Automated Detection and Classification of Desmoplastic Reaction at the Colorectal Tumour Front Using Deep Learning
Abstract
:Simple Summary
Abstract
1. Introduction
2. Results
2.1. Patients’ Characteristics
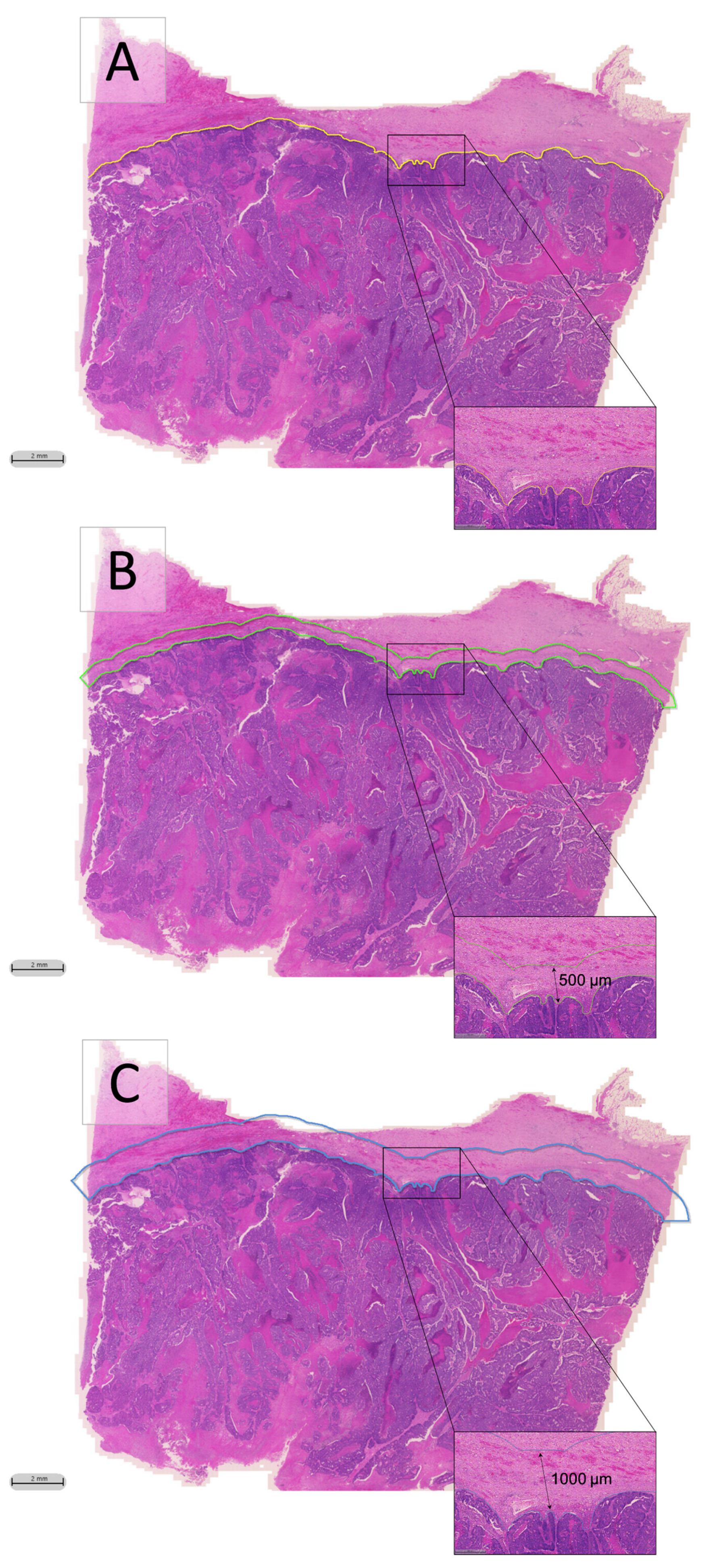
2.2. Classifier Accuracy Evaluation
2.3. Automated DR Classification
2.4. Survival Analysis
3. Discussion
4. Materials and Methods
4.1. Patient Material
4.2. Manual Histologic Evaluation of Desmoplastic Reaction
4.3. Training the Deep Learning Classifier
4.4. Automated DR Classification
4.5. Survival Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Fotheringham, S.; Mozolowski, G.A.; Murray, E.M.A.; Kerr, D.J. Challenges and solutions in patient treatment strategies for stage II colon cancer. Gastroenterol. Rep. 2019, 7, 151–161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, Y.; Jiang, Y.-C.; Sun, C.-K.; Chen, Q.-M. Role of the tumor microenvironment in tumor progression and the clinical applications (Review). Oncol. Rep. 2016, 35, 2499–2515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nearchou, I.P.; Gwyther, B.M.; Georgiakakis, E.C.T.; Gavriel, C.G.; Lillard, K.; Kajiwara, Y.; Ueno, H.; Harrison, D.J.; Caie, P.D. Spatial immune profiling of the colorectal tumor microenvironment predicts good outcome in stage II patients. NPJ Digit. Med. 2020, 3, 1–10. [Google Scholar] [CrossRef]
- Ueno, H.; Sekine, S.; Oshiro, T.; Kanemitsu, Y.; Hamaguchi, T.; Shida, D.; Takashima, A.; Ishiguro, M.; Ito, E.; Hashiguchi, Y.; et al. Disentangling the prognostic heterogeneity of stage III colorectal cancer through histologic stromal categorization. Surgery 2018, 163, 777–783. [Google Scholar] [CrossRef]
- Ueno, H.; Kanemitsu, Y.; Sekine, S.; Ishiguro, M.; Ito, E.; Hashiguchi, Y.; Kondo, F.; Shimazaki, H.; Mochizuki, S.; Kajiwara, Y.; et al. Desmoplastic Pattern at the Tumor Front Defines Poor-prognosis Subtypes of Colorectal Cancer. Am. J. Surg. Pathol. 2017, 41, 1506–1512. [Google Scholar] [CrossRef]
- Nearchou, I.P.; Kajiwara, Y.; Mochizuki, S.; Harrison, D.J.; Caie, P.D.; Ueno, H. Novel Internationally Verified Method Reports Desmoplastic Reaction as the Most Significant Prognostic Feature for Disease-specific Survival in Stage II Colorectal Cancer. Am. J. Surg. Pathol. 2019, 43, 1239–1248. [Google Scholar] [CrossRef]
- Ueno, H.; Shinto, E.; Hashiguchi, Y.; Shimazaki, H.; Kajiwara, Y.; Sueyama, T.; Yamamoto, J.; Hase, K. In rectal cancer, the type of desmoplastic response after preoperative chemoradiotherapy is associated with prognosis. Virchows Arch. 2015, 466, 655–663. [Google Scholar] [CrossRef]
- Konishi, T.; Shimada, Y.; Lee, L.H.; Cavalcanti, M.S.; Hsu, M.; Smith, J.J.; Nash, G.M.; Temple, L.K.; Guillem, J.G.; Paty, P.B.; et al. Poorly Differentiated Clusters Predict Colon Cancer Recurrence. Am. J. Surg. Pathol. 2018, 42, 705–714. [Google Scholar] [CrossRef]
- Nearchou, I.P.; Lillard, K.; Gavriel, C.G.; Ueno, H.; Harrison, D.J.; Caie, P.D. Automated analysis of lymphocytic infiltration, tumor budding, and their spatial relationship improves prognostic accuracy in colorectal cancer. Cancer Immunol. Res. 2019, 7, 609–620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caie, P.D.; Zhou, Y.; Turnbull, A.K.; Oniscu, A.; Harrison, D.J. Novel histopathologic feature identified through image analysis augments stage II colorectal cancer clinical reporting. Oncotarget 2016, 7, 44381–44394. [Google Scholar] [CrossRef] [Green Version]
- Yue, X.; Dimitriou, N.; Caie, P.; Harrison, D.; Arandjelovic, O. Colorectal Cancer Outcome Prediction from H&E Whole Slide Images using Machine Learning and Automatically Inferred Phenotype Profiles. arXiv 2019, arXiv:1902.03582. [Google Scholar]
- Wang, D.; Khosla, A.; Gargeya, R.; Irshad, H.; Beck, A.H. Deep Learning for Identifying Metastatic Breast Cancer. arXiv 2016, arXiv:1606.05718. [Google Scholar]
- Liu, Y.; Gadepalli, K.; Norouzi, M.; Dahl, G.E.; Kohlberger, T.; Boyko, A.; Venugopalan, S.; Timofeev, A.; Nelson, P.Q.; Corrado, G.S. Detecting Cancer Metastases on Gigapixel Pathology Images. arXiv 2017, arXiv:1703.02442. [Google Scholar]
- Lucas, M.; Jansen, I.; Savci-Heijink, C.D.; Meijer, S.L.; De Boer, O.J.; Van Leeuwen, T.G.; De Bruin, D.M.; Marquering, H.A. Deep learning for automatic Gleason pattern classification for grade group determination of prostate biopsies. Virchows Arch. 2019, 475, 77–83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ueno, H.; Jones, A.M.; Wilkinson, K.H.; Jass, J.R.; Talbot, I.C. Histological categorisation of fibrotic cancer stroma in advanced rectal cancer. Gut 2004, 53, 581–586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peddareddigari, V.G.; Wang, D.; Dubois, R.N. The tumor microenvironment in colorectal carcinogenesis. Cancer Microenviron. 2010, 3, 149–166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valkenburg, K.C.; De Groot, A.E.; Pienta, K.J. Targeting the tumour stroma to improve cancer therapy. Nat. Rev. Clin. Oncol. 2018, 15, 366–381. [Google Scholar] [CrossRef]
- Ueno, H.; Shinto, E.; Shimazaki, H.; Kajiwara, Y.; Sueyama, T.; Yamamoto, J.; Hase, K. Histologic Categorization of Desmoplastic Reaction: Its Relevance to the Colorectal Cancer Microenvironment and Prognosis. Ann. Surg. Oncol. 2014, 22, 1504–1512. [Google Scholar] [CrossRef]
- Tellez, D.; Litjens, G.; Bándi, P.; Bulten, W.; Bokhorst, J.-M.; Ciompi, F.; van der Laak, J. Quantifying the effects of data augmentation and stain color normalization in convolutional neural networks for computational pathology. Med Image Anal. 2019, 58, 101544. [Google Scholar] [CrossRef] [Green Version]
- Ueno, H.; Kanemitsu, Y.; Sekine, S.; Ishiguro, M.; Ito, E.; Hashiguchi, Y.; Kondo, F.; Shimazaki, H.; Kajiwara, Y.; Okamoto, K.; et al. A Multicenter Study of the Prognostic Value of Desmoplastic Reaction Categorization in Stage II Colorectal Cancer. Am. J. Surg. Pathol. 2019, 43, 1015–1022. [Google Scholar] [CrossRef] [PubMed]
- Marshall, J. Risk Assessment in Stage II Colorectal Cancer. Oncology 2010, 24, 9–13. [Google Scholar]
- To, K.K.; Tong, C.W.; Wu, M.; Cho, W.C. MicroRNAs in the prognosis and therapy of colorectal cancer: From bench to bedside. World J. Gastroenterol. 2018, 24, 2949–2973. [Google Scholar] [CrossRef] [PubMed]
- JCOG1805: Randomized Controlled Study of Adjuvant Chemotherapy for Stage II Colorectal Cancer Patients at High Risk of Developing Recurrence According to T-Stage and Three Selected Pathological Factors (PanDRa-BD). Available online: https://jrct.niph.go.jp/en-latest-detail/jRCTs031190186 (accessed on 17 July 2020).
- Aeffner, F.; Zarella, M.D.; Buchbinder, N.; Bui, M.M.; Goodman, M.R.; Hartman, D.J.; Lujan, G.M.; Molani, M.A.; Parwani, A.V.; Lillard, K.; et al. Introduction to digital image analysis in whole-slide imaging: A white paper from the digital pathology association. J. Pathol. Inform. 2019, 10, 9. [Google Scholar] [CrossRef]
- FDA Allows Marketing of First Whole Slide Imaging System for Digital Pathology|FDA. Available online: https://www.fda.gov/news-events/press-announcements/fda-allows-marketing-first-whole-slide-imaging-system-digital-pathology (accessed on 20 June 2020).
- Dimitriou, N.; Arandjelović, O.; Caie, P.D. Deep Learning for Whole Slide Image Analysis: An Overview. Front. Med. 2019, 6, 264. [Google Scholar] [CrossRef] [Green Version]
- Puppa, G.; Senore, C.; Sheahan, K.; Vieth, M.; Lugli, A.; Zlobec, I.; Pecori, S.; Wang, L.M.; Langner, C.; Mitomi, H.; et al. Diagnostic Reproducibility of Tumour Budding in Colorectal Cancer: A Multicentre, Multinational Study Using Virtual Microscopy. Histopathology 2012, 61, 562–575. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; van der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the IEEE conference on computer vision and pattern recognition, Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- Aishima, S.-I.; Taguchi, K.-I.; Terashi, T.; Matsuura, S.; Shimada, M.; Tsuneyoshi, M. Tenascin Expression at the Invasive Front Is Associated with Poor Prognosis in Intrahepatic Cholangiocarcinoma. Mod. Pathol. 2003, 16, 1019–1027. [Google Scholar] [CrossRef] [Green Version]
- Kassambara, A.; Kosinski, M.; Biecek, P.; Fabian, S. Package ‘Survminer’ Type Package Title Drawing Survival Curves Using ‘ggplot2’. 2019. Available online: https://rpkgs.datanovia.com/survminer/index.html (accessed on 10 June 2020).
- RStudio Team. RStudio Server: Integrated Development for R; RStudio, Inc.: Boston, MA, USA, 2016; Available online: http://www.rstudio.com/ (accessed on 9 October 2019).
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- IBM Corp. IBM SPSS Statistics for Windows, Version 24.0; IBM Corp: Armonk, NY, USA, 2016. Available online: https://www.ibm.com/support/pages/ibm-spss-statistics-24-documentation (accessed on 10 June 2020).


| Features | Training Set (n = 396) | Test Set (n = 132) |
|---|---|---|
| Freq. (%) | Freq. (%) | |
| Age | ||
| ≤70 | 247 (62.4) | 84 (63.6) |
| 71–79 | 108 (27.3) | 40 (30.3) |
| ≥80 | 41 (10.4) | 8 (6.1) |
| Gender | ||
| Male | 237 (59.8) | 78 (59.1) |
| Female | 159 (40.2) | 54 (40.9) |
| pT Stage | ||
| pT3 | 303 (76.5) | 99 (75.0) |
| pT4 | 93 (23.5) | 33 (25.0) |
| pN Stage | ||
| pN0 | 206 (52.0) | 74 (56.1) |
| pN1 | 132 (33.3) | 34 (25.8) |
| pN2 | 58 (14.6) | 24 (18.2) |
| Tumour Site | ||
| Left | 121 (30.6) | 34 (25.8) |
| Right | 114 (18.8) | 39 (29.5) |
| Rectal | 161 (40.7) | 59 (44.7) |
| Differentiation | ||
| Moderate | 206 (52.0) | 59 (44.7) |
| Poor | 25 (6.3) | 18 (13.6) |
| Well | 165 (41.7) | 55 (41.7) |
| Tumour Type | ||
| Adenocarcinoma | 378 (95.5) | 121 (91.7) |
| Mucinous | 18 (4.5) | 11 (8.3) |
| DR type | ||
| Immature | 94 (23.7) | 31 (23.5) |
| Other | 302 (76.3) | 101 (76.5) |
| Features | Cut-Off Value (mm2) |
|---|---|
| Total myxoid stroma area in Margin 1 | 0.27392 |
| Average myxoid stroma area in Margin 1 | 0.00622 |
| Largest single myxoid stroma area in Margin 1 ACP | 1.04863 |
| Largest single myxoid stroma area in Margin 1 MCP | 0.19600 |
| Total myxoid stroma area in Margin 2 | 0.31949 |
| Average myxoid stroma area in Margin 2 | 0.15859 |
| Largest single myxoid stroma area in Margin 2 ACP | 0.17410 |
| Largest single myxoid stroma area in Margin 2 MCP | 0.19600 |
| Features | Freq. (%) | Univariate | |
|---|---|---|---|
| HR (95% CI) | p | ||
| pT Stage | 1.887 (1.091–3.265) | 0.023 | |
| pT3 | 303 (76.5) | ||
| pT4 | 93 (23.5) | ||
| pN Stage | 1.795 (1.297–2.484) | <0.001 | |
| pN0 | 206 (52.0) | ||
| pN1 | 132 (33.3) | ||
| pN2 | 58 (14.6) | ||
| Differentiation | 0.969 (0.744–1.263) | 0.816 | |
| Moderate | 206 (52.0) | ||
| Poor | 25 (6.3) | ||
| Well | 165 (41.7) | ||
| Tumour Type | 2.813 (1.204–6.573) | 0.017 | |
| Adenocarcinoma | 378 (95.5) | ||
| Mucinous | 18 (4.5) | ||
| Total myxoid stroma area in Margin 1 | 2.742 (1.559–4.821) | <0.001 | |
| High | 197 (49.7) | ||
| Low | 199 (50.3) | ||
| Average myxoid stroma area in Margin 1 | 2.439 (1.293–4.600) | 0.006 | |
| High | 250 (63.1) | ||
| Low | 146 (36.9) | ||
| Largest single myxoid stroma area in Margin 1 ACP | 2.546 (1.395–4.646) | 0.002 | |
| Yes | 51 (12.9) | ||
| No | 345 (87.1) | ||
| Largest single myxoid stroma area in Margin 1 MCP | 1.792 (1.071–2.998) | 0.026 | |
| Yes | 169 (42.7) | ||
| No | 227 (57.3) | ||
| Total myxoid stroma area in Margin 2 | 3.527 (1.784–6.973) | <0.001 | |
| High | 239 (60.4) | ||
| Low | 157 (39.6) | ||
| Average myxoid stroma area in Margin 2 | 2.356 (1.293–4.295) | 0.005 | |
| High | 50 (12.6) | ||
| Low | 346 (87.4) | ||
| Largest single myxoid stroma area in Margin 2 ACP | 2.941 (1.612–5.367) | <0.001 | |
| Yes | 216 (54.5) | ||
| No | 180 (45.5) | ||
| Largest single myxoid stroma area in Margin 2 MCP | 2.671 (1.501–4.752) | <0.001 | |
| Yes | 207 (52.3) | ||
| No | 189 (47.7) | ||
| Manually Assessed DR | 2.588 (1.546–4.331) | <0.001 | |
| Immature | 94 (23.7) | ||
| Other | 302 (76.3) | ||
| Variables in the Equation | Multivariate Cox Regression Model | |||
|---|---|---|---|---|
| HR | 95% CI | p | ||
| Lower | Upper | |||
| Total myxoid stroma area in Margin 2 | 3.527 | 1.784 | 6.973 | <0.001 |
| pN Stage | 1.490 | 1.062 | 2.091 | 0.021 |
| Variables not in equation | ||||
| pT Stage | NS | |||
| Differentiation | NS | |||
| Tumour Type | NS | |||
| Manually Assessed DR | NS | |||
| Total myxoid stroma area in Margin 1 | NS | |||
| Average myxoid stroma area in Margin 1 | NS | |||
| Largest single myxoid stroma area in Margin 1 ACP | NS | |||
| Largest single myxoid stroma area in Margin 1 MCP | NS | |||
| Average myxoid stroma area in Margin 2 | NS | |||
| Largest single myxoid stroma area in Margin 2 ACP | NS | |||
| Largest single myxoid stroma area in Margin 2 MCP | NS | |||
| Features | Freq. (%) | Univariate | |
|---|---|---|---|
| HR (95% CI) | p | ||
| Total myxoid stroma area in Margin 2 | 3.743 (1.091–12.830) | 0.036 | |
| High | 82 (62.1) | ||
| Low | 50 (37.9) | ||
| Manually Assessed DR | 2.635 (1.083–6.408) | 0.033 | |
| Immature | 31 (23.5) | ||
| Other | 101 (76.5) | ||
| Largest single myxoid stroma area in Margin 1 ACP | 4.654 (1.912–11.330) | <0.001 | |
| Yes | 21 (15.9) | ||
| No | 111 (84.1) | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nearchou, I.P.; Ueno, H.; Kajiwara, Y.; Lillard, K.; Mochizuki, S.; Takeuchi, K.; Harrison, D.J.; Caie, P.D. Automated Detection and Classification of Desmoplastic Reaction at the Colorectal Tumour Front Using Deep Learning. Cancers 2021, 13, 1615. https://doi.org/10.3390/cancers13071615
Nearchou IP, Ueno H, Kajiwara Y, Lillard K, Mochizuki S, Takeuchi K, Harrison DJ, Caie PD. Automated Detection and Classification of Desmoplastic Reaction at the Colorectal Tumour Front Using Deep Learning. Cancers. 2021; 13(7):1615. https://doi.org/10.3390/cancers13071615
Chicago/Turabian StyleNearchou, Ines P., Hideki Ueno, Yoshiki Kajiwara, Kate Lillard, Satsuki Mochizuki, Kengo Takeuchi, David J. Harrison, and Peter D. Caie. 2021. "Automated Detection and Classification of Desmoplastic Reaction at the Colorectal Tumour Front Using Deep Learning" Cancers 13, no. 7: 1615. https://doi.org/10.3390/cancers13071615
APA StyleNearchou, I. P., Ueno, H., Kajiwara, Y., Lillard, K., Mochizuki, S., Takeuchi, K., Harrison, D. J., & Caie, P. D. (2021). Automated Detection and Classification of Desmoplastic Reaction at the Colorectal Tumour Front Using Deep Learning. Cancers, 13(7), 1615. https://doi.org/10.3390/cancers13071615






