The microRNA-210-Stathmin1 Axis Decreases Cell Stiffness to Facilitate the Invasiveness of Colorectal Cancer Stem Cells
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmids, shRNA Clones and Synthetic Oligonucleotides
2.2. Cell Culture
2.3. Expansion of Colorectal Cancer Stem Cells
2.4. Transwell Migration Assay
2.5. Two-and-a-Half Dimentional (2.5D) Time-Lapse Trajectory
2.6. Three-Dimensional (3D) Invasion Assay
2.7. Paired Cell Assay
2.8. Elasticity Measurements
2.9. Cell Viability, Clonogenicity and Sphere Formation Assays
2.10. Lentivirus Production and Transduction
2.11. RNA Extraction and Quantitative RT-PCR
2.12. Bioinformatic Analysis
2.13. Immunoblotting
2.14. Immunohistochemical (IHC) Assay
2.15. Preparation of Patient-Derived Xenografts (PDXs)
2.16. Biological Samples
2.17. Statistical Analysis
3. Results
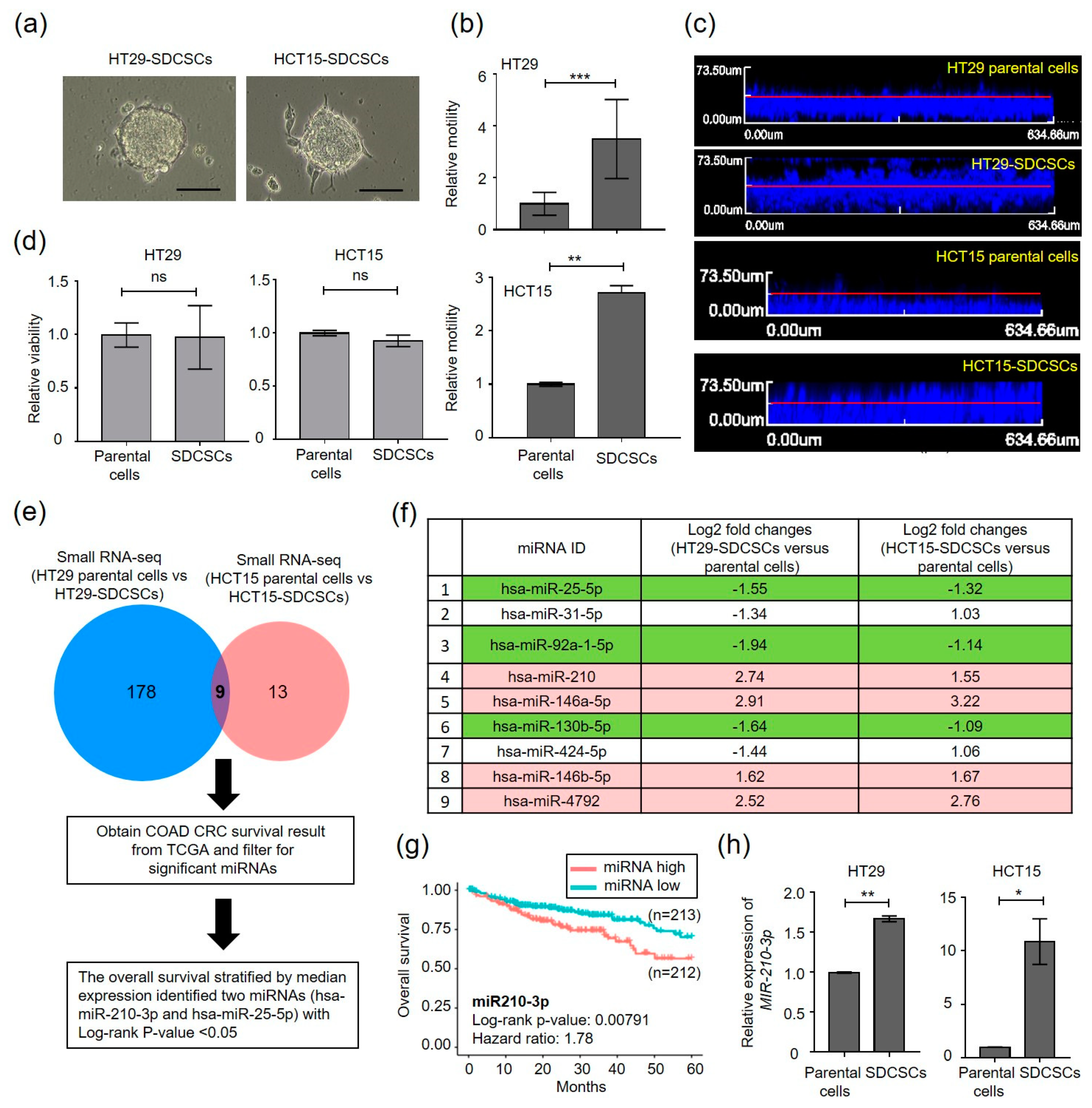
3.1. Small RNA Sequencing (smRNA-seq) Reveals Enhanced miR-210-3p Expression in CRCSCs
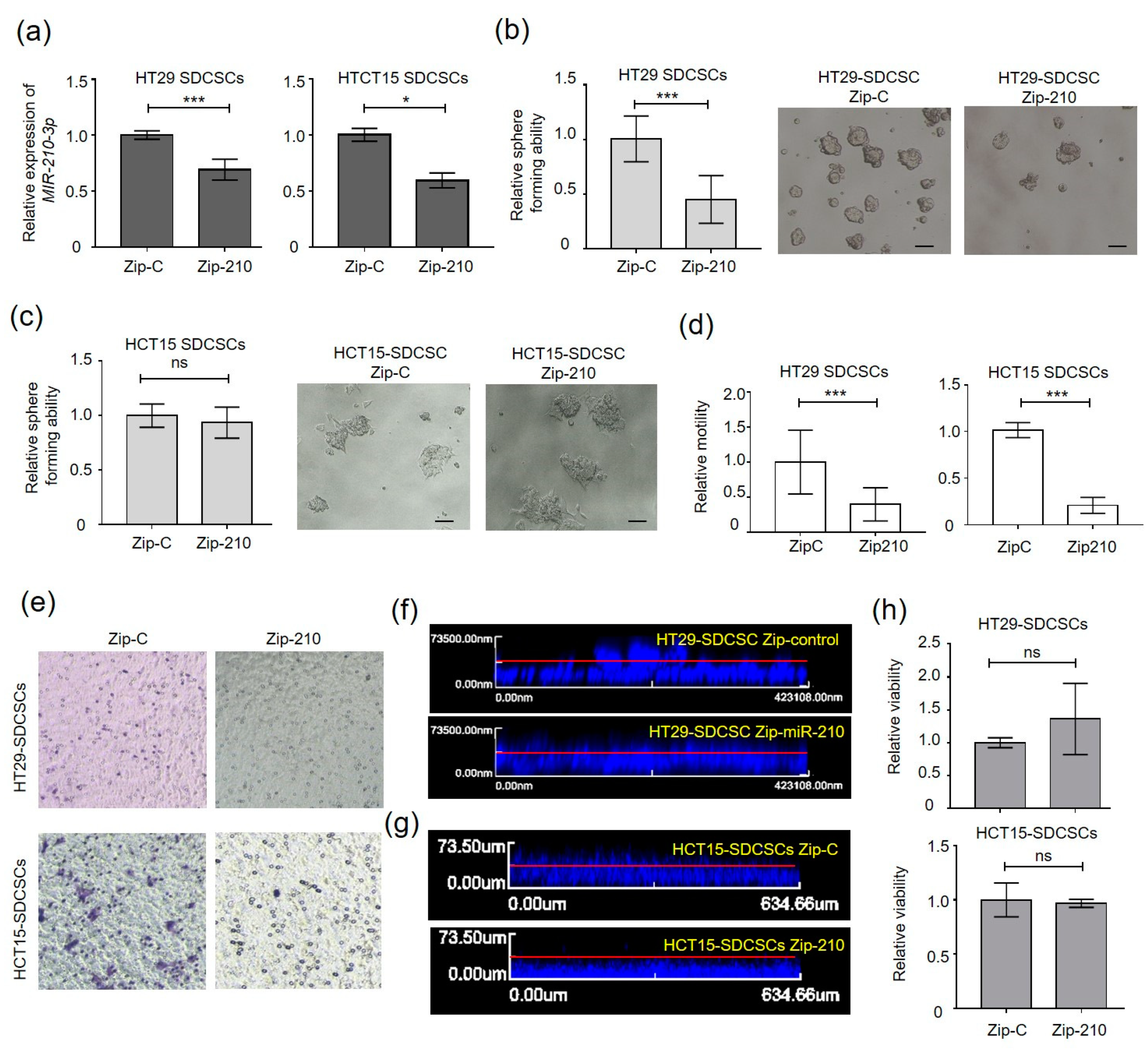
3.2. MiR-210 Is Required for the Migration and Invasiveness of CRCSCs
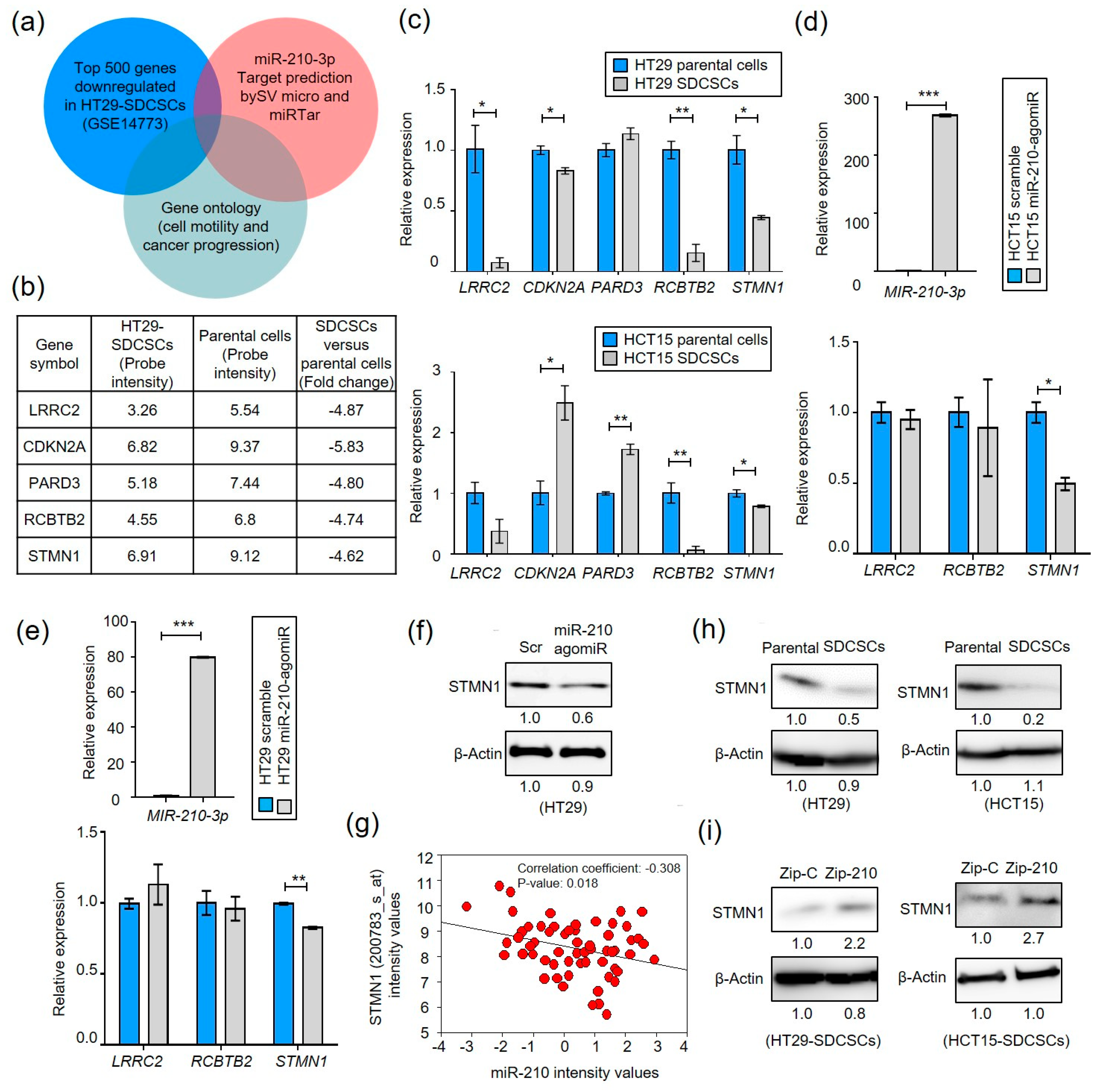
3.3. MiR-210 Suppresses Stathmin1 in CRCSCs
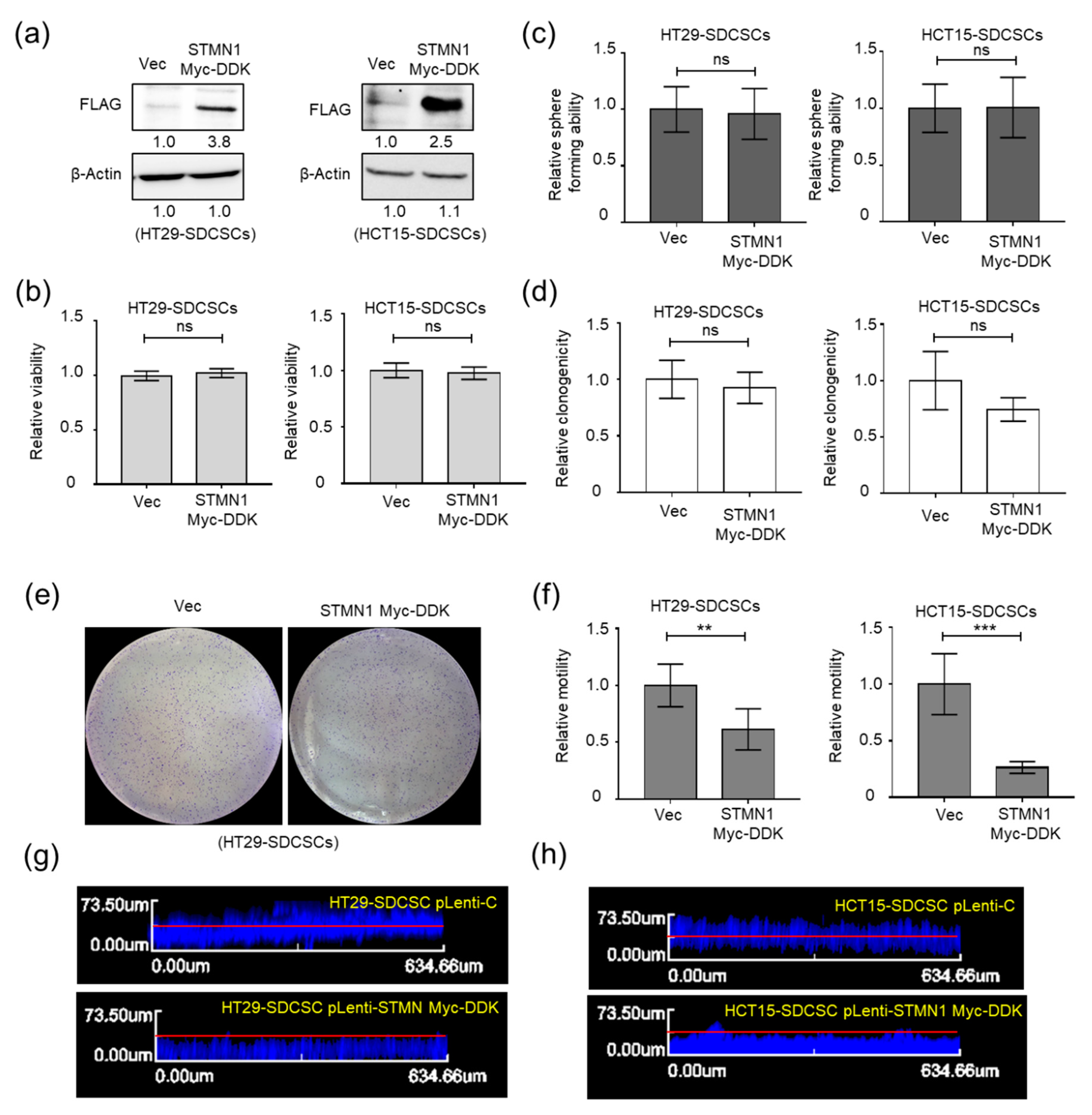
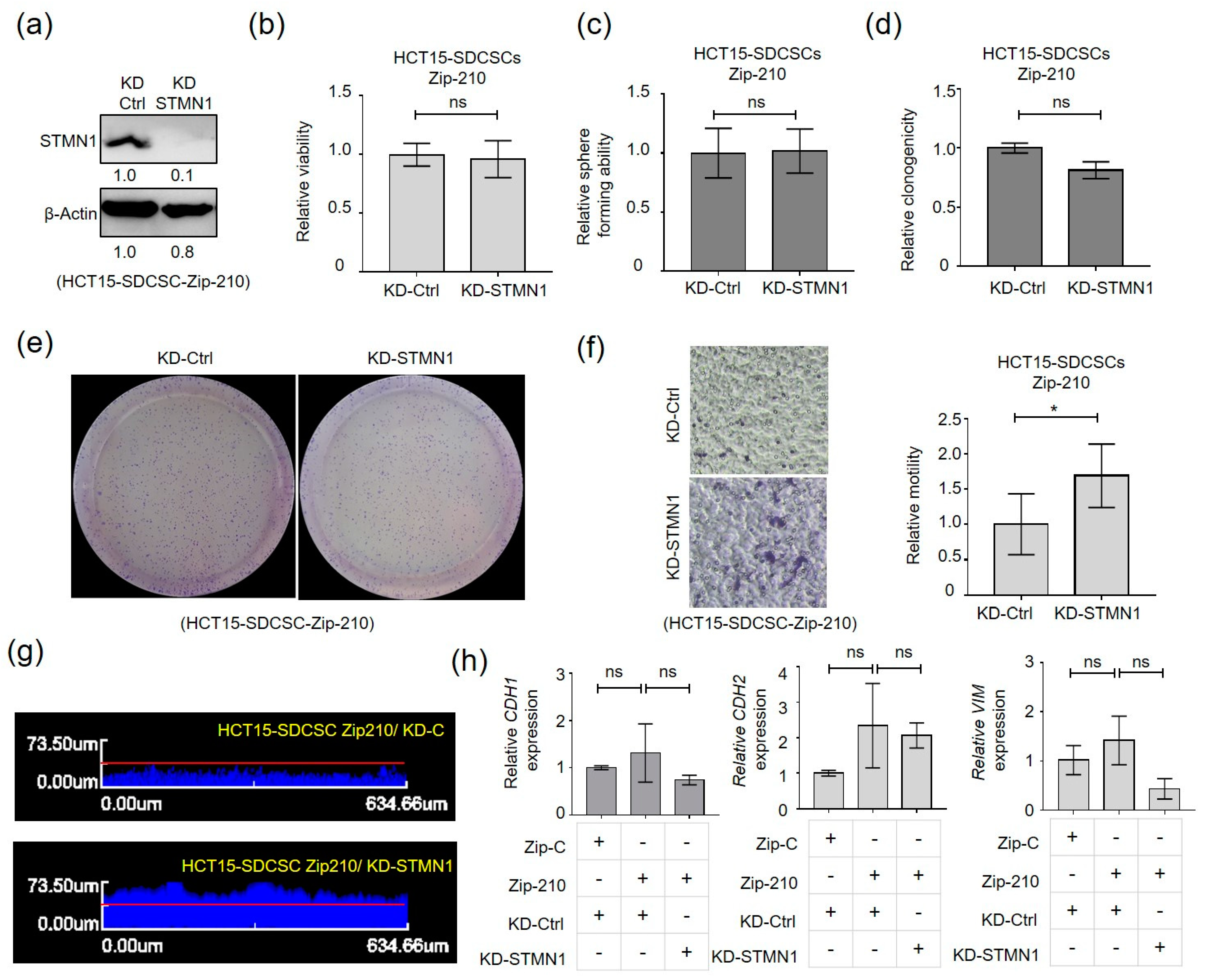
3.4. Stathmin1 Expression Attenuates the Motility of CRCSCs
3.5. The miR-210-STMN1 Axis Determines the Stiffness of CRCSCs
3.6. The miR-210-STMN1 Axis Promotes the Stiffness of CRC Cells
3.7. The miR-210High/STMN1Low Expression Signature Is Associated with Liver Metastasis and Predicts a Poor Clinical Outcome in CRC Patients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chiang, M.Y.; Yangben, Y.; Lin, N.J.; Zhong, J.L.; Yang, L. Relationships among cell morphology, intrinsic cell stiffness and cell-substrate interactions. Biomaterials 2013, 34, 9754–9762. [Google Scholar] [CrossRef]
- Matzke, R.; Jacobson, K.; Radmacher, M. Direct, high-resolution measurement of furrow stiffening during division of adherent cells. Nat. Cell Biol. 2001, 3, 607–610. [Google Scholar] [CrossRef] [PubMed]
- Shimamoto, Y.; Maeda, Y.T.; Ishiwata, S.; Libchaber, A.J.; Kapoor, T.M. Insights into the micromechanical properties of the metaphase spindle. Cell 2011, 145, 1062–1074. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.Q.; Kuo, C.Y.; Wei, M.T.; Wu, K.; Su, P.T.; Huang, C.S.; Chiou, A.E. Intracellular viscoelasticity of HeLa cells during cell division studied by video particle-tracking microrheology. J. Biomed. Opt. 2014, 19, 011008. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.W.; Chen, Y.Q.; Huang, C.M.; Liu, C.Y.; Chiou, A.; Wang, Y.K.; Tang, M.J.; Kuo, J.C. beta-PIX controls intracellular viscoelasticity to regulate lung cancer cell migration. J. Cell. Mol. Med. 2015, 19, 934–947. [Google Scholar] [CrossRef]
- Kole, T.P.; Tseng, Y.; Jiang, I.; Katz, J.L.; Wirtz, D. Intracellular mechanics of migrating fibroblasts. Mol. Biol. Cell 2005, 16, 328–338. [Google Scholar] [CrossRef]
- Winkler, J.; Abisoye-Ogunniyan, A.; Metcalf, K.J.; Werb, Z. Concepts of extracellular matrix remodelling in tumour progression and metastasis. Nat. Commun. 2020, 11, 5120. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, D.; Tardin, C.; Schmidt, C.F.; Mackintosh, F.C. Nonequilibrium mechanics of active cytoskeletal networks. Science 2007, 315, 370–373. [Google Scholar] [CrossRef] [PubMed]
- Wirtz, D. Particle-tracking microrheology of living cells: Principles and applications. Annu. Rev. Biophys. 2009, 38, 301–326. [Google Scholar] [CrossRef] [Green Version]
- Copley, A.L.; King, R.G.; Chien, S.; Usami, S.; Skalak, R.; Huang, C.R. Microscopic observations of viscoelasticity of human blood in steady and oscillatory shear. Biorheology 1975, 12, 257–263. [Google Scholar] [CrossRef]
- Visvader, J.E.; Lindeman, G.J. Cancer stem cells: Current status and evolving complexities. Cell Stem Cell 2012, 10, 717–728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Brien, C.A.; Pollett, A.; Gallinger, S.; Dick, J.E. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature 2006, 445, 106–110. [Google Scholar] [CrossRef]
- Ricci-Vitiani, L.; Lombardi, D.G.; Pilozzi, E.; Biffoni, M.; Todaro, M.; Peschle, C.; De Maria, R. Identification and expansion of human colon-cancer-initiating cells. Nature 2006, 445, 111–115. [Google Scholar] [CrossRef] [PubMed]
- Dylla, S.J.; Beviglia, L.; Park, I.K.; Chartier, C.; Raval, J.; Ngan, L.; Pickell, K.; Aguilar, J.; Lazetic, S.; Smith-Berdan, S.; et al. Colorectal cancer stem cells are enriched in xenogeneic tumors following chemotherapy. PLoS ONE 2008, 3, e2428. [Google Scholar] [CrossRef]
- Pang, R.; Law, W.L.; Chu, A.C.; Poon, J.T.; Lam, C.S.; Chow, A.K.; Ng, L.; Cheung, L.W.; Lan, X.R.; Lan, H.Y.; et al. A subpopulation of CD26+ cancer stem cells with metastatic capacity in human colorectal cancer. Cell Stem Cell 2010, 6, 603–615. [Google Scholar] [CrossRef] [Green Version]
- de Sousa e Melo, F.; Kurtova, A.V.; Harnoss, J.M.; Kljavin, N.; Hoeck, J.D.; Hung, J.; Anderson, J.E.; Storm, E.E.; Modrusan, Z.; Koeppen, H.; et al. A distinct role for Lgr5+ stem cells in primary and metastatic colon cancer. Nature 2017, 543, 676–680. [Google Scholar] [CrossRef]
- Mani, S.A.; Guo, W.; Liao, M.J.; Eaton, E.N.; Ayyanan, A.; Zhou, A.Y.; Brooks, M.; Reinhard, F.; Zhang, C.C.; Shipitsin, M.; et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008, 133, 704–715. [Google Scholar] [CrossRef] [Green Version]
- Vermeulen, L.; De Sousa, E.M.F.; van der Heijden, M.; Cameron, K.; de Jong, J.H.; Borovski, T.; Tuynman, J.B.; Todaro, M.; Merz, C.; Rodermond, H.; et al. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat. Cell Biol. 2010, 12, 468–476. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [Green Version]
- Croce, C.M. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 2009, 10, 704–714. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Tan, X.; Huang, Z.; Chen, Z.; Lin, P.; Fu, S.W. microRNA biomarkers in colorectal cancer liver metastasis. J. Cancer 2018, 9, 3867–3873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, D.; Zhao, S.; Tang, H.; Zhang, D.; Sun, H.; Yu, F.; Jiang, W.; Yue, B.; Wang, J.; Zhang, M.; et al. MicroRNA-20a-5p promotes colorectal cancer invasion and metastasis by downregulating Smad4. Oncotarget 2016, 7, 45199–45213. [Google Scholar] [CrossRef] [Green Version]
- Lam, C.S.; Ng, L.; Chow, A.K.; Wan, T.M.; Yau, S.; Cheng, N.S.; Wong, S.K.M.; Man, J.H.W.; Lo, O.S.H.; Foo, D.C.C.; et al. Identification of microRNA 885-5p as a novel regulator of tumor metastasis by targeting CPEB2 in colorectal cancer. Oncotarget 2017, 8, 26858–26870. [Google Scholar] [CrossRef] [Green Version]
- Hwang, W.L.; Yang, M.H.; Tsai, M.L.; Lan, H.Y.; Su, S.H.; Chang, S.C.; Teng, H.W.; Yang, S.H.; Lan, Y.T.; Chiou, S.H.; et al. SNAIL regulates interleukin-8 expression, stem cell-like activity, and tumorigenicity of human colorectal carcinoma cells. Gastroenterology 2011, 141, 279–291.e5. [Google Scholar] [CrossRef] [Green Version]
- Matsui, Y.; Nakayama, Y.; Okamoto, M.; Fukumoto, Y.; Yamaguchi, N. Enrichment of cell populations in metaphase, anaphase, and telophase by synchronization using nocodazole and blebbistatin: A novel method suitable for examining dynamic changes in proteins during mitotic progression. Eur. J. Cell Biol. 2012, 91, 413–419. [Google Scholar] [CrossRef]
- Wu, P.H.; Hale, C.M.; Chen, W.C.; Lee, J.S.; Tseng, Y.; Wirtz, D. High-throughput ballistic injection nanorheology to measure cell mechanics. Nat. Protoc. 2012, 7, 155–170. [Google Scholar] [CrossRef] [Green Version]
- del Alamo, J.C.; Norwich, G.N.; Li, Y.S.; Lasheras, J.C.; Chien, S. Anisotropic rheology and directional mechanotransduction in vascular endothelial cells. Proc. Natl. Acad. Sci. USA 2008, 105, 15411–15416. [Google Scholar] [CrossRef] [Green Version]
- Baker, E.L.; Bonnecaze, R.T.; Zaman, M.H. Extracellular matrix stiffness and architecture govern intracellular rheology in cancer. Biophys. J. 2009, 97, 1013–1021. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.H.; Shen, P.C.; Chen, C.Y.; Hsu, A.N.; Cho, Y.C.; Lai, Y.L.; Chen, F.H.; Li, C.Y.; Wang, S.C.; Chen, M.; et al. DriverDBv3: A multi-omics database for cancer driver gene research. Nucleic Acids Res. 2020, 48, D863–D870. [Google Scholar] [CrossRef] [Green Version]
- Chung, I.F.; Chang, S.J.; Chen, C.Y.; Liu, S.H.; Li, C.Y.; Chan, C.H.; Shih, C.C.; Cheng, W.C. YM500v3: A database for small RNA sequencing in human cancer research. Nucleic Acids Res. 2017, 45, D925–D931. [Google Scholar] [CrossRef]
- Reinhold, W.C.; Sunshine, M.; Liu, H.; Varma, S.; Kohn, K.W.; Morris, J.; Doroshow, J.; Pommier, Y. CellMiner: A web-based suite of genomic and pharmacologic tools to explore transcript and drug patterns in the NCI-60 cell line set. Cancer Res. 2012, 72, 3499–3511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.; Kotliarova, S.; Kotliarov, Y.; Li, A.; Su, Q.; Donin, N.M.; Pastorino, S.; Purow, B.W.; Christopher, N.; Zhang, W.; et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell 2006, 9, 391–403. [Google Scholar] [CrossRef] [Green Version]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduct Target Ther. 2016, 1, 15004. [Google Scholar] [CrossRef] [Green Version]
- Sugiarto, S.; Persson, A.I.; Munoz, E.G.; Waldhuber, M.; Lamagna, C.; Andor, N.; Hanecker, P.; Ayers-Ringler, J.; Phillips, J.; Siu, J.; et al. Asymmetry-defective oligodendrocyte progenitors are glioma precursors. Cancer Cell 2011, 20, 328–340. [Google Scholar] [CrossRef] [Green Version]
- Hwang, W.L.; Jiang, J.K.; Yang, S.H.; Huang, T.S.; Lan, H.Y.; Teng, H.W.; Yang, C.Y.; Tsai, Y.P.; Lin, C.H.; Wang, H.W.; et al. MicroRNA-146a directs the symmetric division of Snail-dominant colorectal cancer stem cells. Nat. Cell Biol. 2014, 16, 268–280. [Google Scholar] [CrossRef]
- Rando, T.A. The immortal strand hypothesis: Segregation and reconstruction. Cell 2007, 129, 1239–1243. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Yue, D.; Chen, Y.; Gao, S.J.; Huang, Y. Improving performance of mammalian microRNA target prediction. BMC Bioinform. 2010, 11, 476. [Google Scholar] [CrossRef] [Green Version]
- Hsu, J.B.; Chiu, C.M.; Hsu, S.D.; Huang, W.Y.; Chien, C.H.; Lee, T.Y.; Huang, H.D. miRTar: An integrated system for identifying miRNA-target interactions in human. BMC Bioinform. 2011, 12, 300. [Google Scholar] [CrossRef] [Green Version]
- Kiga, K.; Mimuro, H.; Suzuki, M.; Shinozaki-Ushiku, A.; Kobayashi, T.; Sanada, T.; Kim, M.; Ogawa, M.; Iwasaki, Y.W.; Kayo, H.; et al. Epigenetic silencing of miR-210 increases the proliferation of gastric epithelium during chronic Helicobacter pylori infection. Nat. Commun. 2014, 5, 4497. [Google Scholar] [CrossRef] [PubMed]
- Marklund, U.; Larsson, N.; Gradin, H.M.; Brattsand, G.; Gullberg, M. Oncoprotein 18 is a phosphorylation-responsive regulator of microtubule dynamics. EMBO J. 1996, 15, 5290–5298. [Google Scholar] [CrossRef]
- Cassimeris, L. The oncoprotein 18/stathmin family of microtubule destabilizers. Curr. Opin. Cell Biol. 2002, 14, 18–24. [Google Scholar] [CrossRef]
- Williams, K.; Ghosh, R.; Giridhar, P.V.; Gu, G.; Case, T.; Belcher, S.M.; Kasper, S. Inhibition of stathmin1 accelerates the metastatic process. Cancer Res. 2012, 72, 5407–5417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suresh, S. Biomechanics and biophysics of cancer cells. Acta Biomater. 2007, 3, 413–438. [Google Scholar] [CrossRef]
- Belmont, L.D.; Mitchison, T.J. Identification of a protein that interacts with tubulin dimers and increases the catastrophe rate of microtubules. Cell 1996, 84, 623–631. [Google Scholar] [CrossRef] [Green Version]
- Jourdain, L.; Curmi, P.; Sobel, A.; Pantaloni, D.; Carlier, M.F. Stathmin: A tubulin-sequestering protein which forms a ternary T2S complex with two tubulin molecules. Biochemistry 1997, 36, 10817–10821. [Google Scholar] [CrossRef]
- Curmi, P.A.; Andersen, S.S.; Lachkar, S.; Gavet, O.; Karsenti, E.; Knossow, M.; Sobel, A. The stathmin/tubulin interaction in vitro. J. Biol. Chem. 1997, 272, 25029–25036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larsson, N.; Marklund, U.; Gradin, H.M.; Brattsand, G.; Gullberg, M. Control of microtubule dynamics by oncoprotein 18: Dissection of the regulatory role of multisite phosphorylation during mitosis. Mol. Cell Biol. 1997, 17, 5530–5539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wittmann, T.; Bokoch, G.M.; Waterman-Storer, C.M. Regulation of microtubule destabilizing activity of Op18/stathmin downstream of Rac1. J. Biol. Chem. 2004, 279, 6196–6203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iancu, C.; Mistry, S.J.; Arkin, S.; Wallenstein, S.; Atweh, G.F. Effects of stathmin inhibition on the mitotic spindle. J. Cell Sci. 2001, 114 Pt 5, 909–916. [Google Scholar]
- Machado-Neto, J.A.; de Melo Campos, P.; Favaro, P.; Lazarini, M.; Lorand-Metze, I.; Costa, F.F.; Saad, S.T.O.; Traina, F. Stathmin 1 is involved in the highly proliferative phenotype of high-risk myelodysplastic syndromes and acute leukemia cells. Leuk. Res. 2014, 38, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.N.; Arcasoy, M.O.; Brickner, H.E.; Mistry, S.; Schechter, A.D.; Atweh, G.F. Regulated expression of p18, a major phosphoprotein of leukemic cells. J. Biol. Chem. 1991, 266, 21004–21010. [Google Scholar] [CrossRef]
- Rubin, C.I.; French, D.L.; Atweh, G.F. Stathmin expression and megakaryocyte differentiation: A potential role in polyploidy. Exp. Hematol. 2003, 31, 389–397. [Google Scholar] [CrossRef]
- Iancu-Rubin, C.; Gajzer, D.; Tripodi, J.; Najfeld, V.; Gordon, R.E.; Hoffman, R.; Atweh, G.F. Down-regulation of stathmin expression is required for megakaryocyte maturation and platelet production. Blood 2011, 117, 4580–4589. [Google Scholar] [CrossRef] [Green Version]
- Ramlogan-Steel, C.A.; Steel, J.C.; Fathallah, H.; Iancu-Rubin, C.; Soleimani, M.; Dong, Z.; Atweh, G.F. The role of Stathmin, a regulator of mitosis, in hematopoiesis. Blood 2012, 120, 3453. [Google Scholar] [CrossRef]
- Liedtke, W.; Leman, E.E.; Fyffe, R.E.; Raine, C.S.; Schubart, U.K. Stathmin-deficient mice develop an age-dependent axonopathy of the central and peripheral nervous systems. Am. J. Pathol. 2002, 160, 469–480. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, T.B.; Prabhu, V.V.; Piao, Y.H.; Oh, Y.E.; Zahra, R.F.; Chung, Y.C. Effects of Stathmin 1 Gene Knockout on Behaviors and Dopaminergic Markers in Mice Exposed to Social Defeat Stress. Brain Sci. 2019, 9, 215. [Google Scholar] [CrossRef] [Green Version]
- Biaoxue, R.; Xiguang, C.; Hua, L.; Shuanying, Y. Stathmin-dependent molecular targeting therapy for malignant tumor: The latest 5 years’ discoveries and developments. J. Transl. Med. 2016, 14, 279. [Google Scholar] [CrossRef] [Green Version]
- Wu, W.; Tan, X.F.; Tan, H.T.; Lim, T.K.; Chung, M.C. Unbiased proteomic and transcript analyses reveal that stathmin-1 silencing inhibits colorectal cancer metastasis and sensitizes to 5-fluorouracil treatment. Mol. Cancer Res. 2014, 12, 1717–1728. [Google Scholar] [CrossRef] [Green Version]
- Obayashi, S.; Horiguchi, J.; Higuchi, T.; Katayama, A.; Handa, T.; Altan, B.; Bai, T.; Bao, P.; Bao, H.; Yokobori, T.; et al. Stathmin1 expression is associated with aggressive phenotypes and cancer stem cell marker expression in breast cancer patients. Int. J. Oncol. 2017, 51, 781–790. [Google Scholar] [CrossRef] [Green Version]
- Kang, W.; Tong, J.H.; Chan, A.W.; Lung, R.W.; Chau, S.L.; Wong, Q.W.; Wong, N.; Yu, J.; Cheng, A.S.; To, K.F. Stathmin1 plays oncogenic role and is a target of microRNA-223 in gastric cancer. PLoS ONE 2012, 7, e33919. [Google Scholar] [CrossRef]
- Wong, Q.W.; Lung, R.W.; Law, P.T.; Lai, P.B.; Chan, K.Y.; To, K.F.; Wong, N. MicroRNA-223 is commonly repressed in hepatocellular carcinoma and potentiates expression of Stathmin1. Gastroenterology 2008, 135, 257–269. [Google Scholar] [CrossRef]
- Chakravarthi, B.; Chandrashekar, D.S.; Agarwal, S.; Balasubramanya, S.A.H.; Pathi, S.S.; Goswami, M.T.; Jing, X.; Wang, R.; Mehra, R.; Asangani, I.A.; et al. miR-34a Regulates Expression of the Stathmin-1 Oncoprotein and Prostate Cancer Progression. Mol. Cancer Res. 2018, 16, 1125–1137. [Google Scholar] [CrossRef] [Green Version]
- Misek, D.E.; Chang, C.L.; Kuick, R.; Hinderer, R.; Giordano, T.J.; Beer, D.G.; Hanash, S.M. Transforming properties of a Q18-->E mutation of the microtubule regulator Op18. Cancer Cell 2002, 2, 217–228. [Google Scholar] [CrossRef] [Green Version]
- Holmfeldt, P.; Sellin, M.E.; Gullberg, M. Upregulated Op18/stathmin activity causes chromosomal instability through a mechanism that evades the spindle assembly checkpoint. Exp. Cell Res. 2010, 316, 2017–2026. [Google Scholar] [CrossRef]
- Lawrence, M.S.; Stojanov, P.; Mermel, C.H.; Robinson, J.T.; Garraway, L.A.; Golub, T.R.; Meyerson, M.; Gabriel, S.B.; Lander, E.S.; Getz, G. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 2014, 505, 495–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Andrea, S.; Berton, S.; Segatto, I.; Fabris, L.; Canzonieri, V.; Colombatti, A.; Vecchione, A.; Belletti, B.; Baldassarre, G. Stathmin is dispensable for tumor onset in mice. PLoS ONE 2012, 7, e45561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niethammer, P.; Bastiaens, P.; Karsenti, E. Stathmin-tubulin interaction gradients in motile and mitotic cells. Science 2004, 303, 1862–1866. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Huang, X.; Zheng, X.; Wang, X.; Li, S.; Zhang, L.; Yang, Z.; Xia, Z. Enrichment of prostate cancer stem-like cells from human prostate cancer cell lines by culture in serum-free medium and chemoradiotherapy. Int. J. Biol. Sci. 2013, 9, 472–479. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liao, T.-T.; Cheng, W.-C.; Yang, C.-Y.; Chen, Y.-Q.; Su, S.-H.; Yeh, T.-Y.; Lan, H.-Y.; Lee, C.-C.; Lin, H.-H.; Lin, C.-C.; et al. The microRNA-210-Stathmin1 Axis Decreases Cell Stiffness to Facilitate the Invasiveness of Colorectal Cancer Stem Cells. Cancers 2021, 13, 1833. https://doi.org/10.3390/cancers13081833
Liao T-T, Cheng W-C, Yang C-Y, Chen Y-Q, Su S-H, Yeh T-Y, Lan H-Y, Lee C-C, Lin H-H, Lin C-C, et al. The microRNA-210-Stathmin1 Axis Decreases Cell Stiffness to Facilitate the Invasiveness of Colorectal Cancer Stem Cells. Cancers. 2021; 13(8):1833. https://doi.org/10.3390/cancers13081833
Chicago/Turabian StyleLiao, Tsai-Tsen, Wei-Chung Cheng, Chih-Yung Yang, Yin-Quan Chen, Shu-Han Su, Tzu-Yu Yeh, Hsin-Yi Lan, Chih-Chan Lee, Hung-Hsin Lin, Chun-Chi Lin, and et al. 2021. "The microRNA-210-Stathmin1 Axis Decreases Cell Stiffness to Facilitate the Invasiveness of Colorectal Cancer Stem Cells" Cancers 13, no. 8: 1833. https://doi.org/10.3390/cancers13081833
APA StyleLiao, T. -T., Cheng, W. -C., Yang, C. -Y., Chen, Y. -Q., Su, S. -H., Yeh, T. -Y., Lan, H. -Y., Lee, C. -C., Lin, H. -H., Lin, C. -C., Lu, R. -H., Chiou, A. E. -T., Jiang, J. -K., & Hwang, W. -L. (2021). The microRNA-210-Stathmin1 Axis Decreases Cell Stiffness to Facilitate the Invasiveness of Colorectal Cancer Stem Cells. Cancers, 13(8), 1833. https://doi.org/10.3390/cancers13081833







