BTLA/HVEM Axis Induces NK Cell Immunosuppression and Poor Outcome in Chronic Lymphocytic Leukemia
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients’ Samples
2.2. Analysis of BTLA and HVEM Expression on Immune Cells
2.3. Leukemic Cell Depletion Assay
2.4. Intracellular Cytokine Staining
2.5. Evaluation of NK Cell-Mediated Cytotoxicity
2.6. Determination of Soluble BTLA on the Sera of Patients with CLL
2.7. In Silico Analysis of the Role of BTLA in CLL and other Hematological Malignancies
2.8. Statistics
3. Results
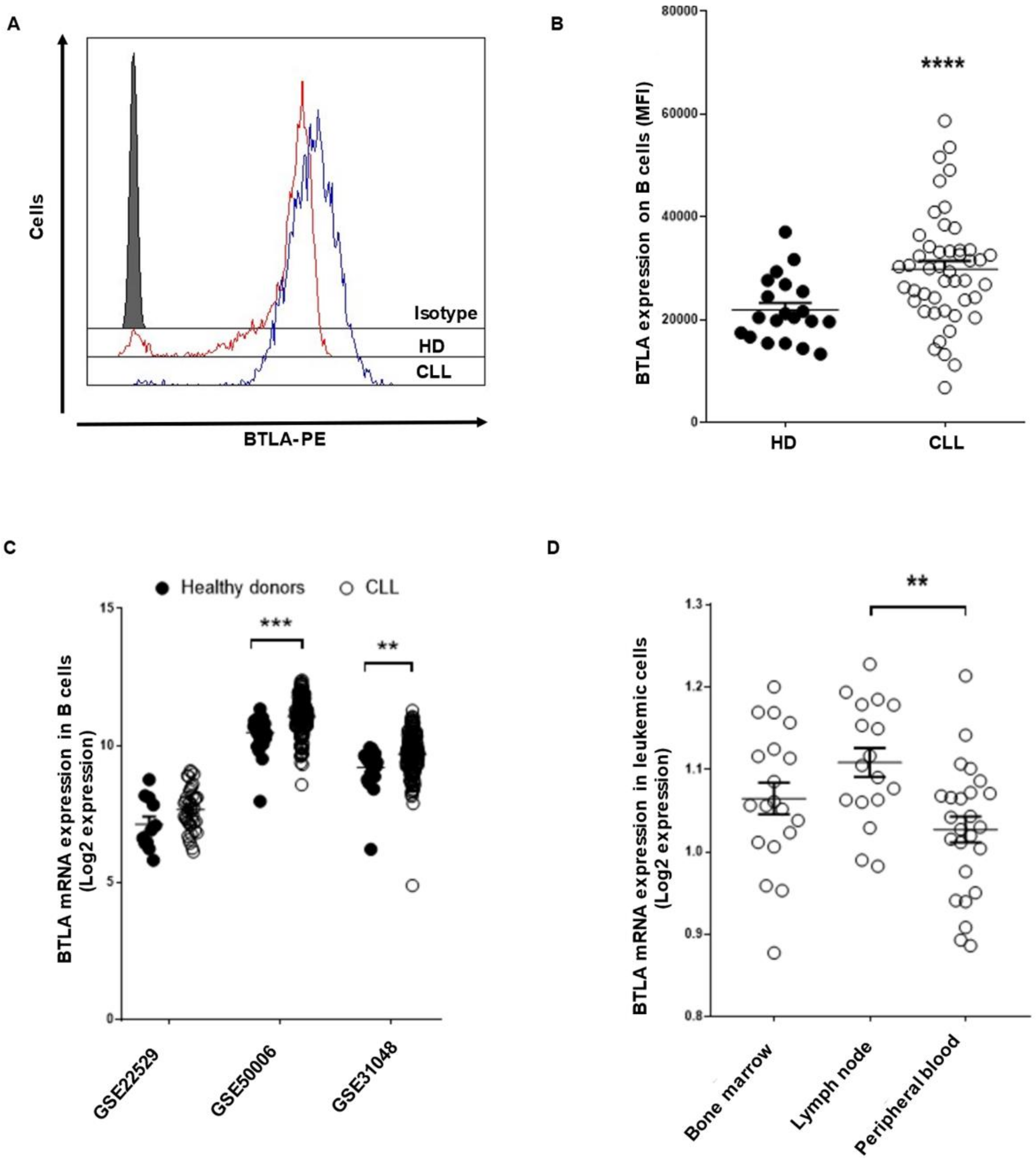
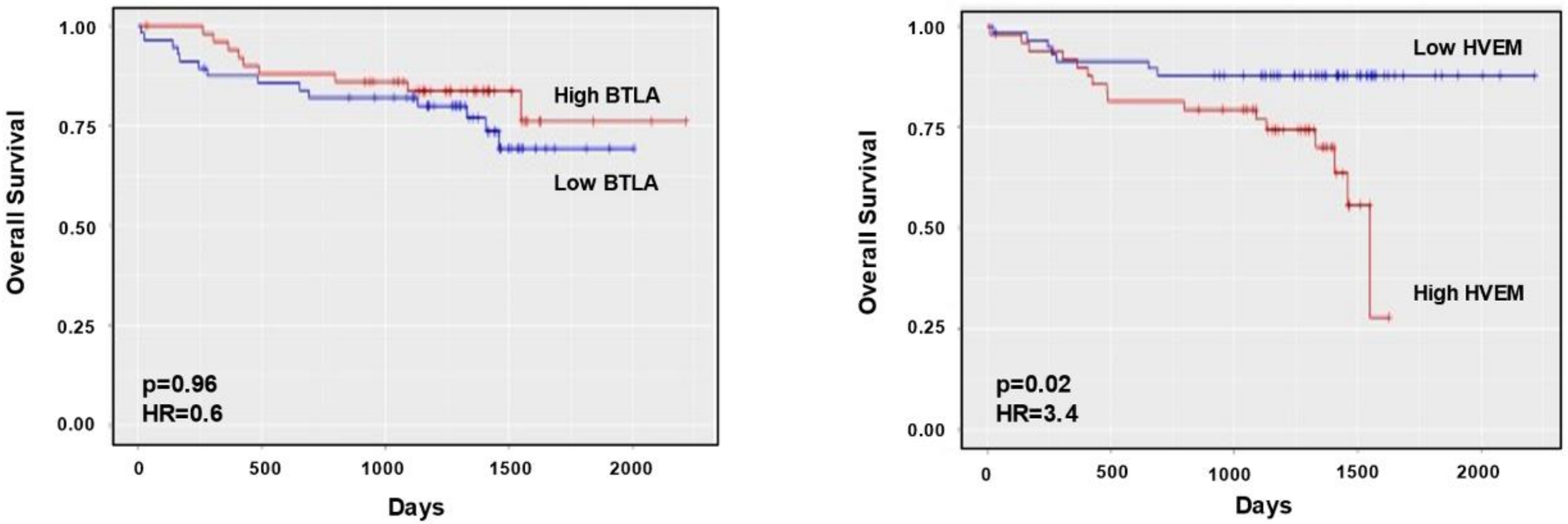
3.1. BTLA/HVEM Axis Is Dysregulated on Leukemic Cells and Impacts on Overall Survival
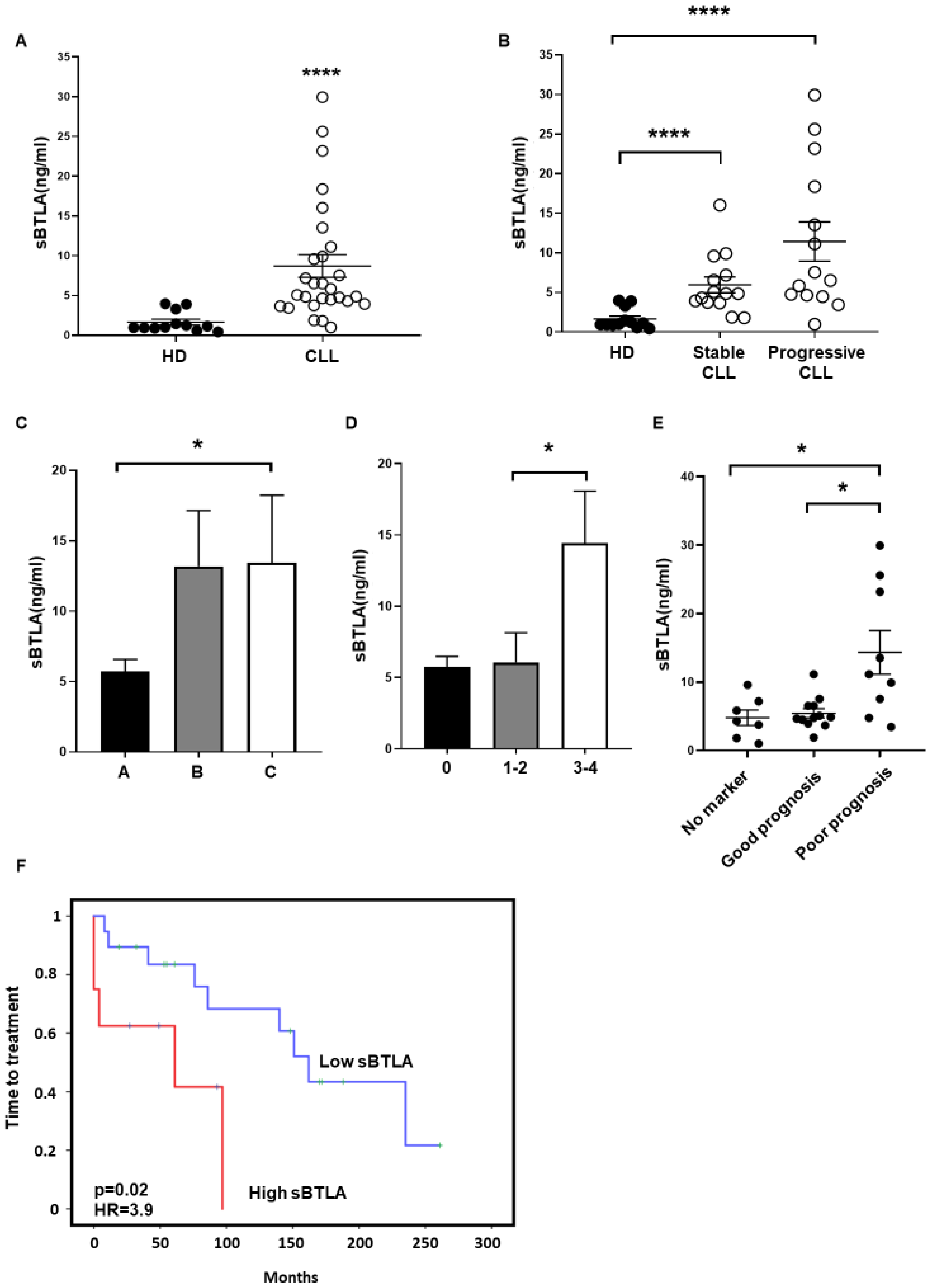
3.2. Soluble BTLA Levels Are Increased and Correlate with Time to Treatment in CLL
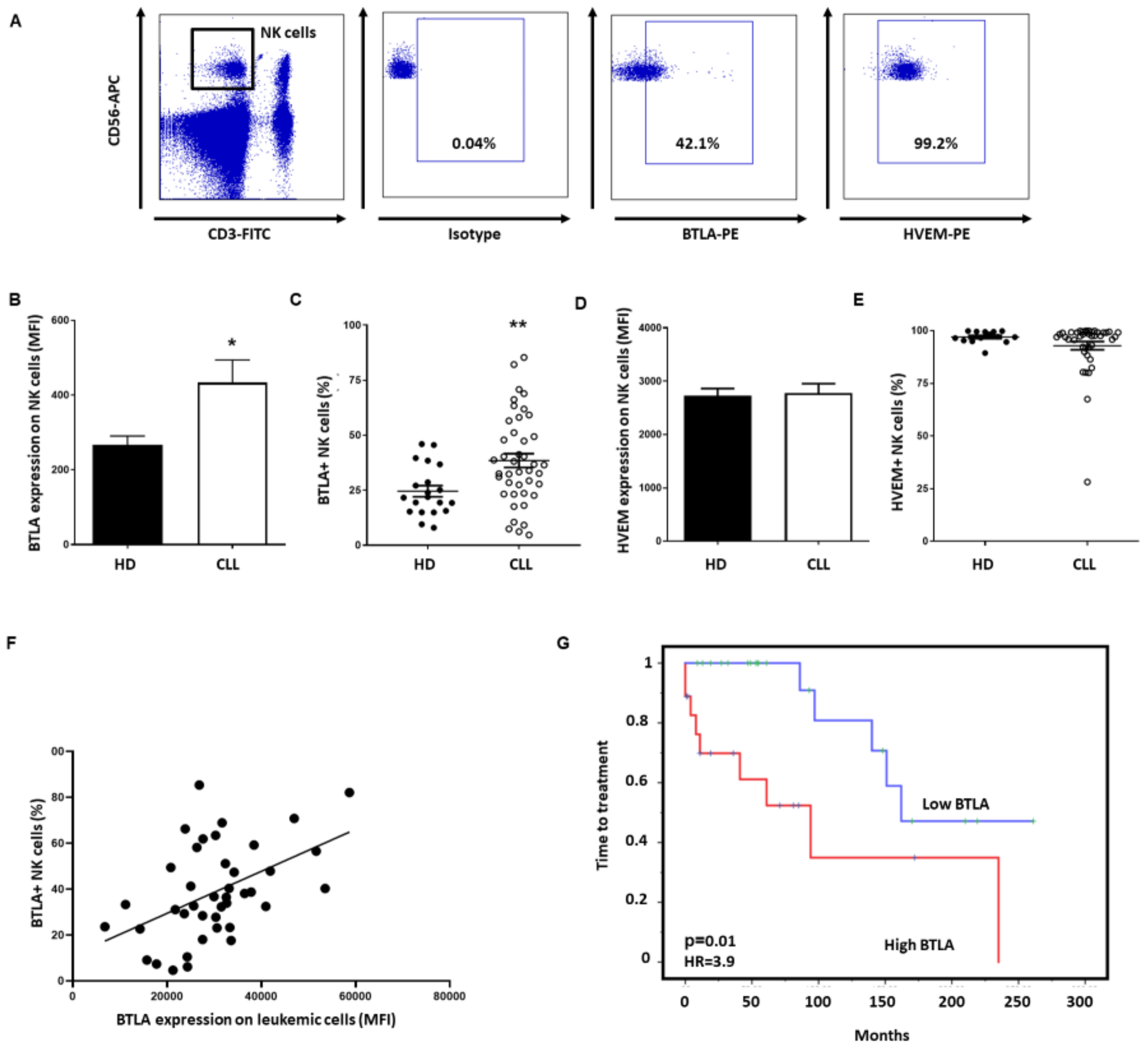
3.3. BTLA Is Upregulated on NK Cells and Is Associated with Poor Outcome
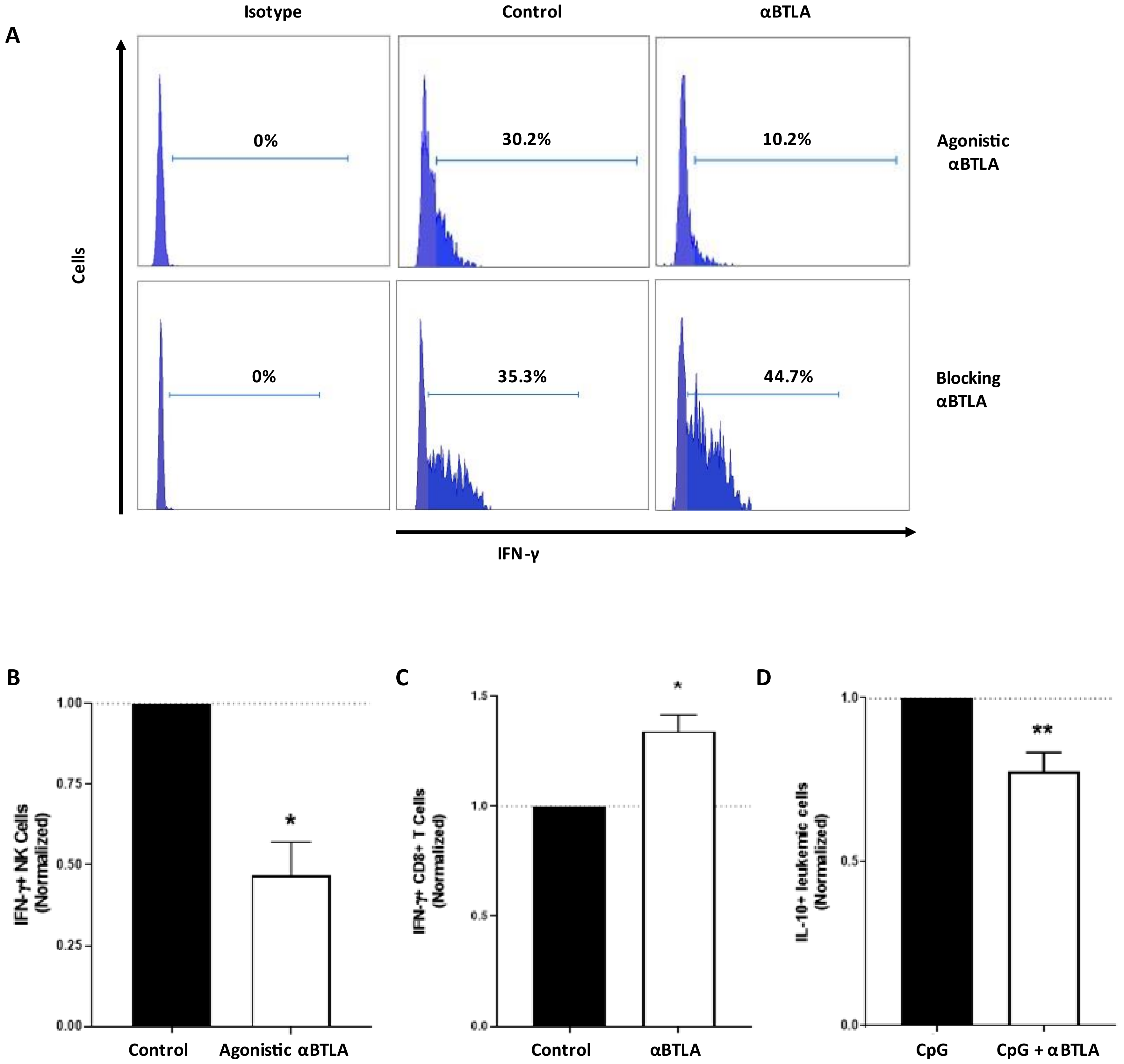
3.4. BTLA Decreases IFN-γ Production and NK Cell-Mediated Cytotoxicity, But It May Be Reversed by BTLA Blockade
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zenz, T.; Mertens, D.; Küppers, R.; Döhner, H.; Stilgenbauer, S. From pathogenesis to treatment of chronic lymphocytic leukaemia. Nat. Rev. Cancer 2009, 10, 37–50. [Google Scholar] [CrossRef] [PubMed]
- Palmer, S.; Hanson, C.A.; Zent, C.S.; Porrata, L.F.; LaPlant, B.; Geyer, S.M.; Markovic, S.N.; Call, T.G.; Bowen, D.A.; Jelinek, D.F.; et al. Prognostic importance of T and NK-cells in a consecutive series of newly diagnosed patients with chronic lymphocytic leukaemia. Br. J. Haematol. 2008, 141, 607–614. [Google Scholar] [CrossRef] [PubMed]
- Riches, J.C.; Gribben, J.G. Understanding the Immunodeficiency in Chronic Lymphocytic Leukemia. Hematol. Clin. North. Am. 2013, 27, 207–235. [Google Scholar] [CrossRef] [PubMed]
- Fürstenau, M.; Hallek, M.; Eichhorst, B. Sequential and combination treatments with novel agents in chronic lymphocytic leukemia. Haematologica 2019, 104, 2144–2154. [Google Scholar] [CrossRef]
- Woyach, J.A. Treatment-naive CLL: Lessons from phase 2 and phase 3 clinical trials. Blood 2019, 134, 1796–1801. [Google Scholar] [CrossRef]
- Smolewski, P.; Robak, T. Current Treatment of Refractory/Relapsed Chronic Lymphocytic Leukemia: A Focus on Novel Drugs. Acta Haematol. 2020, 10, 1–15. [Google Scholar] [CrossRef]
- Sharma, S.; Rai, K.R. Chronic lymphocytic leukemia (CLL) treatment: So many choices, such great options. Cancer 2019, 125, 1432–1440. [Google Scholar] [CrossRef]
- Herling, C.D.; Abedpour, N.; Weiss, J.; Schmitt, A.; Jachimowicz, R.D.; Merkel, O.; Cartolano, M.; Oberbeck, S.; Mayer, P.; Berg, V.; et al. Clonal dynamics towards the development of venetoclax resistance in chronic lymphocytic leukemia. Nat. Commun. 2018, 9, 1–8. [Google Scholar] [CrossRef]
- Woyach, J.A.; Furman, R.R.; Liu, T.-M.; Ozer, H.G.; Zapatka, M.; Ruppert, A.S.; Xue, L.; Li, D.H.-H.; Steggerda, S.M.; Versele, M.; et al. Resistance Mechanisms for the Bruton’s Tyrosine Kinase Inhibitor Ibrutinib. N. Engl. J. Med. 2014, 370, 2286–2294. [Google Scholar] [CrossRef]
- MacFarlane, A.W.; Jillab, M.; Smith, M.R.; Alpaugh, R.K.; Cole, M.E.; Litwin, S.; Millenson, M.M.; Al-Saleem, T.; Cohen, A.D.; Campbell, K.S. NK cell dysfunction in chronic lymphocytic leukemia is associated with loss of the mature cells expressing inhibitory killer cell Ig-like receptors. Oncoimmunology 2017, 6, e1330235. [Google Scholar] [CrossRef]
- Hofland, T.; Endstra, S.; Gomes, C.K.; De Boer, R.; De Weerdt, I.; Bobkov, V.; Riedl, J.A.; Heukers, R.; Smit, M.J.; Eldering, E.; et al. Natural Killer Cell Hypo-responsiveness in Chronic Lymphocytic Leukemia can be Circumvented In Vitro by Adequate Activating Signaling. Hemasphere 2019, 3, e308. [Google Scholar] [CrossRef]
- Riches, J.C.; Davies, J.K.; McClanahan, F.; Fatah, R.; Iqbal, S.; Agrawal, S.; Ramsay, A.G.; Gribben, J.G. T cells from CLL patients exhibit features of T-cell exhaustion but retain capacity for cytokine production. Blood 2013, 121, 1612–1621. [Google Scholar] [CrossRef]
- Hanna, B.S.; Roessner, P.M.; Yazdanparast, H.; Colomer, D.; Campo, E.; Kugler, S.; Yosifov, D.; Stilgenbauer, S.; Schmidt, M.; Gabriel, R.; et al. Control of chronic lymphocytic leukemia development by clonally-expanded CD8+ T-cells that undergo functional exhaustion in secondary lymphoid tissues. Leukemia 2019, 33, 625–637. [Google Scholar] [CrossRef]
- Ramsay, A.G.; Clear, A.J.; Fatah, R.; Gribben, J.G. Multiple inhibitory ligands induce impaired T-cell immunologic synapse function in chronic lymphocytic leukemia that can be blocked with lenalidomide: Establishing a reversible immune evasion mechanism in human cancer. Blood 2012, 120, 1412–1421. [Google Scholar] [CrossRef]
- Moreira, J.; Rabe, K.G.; Cerhan, J.R.; Kay, E.N.; Wilson, J.W.; Call, T.G.; Leis, J.F.; Jelinek, D.F.; Schwager, S.M.; Bowen, A.D.; et al. Infectious complications among individuals with clinical monoclonal B-cell lymphocytosis (MBL): A cohort study of newly diagnosed cases compared to controls. Leukemia 2013, 27, 136–141. [Google Scholar] [CrossRef]
- Hamblin, A.D.; Hamblin, T.J. The immunodeficiency of chronic lymphocytic leukaemia. Br. Med. Bull. 2008, 87, 49–62. [Google Scholar] [CrossRef]
- Gonzalez-Rodriguez, A.P.; Villa-Álvarez, M.; Sordo-Bahamonde, C.; Lorenzo-Herrero, S.; Gonzalez, S.; Rodriguez, G.; Bahamonde, S.; Herrero, L. NK Cells in the Treatment of Hematological Malignancies. J. Clin. Med. 2019, 8, 1557. [Google Scholar] [CrossRef]
- Muntasell, A.; Ochoa, M.C.; Cordeiro, L.; Berraondo, P.; de Cerio, A.L.-D.; Cabo, M.; López-Botet, M.; Melero, I. Targeting NK-cell checkpoints for cancer immunotherapy. Curr. Opin. Immunol. 2017, 45, 73–81. [Google Scholar] [CrossRef]
- Hofland, T.; Eldering, E.; Kater, A.P.; Tonino, S.H. Engaging Cytotoxic T and NK Cells for Immunotherapy in Chronic Lymphocytic Leukemia. Int. J. Mol. Sci. 2019, 20, 4315. [Google Scholar] [CrossRef]
- Sordo-Bahamonde, C.; Vitale, M.; Lorenzo-Herrero, S.; López-Soto, A.; Gonzalez, S. Mechanisms of Resistance to NK Cell Immunotherapy. Cancers 2020, 12, 893. [Google Scholar] [CrossRef]
- Sordo-Bahamonde, C.; Lorenzo-Herrero, S.; Payer, Á.R.; Gonzalez, S.; López-Soto, A. Mechanisms of Apoptosis Resistance to NK Cell-Mediated Cytotoxicity in Cancer. Int. J. Mol. Sci. 2020, 21, 3726. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo-Herrero, S.; López-Soto, A.; Sordo-Bahamonde, C.; Gonzalez-Rodriguez, A.P.; Vitale, M.; Gonzalez, S. NK Cell-Based Immunotherapy in Cancer Metastasis. Cancers 2018, 11, 29. [Google Scholar] [CrossRef] [PubMed]
- López-Soto, A.; Gonzalez, S.; Smyth, M.J.; Galluzzi, L. Control of Metastasis by NK Cells. Cancer Cell 2017, 32, 135–154. [Google Scholar] [CrossRef] [PubMed]
- Huergo-Zapico, L.; Acebes-Huerta, A.; Gonzalez-Rodriguez, A.P.; Contesti, J.; Gonzalez-García, E.; Payer, A.R.; Villa-Alvarez, M.; Fernández-Guizán, A.; López-Soto, A.; Gonzalez, S. Expansion of NK Cells and Reduction of NKG2D Expression in Chronic Lymphocytic Leukemia. Correlation with Progressive Disease. PLoS ONE 2014, 9, e108326. [Google Scholar] [CrossRef]
- Villa-Álvarez, M.; Sordo-Bahamonde, C.; Lorenzo-Herrero, S.; Gonzalez-Rodriguez, A.P.; Payer, A.R.; Gonzalez-Garcia, E.; Villa-Álvarez, M.C.; López-Soto, A.; Gonzalez, S. Ig-Like Transcript 2 (ILT2) Blockade and Lenalidomide Restore NK Cell Function in Chronic Lymphocytic Leukemia. Front. Immunol. 2018, 9, 2917. [Google Scholar] [CrossRef]
- Wang, W.-T.; Zhu, H.-Y.; Wu, Y.-J.; Xia, Y.; Wu, J.-Z.; Wu, W.; Liang, J.-H.; Wang, L.; Fan, L.; Li, J.-Y.; et al. Elevated absolute NK cell counts in peripheral blood predict good prognosis in chronic lymphocytic leukemia. J. Cancer Res. Clin. Oncol. 2018, 144, 449–457. [Google Scholar] [CrossRef]
- Benjamini, O.; Jain, P.; Trinh, L.; Qiao, W.; Strom, S.S.; Lerner, S.; Wang, X.; Burger, J.; Ferrajoli, A.; Kantarjian, H.; et al. Second cancers in patients with chronic lymphocytic leukemia who received frontline fludarabine, cyclophosphamide and rituximab therapy: Distribution and clinical outcomes. Leuk. Lymphoma 2015, 56, 1643–1650. [Google Scholar] [CrossRef]
- Schöllkopf, C.; Rosendahl, D.; Rostgaard, K.; Pipper, C.B.; Hjalgrim, H. Risk of second cancer after chronic lymphocytic leukemia. Int. J. Cancer 2007, 121, 151–156. [Google Scholar] [CrossRef]
- Kumar, V.; Ailawadhi, S.; Bojanini, L.; Mehta, A.; Biswas, S.; Sher, T.; Roy, V.; Vishnu, P.; Marin-Acevedo, J.; Alegria, V.R.; et al. Trends in the risk of second primary malignancies among survivors of chronic lymphocytic leukemia. Blood Cancer J. 2019, 9, 1–10. [Google Scholar] [CrossRef]
- McClanahan, F.; Hanna, B.; Miller, S.; Clear, A.J.; Lichter, P.; Gribben, J.G.; Seiffert, M. PD-L1 checkpoint blockade prevents immune dysfunction and leukemia development in a mouse model of chronic lymphocytic leukemia. Blood 2015, 126, 203–211. [Google Scholar] [CrossRef]
- Ding, W.; LaPlant, B.R.; Call, T.G.; Parikh, S.A.; Leis, J.F.; He, R.; Shanafelt, T.D.; Sinha, S.; Le-Rademacher, J.; Feldman, A.L.; et al. Pembrolizumab in patients with CLL and Richter transformation or with relapsed CLL. Blood 2017, 129, 3419–3427. [Google Scholar] [CrossRef]
- McWilliams, E.M.; Mele, J.M.; Cheney, C.; Timmerman, E.A.; Fiazuddin, F.; Strattan, E.J.; Mo, X.; Byrd, J.C.; Muthusamy, N.; Awan, F.T. Therapeutic CD94/NKG2A blockade improves natural killer cell dysfunction in chronic lymphocytic leukemia. Oncoimmunology 2016, 5, e1226720. [Google Scholar] [CrossRef]
- Mahadevan, D.; Lanasa, M.C.; Farber, C.; Pandey, M.; Whelden, M.; Faas, S.J.; Ulery, T.; Kukreja, A.; Li, L.; Bedrosian, C.L.; et al. Phase I study of samalizumab in chronic lymphocytic leukemia and multiple myeloma: Blockade of the immune checkpoint CD200. J. Immunother. Cancer 2019, 7, 227. [Google Scholar] [CrossRef]
- Khan, M.; Arooj, S.; Wang, H. NK Cell-Based Immune Checkpoint Inhibition. Front. Immunol. 2020, 11, 167. [Google Scholar] [CrossRef]
- Wierz, M.; Pierson, S.; Guyonnet, L.; Viry, E.; Lequeux, A.; Oudin, A.; Niclou, S.P.; Ollert, M.; Berchem, G.; Janji, B.; et al. Dual PD1/LAG3 immune checkpoint blockade limits tumor development in a murine model of chronic lymphocytic leukemia. Blood 2018, 131, 1617–1621. [Google Scholar] [CrossRef]
- Hobo, W.; Norde, W.J.; Schaap, N.; Fredrix, H.; Maas, F.; Schellens, K.; Falkenburg, J.H.F.; Korman, A.J.; Olive, D.; Van Der Voort, R.; et al. B and T Lymphocyte Attenuator Mediates Inhibition of Tumor-Reactive CD8+ T Cells in Patients After Allogeneic Stem Cell Transplantation. J. Immunol. 2012, 189, 39–49. [Google Scholar] [CrossRef]
- Sedy, J.R.; Gavrieli, M.; Potter, K.G.; Hurchla, A.M.; Lindsley, R.C.; Hildner, K.; Scheu, S.; Pfeffer, K.; Ware, C.F.; Murphy, T.L.; et al. B and T lymphocyte attenuator regulates T cell activation through interaction with herpesvirus entry mediator. Nat. Immunol. 2005, 6, 90–98. [Google Scholar] [CrossRef]
- Zhao, Q.; Huang, Z.-L.; He, M.; Gao, Z.; Kuang, D.-M. BTLA identifies dysfunctional PD-1-expressing CD4+ T cells in human hepatocellular carcinoma. Oncoimmunology 2016, 5, e1254855. [Google Scholar] [CrossRef]
- Šedý, J.R.; Ramezani-Rad, P. HVEM network signaling in cancer. Adv. Cancer Res. 2019, 142, 145–186. [Google Scholar] [CrossRef]
- Lan, X.; Li, S.; Gao, H.; Nanding, A.; Quan, L.; Yang, C.; Ding, S.; Xue, Y. Increased BTLA and HVEM in gastric cancer are associated with progression and poor prognosis. OncoTargets Ther. 2017, 10, 919–926. [Google Scholar] [CrossRef]
- Li, X.; Xu, Z.; Cui, G.; Yu, L.; Zhang, X. BTLA Expression in Stage I-III Non-Small-Cell Lung Cancer and Its Correlation with PD-1/PD-L1 and Clinical Outcomes. OncoTargets Ther. 2020, 13, 215–224. [Google Scholar] [CrossRef]
- Quan, L.; Lan, X.; Meng, Y.; Guo, X.; Guo, Y.; Zhao, L.; Chen, X.; Liu, A. BTLA marks a less cytotoxic T-cell subset in diffuse large B-cell lymphoma with high expression of checkpoints. Exp. Hematol. 2018, 60, 47–56.e1. [Google Scholar] [CrossRef]
- Liu, J.; Li, J.; He, M.; Zhang, G.-L.; Zhao, Q. Distinct Changes of BTLA and HVEM Expressions in Circulating CD4+ and CD8+ T Cells in Hepatocellular Carcinoma Patients. J. Immunol. Res. 2018, 2018, 1–8. [Google Scholar] [CrossRef]
- Wang, Q.; Ye, Y.; Yu, H.; Lin, S.H.; Tu, H.; Liang, D.; Chang, D.W.; Huang, M.; Wu, X. Immune check-point-related serum proteins and genetic variants predict outcomes of localized prostate cancer, a cohort study. Cancer Immunol. Immunother. 2020. [Google Scholar] [CrossRef]
- Bian, B.; Fanale, D.; Dusetti, N.; Roque, J.; Pastor, S.; Chretien, A.-S.; Incorvaia, L.; Russo, A.; Olive, D.; Iovanna, J. Prognostic significance of circulating PD-1, PD-L1, pan-BTN3As, BTN3A1 and BTLA in patients with pancreatic adenocarcinoma. OncoImmunology 2019, 8, e1561120. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, J.; Tu, H.; Liang, D.; Chang, D.W.; Ye, Y.; Wu, X. Soluble immune checkpoint-related proteins as predictors of tumor recurrence, survival, and T cell phenotypes in clear cell renal cell carcinoma patients. J. Immunother. Cancer 2019, 7, 334. [Google Scholar] [CrossRef]
- Dong, M.P.; Enomoto, M.; Thuy, L.T.T.; Hai, H.; Hieu, V.N.; Hoang, D.V.; Iida-Ueno, A.; Odagiri, N.; Amano-Teranishi, Y.; Hagihara, A.; et al. Clinical significance of circulating soluble immune checkpoint proteins in sorafenib-treated patients with advanced hepatocellular carcinoma. Sci. Rep. 2020, 10, 1–10. [Google Scholar] [CrossRef]
- Chen, Y.-L.; Lin, H.-W.; Chien, C.-L.; Lai, Y.-L.; Sun, W.-Z.; Chen, C.-A.; Cheng, W.-F. BTLA blockade enhances Cancer therapy by inhibiting IL-6/IL-10-induced CD19high B lymphocytes. J. Immunother. Cancer 2019, 7, 313. [Google Scholar] [CrossRef] [PubMed]
- Fourcade, J.; Sun, Z.; Pagliano, O.; Guillaume, P.; Luescher, I.F.; Sander, C.; Kirkwood, J.M.; Olive, D.; Kuchroo, V.K.; Zarour, H.M. CD8+ T Cells Specific for Tumor Antigens Can Be Rendered Dysfunctional by the Tumor Microenvironment through Upregulation of the Inhibitory Receptors BTLA and PD-1. Cancer Res. 2012, 72, 887–896. [Google Scholar] [CrossRef] [PubMed]
- Šedý, J.R.; Bjordahl, R.L.; Bekiaris, V.; Macauley, M.G.; Ware, B.C.; Norris, P.S.; Lurain, N.S.; Benedict, C.A.; Ware, C.F. CD160 Activation by Herpesvirus Entry Mediator Augments Inflammatory Cytokine Production and Cytolytic Function by NK Cells. J. Immunol. 2013, 191, 828–836. [Google Scholar] [CrossRef] [PubMed]
- Carreras, J.; Lopez-Guillermo, A.; Kikuti, Y.Y.; Itoh, J.; Masashi, M.; Ikoma, H.; Tomita, S.; Hiraiwa, S.; Hamoudi, R.; Rosenwald, A.; et al. High TNFRSF14 and low BTLA are associated with poor prognosis in Follicular Lymphoma and in Diffuse Large B-cell Lymphoma transformation. J. Clin. Exp. Hematop. 2019, 59, 1–16. [Google Scholar] [CrossRef]
- Lesesve, J.-F.; Tardy, S.; Frotscher, B.; Latger-Cannard, V.; Feugier, P.; Bittencourt, M.D.C. Combination of CD160 and CD200 as a useful tool for differential diagnosis between chronic lymphocytic leukemia and other mature B-cell neoplasms. Int. J. Lab. Hematol. 2015, 37, 486–494. [Google Scholar] [CrossRef]
- Liu, F.-T.; Giustiniani, J.; Farren, T.; Jia, L.; Bensussan, A.; Gribben, J.G.; Agrawal, S.G. CD160 signaling mediates PI3K-dependent survival and growth signals in chronic lymphocytic leukemia. Blood 2010, 115, 3079–3088. [Google Scholar] [CrossRef][Green Version]
- Farren, T.W.; Giustiniani, J.; Fanous, M.; Liu, F.; Macey, M.G.; Wright, F.; Prentice, A.M.; Nathwani, A.C.; Agrawal, S.G. Minimal residual disease detection with tumor-specific CD160 correlates with event-free survival in chronic lymphocytic leukemia. Blood Cancer J. 2015, 5, e273. [Google Scholar] [CrossRef][Green Version]
- M’Hidi, H.; Thibult, M.-L.; Chetaille, B.; Rey, F.; Bouadallah, R.; Nicollas, R.; Olive, D.; Xerri, L. High Expression of the Inhibitory Receptor BTLA in T-Follicular Helper Cells and in B-Cell Small Lymphocytic Lymphoma/Chronic Lymphocytic Leukemia. Am. J. Clin. Pathol. 2009, 132, 589–596. [Google Scholar] [CrossRef]
- Trougouboff, P.; Shefer, H.K. B and T lymphocyte attenuator expression in mature B cell lymphomas. J. Hematop. 2012, 6, 57–63. [Google Scholar] [CrossRef]
- Karabon, L.; Partyka, A.; Ciszak, L.; Pawlak-Adamska, E.; Tomkiewicz, A.; Bojarska-Junak, A.; Roliński, J.; Wołowiec, D.; Wrobel, T.; Frydecka, I.; et al. Abnormal Expression of BTLA and CTLA-4 Immune Checkpoint Molecules in Chronic Lymphocytic Leukemia Patients. J. Immunol. Res. 2020, 2020, 1–12. [Google Scholar] [CrossRef]
- Hallek, M.; Cheson, B.D.; Catovsky, D.; Caligaris-Cappio, F.; Dighiero, G.; Döhner, H.; Hillmen, P.; Keating, M.; Montserrat, E.; Chiorazzi, N.; et al. iwCLL guidelines for diagnosis, indications for treatment, response assessment, and supportive management of CLL. Blood 2018, 131, 2745–2760. [Google Scholar] [CrossRef]
- Sordo-Bahamonde, C.; Lorenzo-Herrero, S.; González, S.; López-Soto, A. A cytofluorimetric assay to evaluate intracellular cytokine production by NK cells. Methods Enzymol. 2020, 631, 343–355. [Google Scholar] [CrossRef]
- Lorenzo-Herrero, S.; Sordo-Bahamonde, C.; González, S.; López-Soto, A. Evaluation of NK cell cytotoxic activity against malignant cells by the calcein assay. Methods Enzymol. 2020, 631, 483–495. [Google Scholar] [CrossRef]
- Dumas, J.; Gargano, M.A.; Dancik, G.M. shinyGEO: A web-based application for analyzing gene expression omnibus datasets. Bioinformatics 2016, 32, 3679–3681. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-J.; Yoon, B.-H.; Kim, S.-Y. GENT2: An updated gene expression database for normal and tumor tissues. BMC Med. Genom. 2019, 12, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef] [PubMed]
- Farren, T.W.; Giustiniani, J.; Liu, F.-T.; Tsitsikas, D.A.; Macey, M.G.; Cavenagh, J.D.; Oakervee, H.E.; Taussig, D.; Newland, A.C.; Calaminici, M.; et al. Differential and tumor-specific expression of CD160 in B-cell malignancies. Blood 2011, 118, 2174–2183. [Google Scholar] [CrossRef][Green Version]
- Pasero, C.; Olive, D. Interfering with coinhibitory molecules: BTLA/HVEM as new targets to enhance anti-tumor immunity. Immunol. Lett. 2013, 151, 71–75. [Google Scholar] [CrossRef]
- Steinberg, M.W.; Cheung, T.C.; Ware, C.F. The signaling networks of the herpesvirus entry mediator (TNFRSF14) in immune regulation. Immunol. Rev. 2011, 244, 169–187. [Google Scholar] [CrossRef]
- Wang, J.; Lo, J.C.; Foster, A.; Yu, P.; Chen, H.M.; Wang, Y.; Tamada, K.; Chen, L.; Fu, Y.-X. The regulation of T cell homeostasis and autoimmunity by T cell–derived LIGHT. J. Clin. Investig. 2001, 108, 1771–1780. [Google Scholar] [CrossRef]
- AlHakeem, S.S.; McKenna, M.K.; Oben, K.Z.; Noothi, S.K.; Rivas, J.R.; Hildebrandt, G.C.; Fleischman, R.A.; Rangnekar, V.M.; Muthusamy, N.; Bondada, S. Chronic Lymphocytic Leukemia–Derived IL-10 Suppresses Antitumor Immunity. J. Immunol. 2018, 200, 4180–4189. [Google Scholar] [CrossRef]
- Shaim, H.; Estrov, Z.; Harris, D.; Sanabria, M.H.; Liu, Z.; Ruvolo, P.; Thompson, P.A.; Ferrajoli, A.; Daher, M.; Burger, J.; et al. The CXCR4–STAT3–IL-10 Pathway Controls the Immunoregulatory Function of Chronic Lymphocytic Leukemia and Is Modulated by Lenalidomide. Front. Immunol. 2018, 8, 1773. [Google Scholar] [CrossRef]
- Drennan, S.; D’Avola, A.; Gao, Y.; Weigel, C.; Chrysostomou, E.; Steele, A.J.; Zenz, T.; Plass, C.; Johnson, P.W.; Williams, A.P.; et al. IL-10 production by CLL cells is enhanced in the anergic IGHV mutated subset and associates with reduced DNA methylation of the IL10 locus. Leukemia 2016, 31, 1686–1694. [Google Scholar] [CrossRef]
- Rivas, J.R.; Liu, Y.; Alhakeem, S.S.; Eckenrode, J.M.; Marti, F.; Collard, J.P.; Zhang, Y.; Shaaban, K.A.; Muthu-samy, N.; Hildebrandt, G.C.; et al. Interleukin-10 suppression enhances T-cell antitumor immunity and respons-es to checkpoint blockade in chronic lymphocytic leukemia. Leukemia 2021. [Google Scholar] [CrossRef]
- Gonzalez-Rodriguez, A.P.; Payer, A.R.; Menendez-Suarez, J.J.; Sordo-Bahamonde, C.; Lorenzo-Herrero, S.; Zanabili, J.; Fonseca, A.; Gonzalez-Huerta, A.J.; Palomo, P.; Gonzalez, S. Driver Mutations and Single Copy Number Abnormalities Identify Binet Stage A Patients with Chronic Lymphocytic Leukemia with Aggressive Progression. J. Clin. Med. 2020, 9, 3695. [Google Scholar] [CrossRef]
- Han, L.; Wang, W.; Fang, Y.; Feng, Z.; Liao, S.; Li, W.; Li, Y.; Li, C.; Maitituoheti, M.; Dong, H.; et al. Soluble B and T Lymphocyte Attenuator Possesses Antitumor Effects and Facilitates Heat Shock Protein 70 Vaccine-Triggered Antitumor Immunity against a Murine TC-1 Cervical Cancer Model In Vivo. J. Immunol. 2009, 183, 7842–7850. [Google Scholar] [CrossRef]
- Deng, W.; Gowen, B.G.; Zhang, L.; Wang, L.; Lau, S.; Iannello, A.; Xu, J.; Rovis, T.L.; Xiong, N.; Raulet, D.H. A shed NKG2D ligand that promotes natural killer cell activation and tumor rejection. Science 2015, 348, 136–139. [Google Scholar] [CrossRef]
- Barrow, A.D.; Edeling, M.A.; Trifonov, V.; Luo, J.; Goyal, P.; Bohl, B.; Bando, J.K.; Kim, A.H.; Walker, J.; Andahazy, M.; et al. Natural Killer Cells Control Tumor Growth by Sensing a Growth Factor. Cell 2018, 172, 534–548.e19. [Google Scholar] [CrossRef]
- López-Soto, A.; Gonzalez, S.; Galluzzi, L. Soluble NKG2D ligands limit the efficacy of immune checkpoint blockade. OncoImmunology 2017, 6, e1346766. [Google Scholar] [CrossRef]


| Patients | Patients (n = 46) | % |
|---|---|---|
| Age | ||
| Years (mean) | 72.9 | |
| Sex | ||
| Female | 21 | 45.6 |
| Male | 25 | 54.3 |
| Rai Stage | ||
| 0 | 24 | 54.5 |
| I–II | 14 | 27.2 |
| III–IV | 8 | 18.1 |
| Binet Stage | ||
| A | 34 | 73.9 |
| B | 35 | 10.8 |
| C | 7 | 15.2 |
| Cytogenetic abnormalities (FISH) | ||
| No alterations | 11 | 23.9 |
| del(13q) | 19 | 41.3 |
| del(11q) | 4 | 8.6 |
| del(17p) | 5 | 10.8 |
| Trisomy 12 | 5 | 10.8 |
| Others | 1 | 2.1 |
| Complex karyotype | 6 | 13 |
| IGHV status | ||
| Mutated | 36 | 78.2 |
| Unmutated | 10 | 21.7 |
| Progression | ||
| Stable disease | 29 | 63 |
| Progressive disease | 17 | 36.9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sordo-Bahamonde, C.; Lorenzo-Herrero, S.; Gonzalez-Rodriguez, A.P.; R. Payer, Á.; González-García, E.; López-Soto, A.; Gonzalez, S. BTLA/HVEM Axis Induces NK Cell Immunosuppression and Poor Outcome in Chronic Lymphocytic Leukemia. Cancers 2021, 13, 1766. https://doi.org/10.3390/cancers13081766
Sordo-Bahamonde C, Lorenzo-Herrero S, Gonzalez-Rodriguez AP, R. Payer Á, González-García E, López-Soto A, Gonzalez S. BTLA/HVEM Axis Induces NK Cell Immunosuppression and Poor Outcome in Chronic Lymphocytic Leukemia. Cancers. 2021; 13(8):1766. https://doi.org/10.3390/cancers13081766
Chicago/Turabian StyleSordo-Bahamonde, Christian, Seila Lorenzo-Herrero, Ana P Gonzalez-Rodriguez, Ángel R. Payer, Esther González-García, Alejandro López-Soto, and Segundo Gonzalez. 2021. "BTLA/HVEM Axis Induces NK Cell Immunosuppression and Poor Outcome in Chronic Lymphocytic Leukemia" Cancers 13, no. 8: 1766. https://doi.org/10.3390/cancers13081766
APA StyleSordo-Bahamonde, C., Lorenzo-Herrero, S., Gonzalez-Rodriguez, A. P., R. Payer, Á., González-García, E., López-Soto, A., & Gonzalez, S. (2021). BTLA/HVEM Axis Induces NK Cell Immunosuppression and Poor Outcome in Chronic Lymphocytic Leukemia. Cancers, 13(8), 1766. https://doi.org/10.3390/cancers13081766






