A G316A Polymorphism in the Ornithine Decarboxylase Gene Promoter Modulates MYCN-Driven Childhood Neuroblastoma
Abstract
:Simple Summary
Abstract
1. Introduction
2. Results
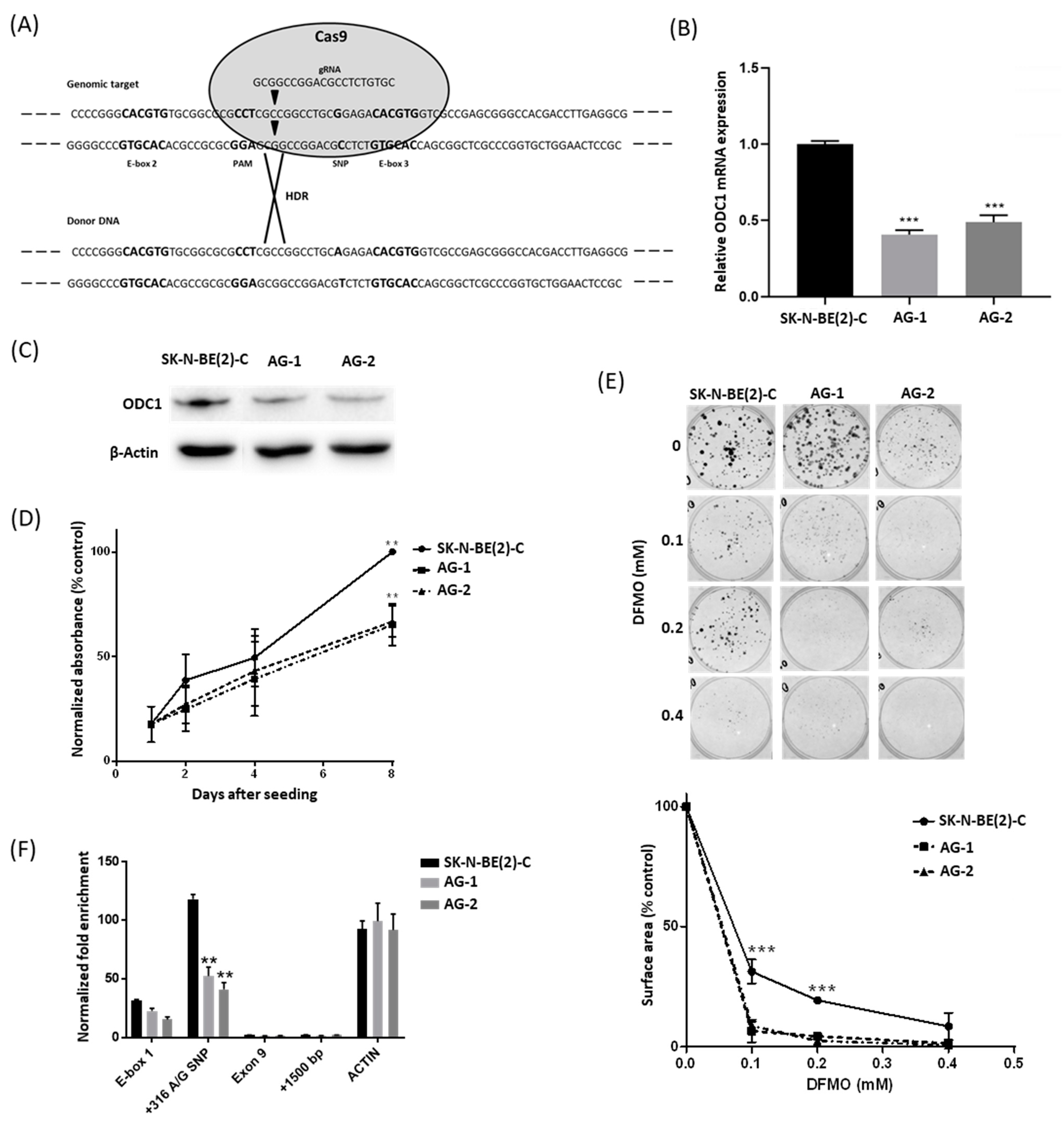
2.1. The 316A Allele Is Associated with Reduced Cell Growth and ODC1 Expression, and Increased Sensitivity to DFMO in Cas9-Edited Neuroblastoma Cells
2.2. The 316A Allele Influences ODC1 Promoter Activity In Vitro
2.3. The ODC1 316A Allele and Outcome in Neuroblastoma Patients
2.4. A Divergent Role for ODC1 in Adult Cancers
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Reagents
4.2. CRISPR-Cas9 Genome Editing
4.3. BrdU Cell Proliferation Assay
4.4. ODC1 Expression in CRISPR-Edited Clones
4.5. Chromatin-Immunoprecipitation (ChIP)
4.6. Clonogenic Assay
4.7. Electrophoretic Mobility Shift Assays (EMSA)
4.8. Luciferase Reporter Assay
4.9. Cohort Descriptions
4.10. ODC1 Expression in Neuroblastoma Patients
4.11. SNP Genotyping
4.12. Statistical Procedures
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cohn, S.L.; Tweddle, D.A. MYCN amplification remains prognostically strong 20 years after its “clinical debut”. Eur. J. Cancer 2004, 40, 2639–2642. [Google Scholar] [CrossRef] [PubMed]
- Thomas, T.; Thomas, T.J. Polyamine metabolism and cancer. J. Cell Mol. Med. 2003, 7, 113–126. [Google Scholar] [CrossRef] [PubMed]
- Bello-Fernandez, C.; Packham, G.; Cleveland, J.L. The ornithine decarboxylase gene is a transcriptional target of c-Myc. Proc. Natl. Acad. Sci. USA 1993, 90, 7804–7808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lutz, W.; Stöhr, M.; Schürmann, J.; Wenzel, A.; Löhr, A.; Schwab, M. Conditional expression of N-MYC in human neuroblastoma cells increases expression of alpha-prothymosin and ornithine decarboxylase and accelerates progression into S-phase early after mitogenic stimulation of quiescent cells. Oncogene 1996, 13, 803–812. [Google Scholar]
- Gerner, E.W.; Meyskens, F.L., Jr. Polyamines and cancer: Old molecules, new understanding. Nat. Rev. Cancer 2004, 4, 781–792. [Google Scholar] [CrossRef] [Green Version]
- Tian, H.; Huang, Q.; Li, L.; Liu, X.-X.; Zhang, Y. Gene expression of ornithine decarboxylase in lung cancers and its clinical significance. Acta Biochim. Biophys. Sin. 2006, 38, 639–645. [Google Scholar] [CrossRef] [Green Version]
- Deng, W.; Jiang, X.; Mei, Y.; Sun, J.; Ma, R.; Liu, X.; Sun, H.; Tian, H.; Sun, X. Role of ornithine decarboxylase in breast cancer. Acta Biochim. Biophys. Sin. 2008, 40, 235–243. [Google Scholar] [CrossRef]
- Hoshino, Y.; Terashima, S.; Teranishi, Y.; Terashima, M.; Kogure, M.; Saitoh, T.; Osuka, F.; Kashimura, S.; Saze, Z.; Gotoh, M. Ornithine decarboxylase activity as a prognostic marker for colorectal cancer. Fukushima J. Med. Sci. 2007, 53, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohan, R.R.; Challa, A.; Gupta, S.; Bostwick, D.G.; Ahmad, N.; Agarwal, R.; Marengo, S.R.; Amini, S.B.; Paras, F.; MacLennan, G.T.; et al. Overexpression of ornithine decarboxylase in prostate cancer and prostatic fluid in humans. Clin. Cancer Res. 1999, 5, 143–147. [Google Scholar] [PubMed]
- Hogarty, M.D.; Norris, M.D.; Davis, K.; Liu, X.; Evageliou, N.F.; Hayes, C.S.; Pawel, B.; Guo, R.; Zhao, H.; Sekyere, E.; et al. ODC1 is a critical determinant of MYCN oncogenesis and a therapeutic target in neuroblastoma. Cancer Res. 2008, 68, 9735–9745. [Google Scholar] [CrossRef] [Green Version]
- Geerts, D.; Koster, J.; Albert, D.; Koomoa, D.L.; Feith, D.J.; Pegg, A.E.; Caron, H.; Bachmann, A. The polyamine metabolism genes ornithine decarboxylase and antizyme 2 predict aggressive behavior in neuroblastomas with and without MYCN amplification. Int. J. Cancer 2010, 126, 2012–2024. [Google Scholar] [PubMed] [Green Version]
- Evageliou, N.F.; Haber, M.; Vu, A.; Laetsch, T.W.; Murray, J.; Gamble, L.D.; Cheng, N.C.; Liu, K.; Reese, M.; Corrigan, K.A.; et al. Polyamine antagonist therapies inhibit neuroblastoma initiation and progression. Clin. Cancer Res. 2016, 22, 4391–4404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gamble, L.D.; Purgato, S.; Murray, J.; Xiao, L.; Yu, D.M.T.; Hanssen, K.M.; Giorgi, F.M.; Carter, D.R.; Gifford, A.J.; Valli, E.; et al. Inhibition of polyamine synthesis and uptake reduces tumor progression and prolongs survival in mouse models of neuroblastoma. Sci. Transl. Med. 2019, 11, 1099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Y.; Harris, R.B.; Rosson, D.; Boorman, D.; O’Brien, T.G. Functional analysis of human ornithine decarboxylase alleles. Cancer Res. 2000, 60, 6314–6317. [Google Scholar] [PubMed]
- Walhout, A.J.M.; Gubbels, M.-J.; Bernards, R.; Van Der Vliet, P.C.; Timmers, H.T.M. C-Myc/Max heterodimers bind cooperatively to the e-box sequences located in the first intron of the rat ornithine decarboxylase (ODC) gene. Nucleic Acids Res. 1997, 25, 1493–1501. [Google Scholar] [CrossRef] [Green Version]
- Solomon, D.L.C.; Amati, B.; Land, H. Distinct DNA binding preferences for the c-Myc/Max and Max/Max dimers. Nucleic Acids Res. 1993, 21, 5372–5376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walhout, A.; Van Der Vliet, P.C.; Timmers, H. Sequences flanking the E-box contribute to cooperative binding by c-Myc/Max heterodimers to adjacent binding sites. Biochim. Biophys. Acta Gene Struct. Expr. 1998, 1397, 189–201. [Google Scholar] [CrossRef]
- Zell, J.A.; Ziogas, A.; Ignatenko, N.; Honda, J.; Qu, N.; Bobbs, A.S.; Neuhausen, S.L.; Gerner, E.W.; Anton-Culver, H. Associations of a polymorphism in the ornithine decarboxylase gene with colorectal cancer survival. Clin. Cancer Res. 2009, 15, 6208–6216. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Long, J.; Wang, P.; Liu, K.; Mai, L.; Guo, Y. Association between the ornithine decarboxylase G316A polymorphism and breast cancer survival. Oncol. Lett. 2015, 10, 485–491. [Google Scholar] [CrossRef]
- Martínez, M.E.; O’Brien, T.G.; Fultz, K.E.; Babbar, N.; Yerushalmi, H.; Qu, N.; Guo, Y.; Boorman, D.; Einspahr, J.; Alberts, D.S.; et al. Pronounced reduction in adenoma recurrence associated with aspirin use and a polymorphism in the ornithine decarboxylase gene. Proc. Natl. Acad. Sci. USA 2003, 100, 7859–7864. [Google Scholar] [CrossRef] [Green Version]
- Hubner, R.A.; Muir, K.R.; Liu, J.-F.; Logan, R.F.; Grainge, M.J.; Houlston, R.S. Ornithine decarboxylase G316A genotype is prognostic for colorectal adenoma recurrence and predicts efficacy of aspirin chemoprevention. Clin. Cancer Res. 2008, 14, 2303–2309. [Google Scholar] [CrossRef] [Green Version]
- Willems, E.; Leyns, L.; Vandesompele, J. Standardization of real-time PCR gene expression data from independent biological replicates. Anal. Biochem. 2008, 379, 127–129. [Google Scholar] [CrossRef]
- Győrffy, B.; Surowiak, P.; Budczies, J.; Lánczky, A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS ONE 2013, 8, e82241. [Google Scholar] [CrossRef] [Green Version]
- Zell, J.A.; McLaren, C.E.; Chen, W.-P.; Thompson, P.A.; Gerner, E.W.; Meyskens, F.L. Ornithine decarboxylase-1 polymorphism, chemoprevention with eflornithine and sulindac, and outcomes among colorectal adenoma patients. J. Natl. Cancer Inst. 2010, 102, 1513–1516. [Google Scholar] [CrossRef] [Green Version]
- Meyskens, F.L., Jr.; Simoneau, A.R.; Gerner, E.W. Chemoprevention of prostate cancer with the polyamine synthesis inhibitor difluoromethylornithine. Recent results in cancer research. Fortschr. Krebsforsch. Prog. Rech. Cancer 2014, 202, 115–120. [Google Scholar]
- Sholler, S.G.L.; Gerner, E.W.; Bergendahl, G.; MacArthur, R.B.; VanderWerff, A.; Ashikaga, T.; Ferguson, W.; Moriya, A. A phase I trial of DFMO targeting polyamine addiction in patients with relapsed/refractory neuroblastoma. PLoS ONE 2015, 10, e0127246. [Google Scholar] [CrossRef] [PubMed]
- Park, J.R.; Kreissman, S.G.; London, W.B.; Naranjo, A.; Cohn, S.L.; Hogarty, M.D.; Kamagutchi, L. Effect of tandem autologous stem cell transplant vs single transplant on event-free survival in patients with high-risk neuroblastoma: A randomized clinical trial. JAMA 2019, 322, 746–755. [Google Scholar] [CrossRef] [PubMed]
- Ladenstein, R.; Pötschger, U.; Pearson, A.D.J.; Brock, P.; Luksch, R.; Castel, V.; Yaniv, I.; Papadakis, V.; Laureys, G.; Malis, J.; et al. Busulfan and melphalan versus carboplatin, etoposide, and melphalan as high-dose chemotherapy for high-risk neuroblastoma (HR-NBL1/SIOPEN): An international, randomised, multi-arm, open-label, phase 3 trial. Lancet Oncol. 2017, 18, 500–514. [Google Scholar] [CrossRef]
- Schneiderman, J.; London, W.B.; Brodeur, G.M.; Castleberry, R.P.; Look, A.T.; Cohn, S.L. Clinical significance of MYCN amplification and ploidy in favorable-stage neuroblastoma: A report from the Children’s Oncology Group. J. Clin. Oncol. 2008, 26, 913–918. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, B.C.; Cheung, A.F.; Simkevich, C.P.; Tam, W.; Ren, X.; Mateyak, M.K.; Sedivy, J.M. A large scale genetic analysis of c-Myc-regulated gene expression patterns. J. Biol. Chem. 2003, 278, 12563–12573. [Google Scholar] [CrossRef] [Green Version]
- Linsalata, M.; Russo, F.; Cavallini, A.; Berloco, P.; Di Leo, A. Polyamines, diamine oxidase, and ornithine decarboxylase activity in colorectal cancer and in normal surrounding mucosa. Dis. Colon Rectum 1993, 36, 662–667. [Google Scholar] [CrossRef]
- Matsubara, N.; Hietala, A.O.; Gilmour, S.K.; Yum, K.Y.; Litwin, S.; Watts, P.; Brennan. O’Brien Association between high levels of ornithine decarboxylase activity and favorable prognosis in human colorectal carcinoma. Clin. Cancer Res. 1995, 1, 665–671. [Google Scholar]
- Manni, A.; Mauger, D.; Gimotty, P.; Badger, B. Prognostic influence on survival of increased ornithine decarboxylase activity in human breast cancer. Clin. Cancer Res. 1996, 2, 1901–1906. [Google Scholar]
- Cañizares, F.; Salinas, J.; Heras, M.D.L.; Diaz, J.; Tovar, I.; Martinez, P.; Peñafiel, R. Prognostic value of ornithine decarboxylase and polyamines in human breast cancer: Correlation with clinicopathologic parameters. Clin. Cancer Res. 1999, 5, 2035–2041. [Google Scholar]
- Love, R.R.; Astrow, S.H.; Cheeks, A.M.; Havighurst, T.C. Ornithine decarboxylase (ODC) as a prognostic factor in operable breast cancer. Breast Cancer Res. Treat. 2003, 79, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Grimminger, P.P.; Schneider, P.M.; Metzger, R.; Vallböhmer, D.; Danenberg, K.D.; Danenberg, P.V.; Hölscher, A.H.; Brabender, J. Ornithine decarboxylase mRNA expression in curatively resected non–small-cell lung cancer. Clin. Lung Cancer 2010, 11, 114–119. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, K.M.; Frank, D.A. CREB in the pathophysiology of cancer: Implications for targeting transcription factors for cancer therapy. Clin. Cancer Res. 2009, 15, 2583–2587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shukla, A.; Bosenberg, M.W.; MacPherson, M.B.; Butnor, K.J.; Heintz, N.H.; Pass, H.I.; Carbone, M.; Testa, J.R.; Mossman, B.T. Activated cAMP response element binding protein is overexpressed in human mesotheliomas and inhibits apoptosis. Am. J. Pathol. 2009, 175, 2197–2206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ni, Y.; Zhou, Y.; Zhou, M.; Zhang, L. Akt and cAMP response element binding protein mediate 17beta-estradiol regulation of glucose transporter 3 expression in human SH-SY5Y neuroblastoma cell line. Neurosci. Lett. 2015, 604, 58–63. [Google Scholar] [CrossRef]
- Zamarbide, M.; Etayo-Labiano, I.; Ricobaraza, A.; Martínez-Pinilla, E.; Aymerich, M.S.; Lanciego, J.L.; Pérez-Mediavilla, A.; Franco, R. GPR40 activation leads to CREB and ERK phosphorylation in primary cultures of neurons from the mouse CNS and in human neuroblastoma cells. Hippocampus 2014, 24, 733–739. [Google Scholar] [CrossRef] [PubMed]
- Sapio, L.; Salzillo, A.; Ragone, A.; Illiano, M.; Spina, A.; Naviglio, S. Targeting CREB in cancer therapy: A key candidate or one of many? An update. Cancers 2020, 12, 3166. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Jin, L.; Nie, S.; Han, L.; Lu, N.; Zhou, Y. miR-205 inhibits neuroblastoma growth by targeting cAMP-responsive element-binding protein. Oncol. Res. 2018, 26, 445–455. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Xue, W.-J.; Feng, Y.; Mao, Q.-S. MicroRNA-205 functions as a tumor suppressor in colorectal cancer by targeting cAMP responsive element binding protein 1 (CREB1). Am. J. Transl. Res. 2015, 7, 2053–2059. [Google Scholar] [PubMed]
- Inoue, K.; Sugiyama, H.; Ogawa, H.; Nakagawa, M.; Yamagami, T.; Miwa, H.; Kita, K.; Hiraoka, A.; Masaoka, T.; Nasu, K. WT1 as a new prognostic factor and a new marker for the detection of minimal residual disease in acute leukemia. Blood 1994, 84, 3071–3079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyoshi, Y.; Ando, A.; Egawa, C.; Taguchi, T.; Tamaki, Y.; Tamaki, H.; Sugiyama, H.; Noguchi, S. High expression of Wilms’ tumor suppressor gene predicts poor prognosis in breast cancer patients. Clin. Cancer Res. 2002, 8, 1167–1171. [Google Scholar]
- Dennis, S.L.; Manji, S.S.; Carrington, D.P.; Scarcella, D.L.; Ashley, D.M.; Smith, P.J.; Algar, E.M. Expression and mutation analysis of the Wilms’ tumor 1 gene in human neural tumors. Int. J. Cancer 2002, 97, 713–715. [Google Scholar] [CrossRef]
- Wang, J.; Oue, T.; Uehara, S.; Yamanaka, H.; Oji, Y.; Fukuzawa, M. The role of WT1 gene in neuroblastoma. J. Pediatr. Surg. 2011, 46, 326–331. [Google Scholar] [CrossRef]
- Masserot, C.; Liu, Q.; Nguyen, E.; Gattolliat, C.-H.; Valteau-Couanet, D.; Bénard, J.; Huber, C.; Ségal-Bendirdjian, E. WT1 expression is inversely correlated with MYCN amplification or expression and associated with poor survival in non-MYCN-amplified neuroblastoma. Mol. Oncol. 2015, 10, 240–252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, A.; Cheng, Z.; Kong, L.; Zhu, Z.; Lin, S.; Gao, G.; Zhang, B. CasOT: A genome-wide Cas9/gRNA off-target searching tool. Bioinformatics 2014, 30, 1180–1182. [Google Scholar] [CrossRef] [Green Version]
- Iraci, N.; Diolaiti, D.; Papa, A.; Porro, A.; Valli, E.; Gherardi, S.; Smith, A.; Diollin, P. A SP1/MIZ1/MYCN repression complex recruits HDAC1 at the TRKA and p75NTR promoters and affects neuroblastoma malignancy by inhibiting the cell response to NGF. Cancer Res. 2011, 71, 404–412. [Google Scholar] [CrossRef] [Green Version]
- Perini, G.; Diolaiti, D.; Porro, A.; Della Valle, G. In vivo transcriptional regulation of N-Myc target genes is controlled by E-box methylation. Proc. Natl. Acad. Sci. USA 2005, 102, 12117–12122. [Google Scholar] [CrossRef] [Green Version]
- Dignam, J.D.; Lebovitz, R.M.; Roeder, R.G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983, 11, 1475–1489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haber, M.; Smith, J.; Bordow, S.B.; Flemming, C.; Cohn, S.L.; London, W.B.; Marshall, G.M.; Norris, M.D. Association of high-level MRP1 expression with poor clinical outcome in a large prospective study of primary neuroblastoma. J. Clin. Oncol. 2006, 24, 1546–1553. [Google Scholar] [CrossRef] [PubMed]
- Pajic, M.; Murray, J.; Marshall, G.M.; Cole, S.P.; Norris, M.D.; Haber, M. ABCC1 G2012T single nucleotide polymorphism is associated with patient outcome in primary neuroblastoma and altered stability of the ABCC1 gene transcript. Pharm. Genom. 2011, 21, 270–279. [Google Scholar] [CrossRef]
- Bordow, S.B.; Haber, M.; Madafiglio, J.; Cheung, B.; Marshall, G.M.; Norris, M.D. Expression of the multidrug resistance-associated protein (MRP) gene correlates with amplification and overexpression of the N-myc oncogene in childhood neuroblastoma. Cancer Res. 1994, 54, 5036–5040. [Google Scholar]
- Norris, M.D.; Haber, M.; Gilbert, J.; Kavallaris, M.; Marshall, G.M.; Stewart, B.W. N-myc gene amplification in neuroblastoma determined by the polymerase chain reaction. Prog. Clin. Biol. Res. 1994, 385, 27–33. [Google Scholar]
- Vermeulen, J.; De Preter, K.; Naranjo, A.; Vercruysse, L.; Van Roy, N.; Hellemans, J.; Swerts, K.; Bravo, S.; Scaruffi, P.; Tonini, G.P.; et al. Predicting outcomes for children with neuroblastoma using a multigene-expression signature: A retrospective SIOPEN/COG/GPOH study. Lancet Oncol. 2009, 10, 663–671. [Google Scholar] [CrossRef] [Green Version]
- Rihani, A.; De Wilde, B.; Zeka, F.; Laureys, G.; Francotte, N.; Tonini, G.P.; Coco, S.; Versteeg, R.; Noguera, R.; Schulte, J.H.; et al. CASP8 SNP D302H (rs1045485) is associated with worse survival in MYCN-amplified neuroblastoma patients. PLoS ONE 2014, 9, e114696. [Google Scholar] [CrossRef] [Green Version]
- Diskin, S.J.; Capasso, M.; Schnepp, R.W.; Cole, A.K.; Attiyeh, E.F.; Hou, C.; Diamond, M.; Carpenter, E.L.; Winter, C.; Lee, H.; et al. Common variation at 6q16 within HACE1 and LIN28B influences susceptibility to neuroblastoma. Nat. Genet. 2012, 44, 1126–1130. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [Green Version]
- Delaneau, O.; Marchini, J.; Zagury, J.-F. A linear complexity phasing method for thousands of genomes. Nat. Methods 2011, 9, 179–181. [Google Scholar] [CrossRef] [PubMed]
- Howie, B.N.; Donnelly, P.; Marchini, J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009, 5, e1000529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henderson, M.J.; Haber, M.; Porro, A.; Munoz, M.A.; Iraci, N.; Xue, C.; Murray, J.; Flemming, C.L.; Smith, J.; Fletcher, J.I.; et al. ABCC multidrug transporters in childhood neuroblastoma: Clinical and biological effects independent of cytotoxic drug efflux. J. Natl. Cancer Inst. 2011, 103, 1236–1251. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Factors | Genotype | Total | p | ||||
|---|---|---|---|---|---|---|---|
| AA | AG | GG | |||||
| Study cohort | Age | ≤18 months | 36 (7.9%) | 141 (31.0%) | 278 (61.1%) | 455 | 0.551 |
| >18 months | 25 (6.5%) | 130 (33.9%) | 228 (59.5%) | 383 | |||
| Tumor stage | Favourable | 31 (8.9%) | 96 (27.5%) | 223 (63.7%) | 350 | 0.049 | |
| Unfavourable | 28 (6.2%) | 158 (35.2%) | 263 (58.6%) | 449 | |||
| MYCN status | Non-amplified | 50 (7.2%) | 224 (32.2%) | 421 (60.6%) | 695 | 0.970 | |
| Amplified | 11 (7.7%) | 45 (31.7%) | 86 (60.6%) | 142 | |||
| GWAS cohort | Age | ≤18 months | 187 (8.3%) | 887 (39.4%) | 1177 (52.3%) | 2251 | 0.328 |
| >18 months | 238 (9.0%) | 1076 (40.7%) | 1327 (50.2%) | 2641 | |||
| Risk group | Low | 144 (9.2%) | 622 (39.9%) | 794 (50.9%) | 1560 | 0.826 | |
| Intermediate | 100 (8.8%) | 451 (39.9%) | 580 (51.3%) | 1131 | |||
| High | 171 (8.2%) | 855 (40.8%) | 1070 (51.0%) | 2096 | |||
| MYCN status | Non-amplified | 341 (9.2%) | 1469 (39.7%) | 1889 (51.1%) | 3699 | 0.033 | |
| Amplified | 54 (6.4%) | 349 (41.5%) | 437 (52.0%) | 840 | |||
| Sample | Genotype | Event-Free Survival | Overall Survival | ||
|---|---|---|---|---|---|
| Relative Hazard (95% CI) | p | Relative Hazard (95% CI) | p | ||
| Study cohort | |||||
| All patients | GG | 0.98 (0.77–1.24) | 0.847 | 0.99 (0.75–1.29) | 0.914 |
| AG/GG | 1.63 (0.95–2.78) | 0.074 | 1.45 (0.81–2.59) | 0.212 | |
| MYCN non-amplified | GG | 0.79 (0.59–1.06) | 0.120 | 0.76 (0.54–1.07) | 0.115 |
| AG/GG | 2.46 (1.09–5.45) | 0.025 | 2.05 (0.84–5.01) | 0.103 | |
| MYCN amplified | GG | 1.93 (1.27–2.93) | 0.001 | 1.91 (1.22–2.97) | 0.003 |
| AG/GG | 1.07 (0.52–2.21) | 0.846 | 1.13 (0.52–2.45) | 0.752 | |
| GWAS cohort | |||||
| All patients | GG | 0.96 (0.87–1.07) | 0.469 | 0.98 (0.87–1.11) | 0.792 |
| AG/GG | 1.10 (0.91–1.32) | 0.323 | 1.14 (0.92–1.43) | 0.237 | |
| MYCN non-amplified | GG | 0.88 (0.77–1.01) | 0.063 | 0.87 (0.74–1.03) | 0.107 |
| AG/GG | 1.02 (0.81–1.29) | 0.858 | 1.03 (0.78–1.37) | 0.828 | |
| MYCN amplified | GG | 1.13 (0.94–1.36) | 0.197 | 1.15 (0.95–1.40) | 0.152 |
| AG/GG | 0.93 (0.65–1.34) | 0.711 | 1.12 (0.74–1.69) | 0.597 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gamble, L.D.; Purgato, S.; Henderson, M.J.; Di Giacomo, S.; Russell, A.J.; Pigini, P.; Murray, J.; Valli, E.; Milazzo, G.; Giorgi, F.M.; et al. A G316A Polymorphism in the Ornithine Decarboxylase Gene Promoter Modulates MYCN-Driven Childhood Neuroblastoma. Cancers 2021, 13, 1807. https://doi.org/10.3390/cancers13081807
Gamble LD, Purgato S, Henderson MJ, Di Giacomo S, Russell AJ, Pigini P, Murray J, Valli E, Milazzo G, Giorgi FM, et al. A G316A Polymorphism in the Ornithine Decarboxylase Gene Promoter Modulates MYCN-Driven Childhood Neuroblastoma. Cancers. 2021; 13(8):1807. https://doi.org/10.3390/cancers13081807
Chicago/Turabian StyleGamble, Laura D., Stefania Purgato, Michelle J. Henderson, Simone Di Giacomo, Amanda J. Russell, Paolo Pigini, Jayne Murray, Emanuele Valli, Giorgio Milazzo, Federico M. Giorgi, and et al. 2021. "A G316A Polymorphism in the Ornithine Decarboxylase Gene Promoter Modulates MYCN-Driven Childhood Neuroblastoma" Cancers 13, no. 8: 1807. https://doi.org/10.3390/cancers13081807
APA StyleGamble, L. D., Purgato, S., Henderson, M. J., Di Giacomo, S., Russell, A. J., Pigini, P., Murray, J., Valli, E., Milazzo, G., Giorgi, F. M., Cowley, M., Ashton, L. J., Bhalshankar, J., Schleiermacher, G., Rihani, A., Van Maerken, T., Vandesompele, J., Speleman, F., Versteeg, R., ... Haber, M. (2021). A G316A Polymorphism in the Ornithine Decarboxylase Gene Promoter Modulates MYCN-Driven Childhood Neuroblastoma. Cancers, 13(8), 1807. https://doi.org/10.3390/cancers13081807










