Expression of ERBB Family Members as Predictive Markers of Prostate Cancer Progression and Mortality
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Tissue Culture Conditions
2.2. Creation of Cell Line Pellets
2.3. Western Blot Analysis
2.4. Patient Cohort
2.5. Construction of TMA
2.6. Immunofluorescence
2.7. Digital Image Analyses and Pre-Processing of Scoring Data
2.8. Statistical Analysis
3. Results
3.1. Antibody Validation in PC Cell Lines
3.2. Patient Characteristics and Clinical Parameters
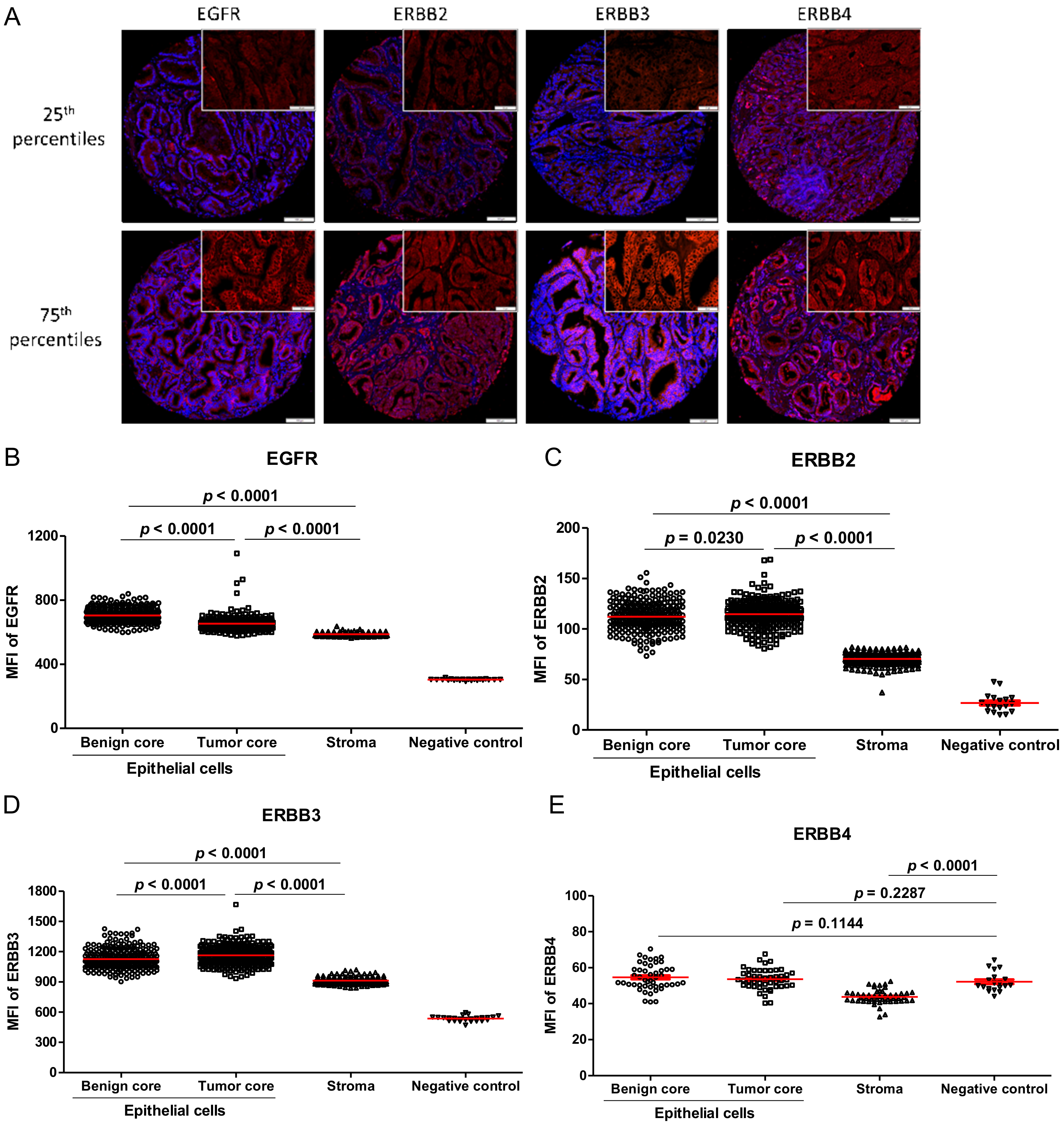
3.3. ERBB Family Member’s Expression in Human PC Specimens
3.4. EGFR, ERBB2, and ERBB3 Expression Is Associated with an Increased Risk of BCR at 5 Years
3.5. Combining the Expression Levels of EGFR, ERBB2, and ERBB3 Predicts BCR at 5 Years
3.6. Expression of EGFR, ERBB2, and ERBB3 Can Predict Bone Metastasis Development at 10 Years
3.7. Combining the Expression Levels of EGFR, ERBB2, and ERBB3 Can Predict Bone Metastasis Development
3.8. Expression of EGFR, ERBB2, and ERBB3 Can Predict PC-Specific Mortality
3.9. Combining the Expression Levels of EGFR, ERBB2, and ERBB3 Can Predict PC Mortality
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Quinlan, D.M.; Partin, A.W.; Walsh, P.C. Can aggressive prostatic carcinomas be identified and can their natural history be altered by treatment? Urology 1995, 46, 77–82. [Google Scholar] [CrossRef]
- Epstein, J.I. An Update of the Gleason Grading System. J. Urol. 2010, 183, 433–440. [Google Scholar] [CrossRef]
- Hoogland, A.M.; Kweldam, C.F.; Van Leenders, G.J.L.H. Prognostic Histopathological and Molecular Markers on Prostate Cancer Needle-Biopsies: A Review. BioMed Res. Int. 2014, 2014, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Oldenhuis, C.; Oosting, S.; Gietema, J.; de Vries, E. Prognostic versus predictive value of biomarkers in oncology. Eur. J. Cancer 2008, 44, 946–953. [Google Scholar] [CrossRef]
- Gannon, P.O.; Lessard, L.; Stevens, L.-M.; Forest, V.; Bégin, L.R.; Minner, S.; Tennstedt, P.; Schlomm, T.; Mes-Masson, A.-M.; Saad, F. Large-scale independent validation of the nuclear factor-kappa B p65 prognostic biomarker in prostate cancer. Eur. J. Cancer 2013, 49, 2441–2448. [Google Scholar] [CrossRef] [PubMed]
- Labouba, I.; Page, C.L.; Communal, L.; Kristessen, T.; You, X.; Peant, B.; Barres, V.; Gannon, P.O.; Mes-Masson, A.-M.; Saad, F. Potential Cross-Talk between Alternative and Classical NF-kappaB Pathways in Prostate Cancer Tissues as Measured by a Multi-Staining Immunofluorescence Co-Localization Assay. PLoS ONE 2015, 10, e0131024. [Google Scholar] [CrossRef] [PubMed]
- Jamaspishvili, T.; Patel, P.G.; Niu, Y.; Vidotto, T.; Caven, I.; Livergant, R.; Fu, W.; Kawashima, A.; How, N.; Okello, J.B.; et al. Risk Stratification of Prostate Cancer Through Quantitative Assessment of PTEN Loss (qPTEN). J. Natl. Cancer Inst. 2020, 112, 1098–1104. [Google Scholar] [CrossRef] [PubMed]
- Leclerc, B.G.; Charlebois, R.; Chouinard, G.; Allard, B.; Pommey, S.; Saad, F.; Stagg, J. CD73 Expression Is an Independent Prognostic Factor in Prostate Cancer. Clin. Cancer Res. 2016, 22, 158–166. [Google Scholar] [CrossRef] [Green Version]
- Clairefond, S.; Péant, B.; Ouellet, V.; Barrès, V.; Tian, Z.; Trudel, D.; Karakiewicz, P.I.; Mes-Masson, A.-M.; Saad, F. PUMA and NOXA Expression in Tumor-Associated Benign Prostatic Epithelial Cells Are Predictive of Prostate Cancer Biochemical Recurrence. Cancers 2020, 12, 3187. [Google Scholar] [CrossRef] [PubMed]
- Grosset, A.A.; Ouellet, V.; Caron, C.; Fragoso, G.; Barres, V.; Delvoye, N.; Latour, M.; Aprikian, A.; Bergeron, A.; Chevalier, S.; et al. Canadian Prostate Cancer Biomarker. Validation of the prognostic value of NF-kappaB p65 in prostate cancer: A retrospective study using a large multi-institutional cohort of the Canadian Prostate Cancer Biomarker Network. PLoS Med. 2019, 16, e1002847. [Google Scholar] [CrossRef]
- Dankner, M.; Ouellet, V.; Communal, L.; Schmitt, E.; Perkins, D.; Annis, M.G.; Barrès, V.; Caron, C.; Mes-Masson, A.-M.; Saad, F.; et al. CCN3/Nephroblastoma Overexpressed Is a Functional Mediator of Prostate Cancer Bone Metastasis That Is Associated with Poor Patient Prognosis. Am. J. Pathol. 2019, 189, 1451–1461. [Google Scholar] [CrossRef] [Green Version]
- Fleischmann, A.; Rocha, C.; Saxer-Sekulic, N.; Zlobec, I.; Sauter, G.; Thalmann, G.N. High CD10 expression in lymph node metastases from surgically treated prostate cancer independently predicts early death. Virchows Archiv 2011, 458, 741–748. [Google Scholar] [CrossRef]
- Dunn, K.L.; Espino, P.S.; Drobic, B.; He, S.; Davie, J.R. The Ras-MAPK signal transduction pathway, cancer and chromatin remodeling. Biochem. Cell Biol. 2005, 83, 1–14. [Google Scholar] [CrossRef]
- Normanno, N.; Bianco, C.; De Luca, A.; Maiello, M.R.; Salomon, D.S. Target-based agents against ErbB receptors and their ligands: A novel approach to cancer treatment. Endocrine-Related Cancer 2003, 10, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Gschwind, A.; Fischer, O.M.; Ullrich, A. The discovery of receptor tyrosine kinases: Targets for cancer therapy. Nat. Rev. Cancer 2004, 4, 361–370. [Google Scholar] [CrossRef]
- Holbro, T.; Civenni, G.; Hynes, N.E. The ErbB receptors and their role in cancer progression. EGF Recept. Fam. 2003, 284, 103–114. [Google Scholar] [CrossRef]
- Hynes, N.E.; MacDonald, G. ErbB receptors and signaling pathways in cancer. Curr. Opin. Cell Biol. 2009, 21, 177–184. [Google Scholar] [CrossRef]
- Roskoski, R. The ErbB/HER family of protein-tyrosine kinases and cancer. Pharmacol. Res. 2014, 79, 34–74. [Google Scholar] [CrossRef]
- Ramieri, M.T.; Murari, R.; Botti, C.; Pica, E.; Zotti, G.; Alo, P.L. Detection of HER2 amplification using the SISH technique in breast, colon, prostate, lung and ovarian carcinoma. Anticancer. Res. 2010, 30, 1287–1292. [Google Scholar]
- Huang, Y.; Chang, Y. Epidermal Growth Factor Receptor (EGFR) Phosphorylation, Signaling and Trafficking in Prostate Cancer. Prostate Cancer-From Bench Bedside 2011, 143, 72. [Google Scholar] [CrossRef] [Green Version]
- Di Lorenzo, G.; Bianco, R.; Tortora, G.; Ciardiello, F. Involvement of growth factor receptors of the epidermal growth factor receptor family in prostate cancer development and progression to androgen independence. Clin. Prostate Cancer 2003, 2, 50–57. [Google Scholar] [CrossRef]
- Koumakpayi, I.H.; Page, C.L.; Mes-Masson, A.M.; Saad, F. Hierarchical clustering of immunohistochemical analysis of the activated ErbB/PI3K/Akt/NF-kappaB signalling pathway and prognostic significance in prostate cancer. Br. J. Cancer 2010, 102, 1163–1173. [Google Scholar] [CrossRef] [Green Version]
- Di Lorenzo, G.; Tortora, G.; D’Armiento, F.P.; De Rosa, G.; Staibano, S.; Autorino, R.; D’Armiento, M.; De Laurentiis, M.; De Placido, S.; Catalano, G.; et al. Expression of epidermal growth factor receptor correlates with disease relapse and progression to androgen-independence in human prostate cancer. Clin. Cancer Res. 2002, 8, 3438–3444. [Google Scholar] [PubMed]
- Peraldo-Neia, C.; Migliardi, G.; Mello-Grand, M.; Montemurro, F.; Segir, R.; Pignochino, Y.; Cavalloni, G.; Torchio, B.; Mosso, L.; Chiorino, G.; et al. Epidermal Growth Factor Receptor (EGFR) mutation analysis, gene expression profiling and EGFR protein expression in primary prostate cancer. BMC Cancer 2011, 11, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Tambo, M.; Higashihara, E.; Terado, Y.; Nutahara, K.; Okegawa, T. Comparison of serum HER2/neu with immunohistochemical HER2/neu expression for the prediction of biochemical progression in metastatic prostate cancer. Int. J. Urol. 2009, 16, 369–374. [Google Scholar] [CrossRef]
- Edwards, J.; Traynor, P.; Munro, A.F.; Pirret, C.F.; Dunne, B.; Bartlett, J.M. The Role of HER1-HER4 and EGFRvIII in Hormone-Refractory Prostate Cancer. Clin. Cancer Res. 2006, 12, 123–130. [Google Scholar] [CrossRef] [Green Version]
- Koumakpayi, I.H.; Diallo, J.-S.; Le Page, C.; Lessard, L.; Filali-Mouhim, A.; Bégin, L.R.; Mes-Masson, A.-M.; Saad, F. Low nuclear ErbB3 predicts biochemical recurrence in patients with prostate cancer. BJU Int. 2007, 100, 303–309. [Google Scholar] [CrossRef]
- Hashemi, M.; Moradi, N.; Rezaei, M.; Sanaei, S.; Ziaee, S.A.M.; Narouie, B.; Sotoudeh, M.; Bahari, G.; Ghavami, S. ERBB4 gene polymorphisms and the risk of prostate cancer in a sample of Iranian Population. Cell. Mol. Boil. 2016, 62, 43–48. [Google Scholar]
- Robinson, D.; He, F.; Pretlow, T.; Kung, H.J. A tyrosine kinase profile of prostate carcinoma. Proc. Natl. Acad. Sci. USA 1996, 93, 5958–5962. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Appert-Collin, A.; Hubert, P.; Crémel, G.; Bennasroune, A. Role of ErbB Receptors in Cancer Cell Migration and Invasion. Front. Pharmacol. 2015, 6, 283. [Google Scholar] [CrossRef] [Green Version]
- Zietarska, M.; Madore, J.; Diallo, J.-S.; Delvoye, N.; Saad, F.; Provencher, D.; Mes-Masson, A.-M. A novel method of cell embedding for tissue microarrays. Histopathology 2010, 57, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Schlomm, T.; Kirstein, P.; Iwers, L.; Daniel, B.; Steuber, T.; Walz, J.; Chun, F.H.; Haese, A.; Kollermann, J.; Graefen, M.; et al. Clinical Significance of Epidermal Growth Factor Receptor Protein Overexpression and Gene Copy Number Gains in Prostate Cancer. Clin. Cancer Res. 2007, 13, 6579–6584. [Google Scholar] [CrossRef] [Green Version]
- Mandel, A.; Larsson, P.; Sarwar, M.; Semenas, J.; Khaja, A.S.S.; Persson, J.L. The interplay between AR, EGF receptor and MMP-9 signaling pathways in invasive prostate cancer. Mol. Med. 2018, 24, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Koumakpayi, I.H.; Diallo, J.-S.; Le Page, C.; Lessard, L.; Gleave, M.; Bégin, L.R.; Mes-Masson, A.-M.; Saad, F. Expression and Nuclear Localization of ErbB3 in Prostate Cancer. Clin. Cancer Res. 2006, 12, 2730–2737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Page, C.L.; Koumakpayi, I.H.; Lessard, L.; Mes-Masson, A.-M. and Saad, F. EGFR and Her-2 regulate the constitutive activation of NF-kappaB in PC-3 prostate cancer cells. Prostate 2005, 65, 130–140. [Google Scholar] [CrossRef]
- Minner, S.; Jessen, B.; Stiedenroth, L.; Burandt, E.; Köllermann, J.; Mirlacher, M.; Erbersdobler, A.; Eichelberg, C.; Fisch, M.; Brümmendorf, T.H.; et al. Low Level Her2 Overexpression Is Associated with Rapid Tumor Cell Proliferation and Poor Prognosis in Prostate Cancer. Clin. Cancer Res. 2010, 16, 1553–1560. [Google Scholar] [CrossRef] [Green Version]
- Baek, K.H.; Hong, M.E.; Jung, Y.Y.; Lee, C.H.; Lee, T.J.; Park, E.S.; Kim, M.K.; Yoo, J.H.; Lee, S.W. Correlation of AR, EGFR, and HER2 Expression Levels in Prostate Cancer: Immunohistochemical Analysis and Chromogenic In Situ Hybridization. Cancer Res. Treat. 2012, 44, 50–56. [Google Scholar] [CrossRef]
- Xiong, Z.; Deng, G.; Huang, X.; Li, X.; Xie, X.; Wang, J.; Shuang, Z.; Wang, X. Bone metastasis pattern in initial metastatic breast cancer: A population-based study. Cancer Manag. Res. 2018, 10, 287. [Google Scholar] [CrossRef] [Green Version]
- Muniyan, S.; Chen, S.-J.; Lin, F.-F.; Wang, Z.; Mehta, P.P.; Batra, S.K.; Lin, M.-F. ErbB-2 signaling plays a critical role in regulating androgen-sensitive and castration-resistant androgen receptor-positive prostate cancer cells. Cell. Signal. 2015, 27, 2261–2271. [Google Scholar] [CrossRef] [Green Version]
- Day, K.C.; Hiles, G.L.; Kozminsky, M.; Dawsey, S.J.; Paul, A.; Broses, L.J.; Shah, R.; Kunja, L.P.; Hall, C.; Palanisamy, N.; et al. HER2 and EGFR Overexpression Support Metastatic Progression of Prostate Cancer to Bone. Cancer Res. 2017, 77, 74–85. [Google Scholar] [CrossRef] [Green Version]
- Murray, N.P.; Reyes, E.; Fuentealba, C.; Jacob, O.; Orellana, N. Possible Role of HER-2 in the Progression of Prostate Cancer from Primary Tumor to Androgen Independence. Asian Pac. J. Cancer Prev. 2015, 16, 6615–6619. [Google Scholar] [CrossRef] [Green Version]
- Tome-Garcia, J.; Li, D.; Ghazaryan, S.; Shu, L.; Wu, L. ERBB2 Increases Metastatic Potentials Specifically in Androgen-Insensitive Prostate Cancer Cells. PLoS ONE 2014, 9, e99525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ziada, A.; Barqawi, A.; Glode, L.M.; Varella-Garcia, M.; Crighton, F.; Majeski, S.; Rosenblum, M.; Kane, M.; Chen, L.; Crawford, E.D. The use of trastuzumab in the treatment of hormone refractory prostate cancer; phase II trial. Prostate 2004, 60, 332–337. [Google Scholar] [CrossRef] [PubMed]
- Festuccia, C.; Gravina, G.L.; Biordi, L.; D’Ascenzo, S.; Dolo, V.; Ficorella, C.; Ricevuto, E.; Tombolini, V.; Biordi, A.L. Effects of EGFR tyrosine kinase inhibitor erlotinib in prostate cancer cells in vitro. Prostate 2009, 69, 1529–1537. [Google Scholar] [CrossRef]
- Whang, Y.E.; Armstrong, A.J.; Rathmell, W.K.; Godley, P.A.; Kim, W.Y.; Pruthi, R.S.; Wallen, E.M.; Crane, J.M.; Moore, D.T.; Grigson, G.; et al. A phase II study of lapatinib, a dual EGFR and HER-2 tyrosine kinase inhibitor, in patients with castration-resistant prostate cancer. Urol. Oncol. Semin. Orig. Investig. 2013, 31, 82–86. [Google Scholar] [CrossRef]



| Number of patients | 285 |
| Median age at RP, years (IQR) | 63 (59–67) |
| Median PSA at diagnosis, ng/mL (IQR) | 7.0 (5.0–10.8) |
| Pathological TNM | |
| 2 | 201 |
| 3 | 75 |
| 4 | 9 |
| Gleason score at RP | |
| ≤3 + 3 | 140 |
| 3 + 4 | 93 |
| 4 + 3 | 19 |
| ≥4 + 4 | 29 |
| Unknown | 4 |
| Positive margin | 95 |
| Median follow-up, months (IQR) | 129 (76–174) |
| Biochemical recurrence at 5 years | |
| Number | 94 |
| Median time to BCR, months (IQR) | 11 (3–26) |
| Bone Metastasis at 10 years | |
| Number | 18 |
| Median time to bone metastasis, months (IQR) | 42 (20–83) |
| Overall survival | |
| Alive | 236 |
| Death from other cause | 31 |
| Death from PC | 18 |
| Median time to death specific PC mortality, months (IQR) | 68 (49–150) |
| Univariate | Multivariate with Dichotomized EGFR Expression | Multivariate with Dichotomized ERBB2 Expression | Multivariate with Dichotomized ERBB3 Expression | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HR [95% CI] | p-Value | HR [95% CI] | p-Value | HR [95% CI] | p-Value | HR [95% CI] | p-Value | |||||
| Age at Dx | 0.999 | [0.963–1.035] | 0.942 | - | - | - | - | - | - | - | - | - |
| PSA at Dx | 1.061 | [1.033–1.089] | 0.001 | 1.032 | [0.996–1.069] | 0.086 | 1.036 | [1.000–1.073] | 0.048 | 1.036 | [1.001–1.073] | 0.043 |
| Gleason score (4 categories) | 1.852 | [1.549–2.214] | 0.001 | 1.530 | [1.237–1.891] | 0.001 | 1.464 | [1.179–1.819] | 0.001 | 1.517 | [1.231–1.869] | 0.001 |
| Margin | 3.349 | [2.216–5.062] | 0.001 | 2.617 | [1.655–4.139] | 0.001 | 2.479 | [1.560–3.939] | 0.001 | 2.531 | [1.596–4.015] | 0.001 |
| pTNM (4 categories) | 2.884 | [2.133–3.900] | 0.001 | 1.555 | [1.047–2.309] | 0.029 | 1.683 | [1.131–2.503] | 0.010 | 1.578 | [1.065–2.340] | 0.023 |
| EGFR_Tumor_Continuous | 1.006 | [1.003–1.009] | 0.001 | - | - | - | - | - | - | - | - | - |
| EGFR_Tumor_Dichotomized_75th | 1.703 | [1.097–2.644] | 0.018 | 1.267 | [0.781–2.057] | 0.337 | - | - | - | - | - | - |
| ERBB2_Tumor_Continuous | 0.987 | [0.970–1.004] | 0.122 | - | - | - | - | - | - | - | - | - |
| ERBB2_Tumor_Dichotomized_50th * | 0.716 | [0.468–1.095] | 0.123 | - | - | - | 0.787 | [0.503–1.232] | 0.295 | - | - | - |
| ERBB3_Tumor_Continuous | 0.999 | [0.996–1.001] | 0.196 | - | - | - | - | - | - | - | - | - |
| ERBB3_Tumor_Dichotomized_25th | 0.650 | [0.418–1.011] | 0.056 | - | - | - | - | - | - | 0.856 | [0.856–1.367] | 0.515 |
| Univariate | Multivariate | ||||||
|---|---|---|---|---|---|---|---|
| HR [95% CI] | p-Value | HR [95% CI] | p-Value | ||||
| Age at Dx | 0.999 | [0.963–1.035] | 0.942 | - | - | - | |
| PSA at Dx | 1.061 | [1.033–1.089] | 0.001 | 1.029 | [0.993–1.067] | 0.113 | |
| Gleason score | 1.852 | [1.549–2.214] | 0.001 | 1.495 | [1.205–1.856] | 0.001 | |
| Margin | 3.349 | [2.216–5.062] | 0.001 | 2.508 | [1.582–3.976] | 0.001 | |
| pTNM (category) | 2.884 | [2.133–3.900] | 0.001 | 1.559 | [1.044–2.329] | 0.030 | |
| Category | Combined all ERBB members | 1.609 | [1.276–2.027] | 0.001 | 1.204 | [0.930–1.559] | 0.158 |
| EGFRlow | 1.000 | - | - | 1.000 | - | - | |
| EGFRhigh/ERBB3high/ERBB2high | 1.231 | [0.675–2.244] | 0.498 | 1.197 | [0.622–2.303] | 0.591 | |
| EGFRhigh/ERBB3high/ERBB2low | 2.189 | [1.085–4.417] | 0.029 | 1.122 | [0.514–2.449] | 0.773 | |
| EGFRhigh/ERBB3low | 5.455 | [2.478–12.011] | 0.001 | 2.202 | [0.915–5.297] | 0.078 | |
| Univariate | ||||
|---|---|---|---|---|
| HR [95% CI] | p-Value | |||
| Age at Dx | 0.987 | [0.907–1.073] | 0.757 | |
| PSA at Dx | 1.059 | [1.002–1.119] | 0.043 | |
| Gleason score | 3.704 | [2.304–5.955] | 0.001 | |
| Margin | 3.290 | [1.275–8.489] | 0.014 | |
| pTNM (category) | 7.490 | [3.830–14.647] | 0.001 | |
| EGFR_Continuous | 1.012 | [1.007–1.016] | 0.001 | |
| EGFR_Dichotomized | 3.642 | [1.404–9.444] | 0.008 | |
| ERBB2_Continuous | 0.976 | [0.939–1.016] | 0.234 | |
| ERBB2_Dichotomized | 0.312 | [0.101–0.969] | 0.044 | |
| ERBB3_Continuous | 0.994 | [0.989–1.000] | 0.037 | |
| ERBB3_Dichotomized | 0.622 | [0.230–1.683] | 0.350 | |
| Category | Combined all ERBB members | 2.036 | [1.438–2.881] | 0.001 |
| ERBB2high | 1.000 | - | 0.001 | |
| ERBB2low/EGFRlow | 1.897 | [0.535–6.722] | 0.321 | |
| ERBB2low/EGFRhigh/ERBB3high | 9.273 | [2.311–37.197] | 0.002 | |
| ERBB2low/EGFRhigh/ERBB3low | 14.774 | [2.687–81.237] | 0.002 | |
| Univariate | ||||
|---|---|---|---|---|
| HR [95% CI] | p-Value | |||
| Age at Dx | 0.976 | [0.896–1.062] | 0.569 | |
| PSA at Dx | 1.062 | [1.009–1.117] | 0.021 | |
| Gleason score | 3.968 | [2.395–6.573] | 0.001 | |
| Margin | 1.731 | [0.682–4.391] | 0.248 | |
| pTNM (category) | 5.655 | [2.928–10.923] | 0.001 | |
| EGFR_Continuous | 1.017 | [1.010–1.023] | 0.001 | |
| EGFR_Dichotomized | 2.215 | [0.843–5.824] | 0.107 | |
| ERBB2_Continuous | 0.969 | [0.933–1.007] | 0.107 | |
| ERBB2_Dichotomized | 0.312 | [0.101–0.968] | 0.044 | |
| ERBB3_Continuous | 0.991 | [0.985–0.997] | 0.002 | |
| ERBB3_Dichotomized | 0.373 | [0.144–0.969] | 0.043 | |
| Category | Combined all ERBB members | 1.865 | [1.256–2.768] | 0.002 |
| ERBB3high/ERBB2high | 1.000 | - | 0.004 | |
| ERBB3high/ERBB2low/EGFRlow | 2.915 | [0.487–17.465] | 0.241 | |
| ERBB3high/ERBB2low/EGFRhigh | 11.755 | [1.958–70.566] | 0.007 | |
| ERBB3low/EGFRlow | 5.671 | [1.143–28.129] | 0.034 | |
| ERBB3low/EGFRhigh | 36.732 | [4.901–275.271] | 0.001 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Clairefond, S.; Ouellet, V.; Péant, B.; Barrès, V.; Karakiewicz, P.I.; Mes-Masson, A.-M.; Saad, F. Expression of ERBB Family Members as Predictive Markers of Prostate Cancer Progression and Mortality. Cancers 2021, 13, 1688. https://doi.org/10.3390/cancers13071688
Clairefond S, Ouellet V, Péant B, Barrès V, Karakiewicz PI, Mes-Masson A-M, Saad F. Expression of ERBB Family Members as Predictive Markers of Prostate Cancer Progression and Mortality. Cancers. 2021; 13(7):1688. https://doi.org/10.3390/cancers13071688
Chicago/Turabian StyleClairefond, Sylvie, Véronique Ouellet, Benjamin Péant, Véronique Barrès, Pierre I. Karakiewicz, Anne-Marie Mes-Masson, and Fred Saad. 2021. "Expression of ERBB Family Members as Predictive Markers of Prostate Cancer Progression and Mortality" Cancers 13, no. 7: 1688. https://doi.org/10.3390/cancers13071688
APA StyleClairefond, S., Ouellet, V., Péant, B., Barrès, V., Karakiewicz, P. I., Mes-Masson, A.-M., & Saad, F. (2021). Expression of ERBB Family Members as Predictive Markers of Prostate Cancer Progression and Mortality. Cancers, 13(7), 1688. https://doi.org/10.3390/cancers13071688







