneslaw, L.; Miraflor, A.; Werneck Krauss Silva, V.; Busam, K.J.; Brogi, E.; Reuter, V.E.; Klimstra, D.S.; Fuchs, T.J. Clinical-Grade Computational Pathology Using Weakly Supervised Deep Learning on Whole Slide Images. Nat. Med. 2019, 25, 1301–1309. [Google Scholar] [CrossRef]
- Turkki, R.; Byckhov, D.; Lundin, M.; Isola, J.; Nordling, S.; Kovanen, P.E.; Verrill, C.; von Smitten, K.; Joensuu, H.; Lundin, J.; et al. Breast Cancer Outcome Prediction with Tumour Tissue Images and Machine Learning. Breast Cancer Res. Treat. 2019, 177, 41–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bychkov, D.; Linder, N.; Turkki, R.; Nordling, S.; Kovanen, P.E.; Verrill, C.; Walliander, M.; Lundin, M.; Haglund, C.; Lundin, J. Deep Learning Based Tissue Analysis Predicts Outcome in Colorectal Cancer. Sci. Rep. 2018, 8, 3395. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Shi, J.; Ye, Z.; Dong, D.; Yu, D.; Zhou, M.; Liu, Y.; Gevaert, O.; Wang, K.; Zhu, Y.; et al. Predicting EGFR Mutation Status in Lung Adenocarcinoma on Computed Tomography Image Using Deep Learning. Eur. Respir. J. 2019, 53, 1800986. [Google Scholar] [CrossRef]
- Sirinukunwattana, K.; Domingo, E.; Richman, S.D.; Redmond, K.L.; Blake, A.; Verrill, C.; Leedham, S.J.; Chatzipli, A.; Hardy, C.; Whalley, C.M.; et al. Image-Based Consensus Molecular Subtype (ImCMS) Classification of Colorectal Cancer Using Deep Learning. Gut 2020, 70, 544–554. [Google Scholar] [CrossRef] [PubMed]
- Steiner, D.F.; MacDonald, R.; Liu, Y.; Truszkowski, P.; Hipp, J.D.; Gammage, C.; Thng, F.; Peng, L.; Stumpe, M.C. Impact of Deep Learning Assistance on the Histopathologic Review of Lymph Nodes for Metastatic Breast Cancer. Am. J. Surg. Pathol. 2018, 42, 1636–1646. [Google Scholar] [CrossRef]
- Ehteshami Bejnordi, B.; Veta, M.; Johannes van Diest, P.; van Ginneken, B.; Karssemeijer, N.; Litjens, G.; van der Laak, J.A.W.M.; the CAMELYON16 Consortium; Hermsen, M.; Manson, Q.F.; et al. Diagnostic Assessment of Deep Learning Algorithms for Detection of Lymph Node Metastases in Women With Breast Cancer. JAMA 2017, 318, 2199. [Google Scholar] [CrossRef]
- Linder, N.; Taylor, J.C.; Colling, R.; Pell, R.; Alveyn, E.; Joseph, J.; Protheroe, A.; Lundin, M.; Lundin, J.; Verrill, C. Deep Learning for Detecting Tumour-Infiltrating Lymphocytes in Testicular Germ Cell Tumours. J. Clin. Pathol. 2019, 72, 157–164. [Google Scholar] [CrossRef]
- Aberdeen, Alan Annotation of Image Data by Assignment. Available online: https://github.com/alanaberdeen/AIDA (accessed on 24 January 2021).
- Chen, L.-C.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-Decoder with Atrous Separable Convolution for Semantic Image Segmentation. In Proceedings of the Computer Vision—ECCV 2018, Munich, Germany, 8–14 September 2018; Ferrari, V., Hebert, M., Sminchisescu, C., Weiss, Y., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 833–851. [Google Scholar]
- Landis, J.R.; Koch, G.G. The Measurement of Observer Agreement for Categorical Data. Biometrics 1977, 33, 159. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.; James, J.; Thomas, G.; West, N.; Jones, L.; Lee, J.; Oien, K.; Freeman, A.; Craig, C.; Sloan, P.; et al. Quality Assurance Guidance for Scoring and Reporting for Pathologists and Laboratories Undertaking Clinical Trial Work. J. Pathol. Clin. Res. 2019, 5, 91–99. [Google Scholar] [CrossRef] [Green Version]
- Bryant, R.J.; Schmitt, A.J.; Roberts, I.S.D.; Gill, P.S.; Browning, L.; Brewster, S.F.; Hamdy, F.C.; Verrill, C. Variation between Specialist Uropathologists in Reporting Extraprostatic Extension after Radical Prostatectomy. J. Clin. Pathol. 2015, 68, 465–472. [Google Scholar] [CrossRef]
- Evans, A.J.; Henry, P.C.; Van der Kwast, T.H.; Tkachuk, D.C.; Watson, K.; Lockwood, G.A.; Fleshner, N.E.; Cheung, C.; Belanger, E.C.; Amin, M.B.; et al. Interobserver Variability Between Expert Urologic Pathologists for Extraprostatic Extension and Surgical Margin Status in Radical Prostatectomy Specimens. Am. J. Surg. Pathol. 2008, 32, 1503–1512. [Google Scholar] [CrossRef]
- Sirinukunwattana, K.; Aberdeen, A.; Theissen, H.; Sousos, N.; Psaila, B.; Mead, A.J.; Turner, G.D.H.; Rees, G.; Rittscher, J.; Royston, D. Artificial Intelligence–Based Morphological Fingerprinting of Megakaryocytes: A New Tool for Assessing Disease in MPN Patients. Blood Adv. 2020, 4, 3284–3294. [Google Scholar] [CrossRef]
- Colling, R.; Pitman, H.; Oien, K.; Rajpoot, N.; Macklin, P.; CM-Path AI in Histopathology Working Group; Bachtiar, V.; Booth, R.; Bryant, A.; Bull, J.; et al. Artificial Intelligence in Digital Pathology: A Roadmap to Routine Use in Clinical Practice. J. Pathol. 2019, 249, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Litchfield, K.; Summersgill, B.; Yost, S.; Sultana, R.; Labreche, K.; Dudakia, D.; Renwick, A.; Seal, S.; Al-Saadi, R.; Broderick, P.; et al. Whole-Exome Sequencing Reveals the Mutational Spectrum of Testicular Germ Cell Tumours. Nat. Commun. 2015, 6, 5973. [Google Scholar] [CrossRef]
- Shen, H.; Shih, J.; Hollern, D.P.; Wang, L.; Bowlby, R.; Tickoo, S.K.; Thorsson, V.; Mungall, A.J.; Newton, Y.; Hegde, A.M.; et al. Integrated Molecular Characterization of Testicular Germ Cell Tumors. Cell Rep. 2018, 23, 3392–3406. [Google Scholar] [CrossRef]
- Taylor-Weiner, A.; Zack, T.; O’Donnell, E.; Guerriero, J.L.; Bernard, B.; Reddy, A.; Han, G.C.; AlDubayan, S.; Amin-Mansour, A.; Schumacher, S.E.; et al. Genomic Evolution and Chemoresistance in Germ-Cell Tumours. Nature 2016, 540, 114–118. [Google Scholar] [CrossRef] [PubMed]
- Looijenga, L.H.J.; Zafarana, G.; Grygalewicz, B.; Summersgill, B.; Debiec-Rychter, M.; Veltman, J.; Schoenmakers, E.F.P.M.; Rodriguez, S.; Jafer, O.; Clark, J.; et al. Role of Gain of 12p in Germ Cell Tumour Development. APMIS 2003, 111, 161–173. [Google Scholar] [CrossRef] [PubMed]
- Chieffi, P.; De Martino, M.; Esposito, F. New Anti-Cancer Strategies in Testicular Germ Cell Tumors. Recent Patents Anticancer Drug Discov. 2019, 14, 53–59. [Google Scholar] [CrossRef] [PubMed]



| Cohort Summary | Training Set | Validation Set |
|---|---|---|
| Cases | 19 | 10 |
| Seminoma | 11 | 5 |
| Non-seminoma | 8 | 5 |
| Whole slide images | 184 | 118 |
| Round 1 | 141 | - |
| Round 2 | 43 | |
| Total initially annotated LVI Candidate foci | 471 | - |
| Round 1 | 350 | |
| Round 2 | 121 | |
| Total foci used for training (after consensus review) | 272 | - |
| Round 1 | 196 | |
| Round 2 | 76 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghosh, A.; Sirinukunwattana, K.; Khalid Alham, N.; Browning, L.; Colling, R.; Protheroe, A.; Protheroe, E.; Jones, S.; Aberdeen, A.; Rittscher, J.; et al. The Potential of Artificial Intelligence to Detect Lymphovascular Invasion in Testicular Cancer. Cancers 2021, 13, 1325. https://doi.org/10.3390/cancers13061325
Ghosh A, Sirinukunwattana K, Khalid Alham N, Browning L, Colling R, Protheroe A, Protheroe E, Jones S, Aberdeen A, Rittscher J, et al. The Potential of Artificial Intelligence to Detect Lymphovascular Invasion in Testicular Cancer. Cancers. 2021; 13(6):1325. https://doi.org/10.3390/cancers13061325
Chicago/Turabian StyleGhosh, Abhisek, Korsuk Sirinukunwattana, Nasullah Khalid Alham, Lisa Browning, Richard Colling, Andrew Protheroe, Emily Protheroe, Stephanie Jones, Alan Aberdeen, Jens Rittscher, and et al. 2021. "The Potential of Artificial Intelligence to Detect Lymphovascular Invasion in Testicular Cancer" Cancers 13, no. 6: 1325. https://doi.org/10.3390/cancers13061325
APA StyleGhosh, A., Sirinukunwattana, K., Khalid Alham, N., Browning, L., Colling, R., Protheroe, A., Protheroe, E., Jones, S., Aberdeen, A., Rittscher, J., & Verrill, C. (2021). The Potential of Artificial Intelligence to Detect Lymphovascular Invasion in Testicular Cancer. Cancers, 13(6), 1325. https://doi.org/10.3390/cancers13061325







