Minimal Disease Monitoring in Pediatric Non-Hodgkin’s Lymphoma: Current Clinical Application and Future Challenges
Abstract
:Simple Summary
Abstract
1. Introduction
2. Current Status of MDD and MRD in Lymphoblastic Lymphoma (LBL)
3. Current Status of MDD and MRD in Burkitt Lymphoma/Leukemia (BL/B-AL)
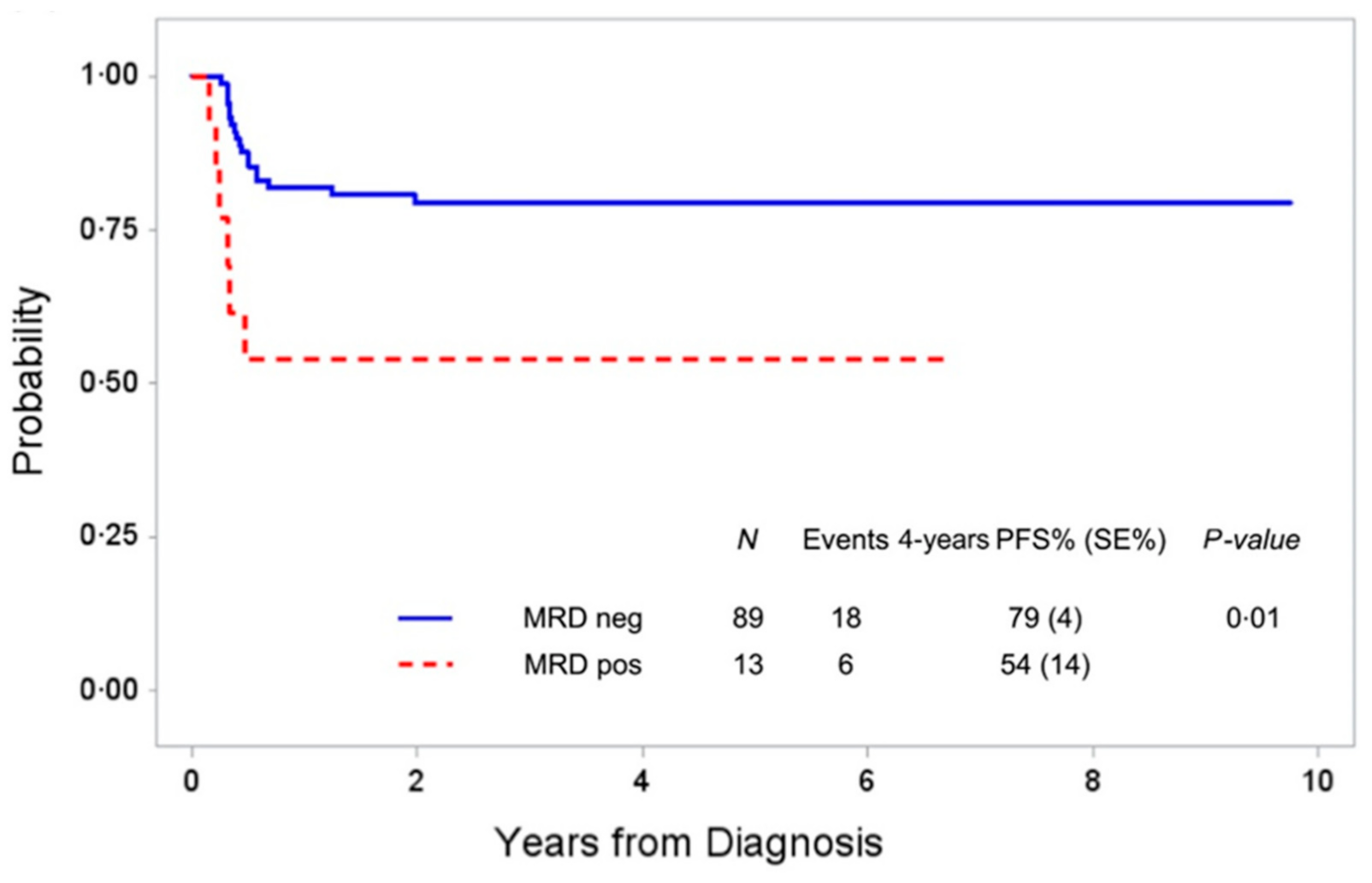
4. Current Status of MDD and MRD in Anaplastic Large Cell Lymphoma (ALCL)
5. Data on Minimal Disease in Rarer Childhood NHL
6. Future Outlook of Minimal Disease Evaluation in Childhood NHL
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Burkhardt, B.; Zimmermann, M.; Oschlies, I.; Niggli, F.; Mann, G.; Parwaresch, R.; Riehm, H.; Schrappe, M.; Reiter, A.; BFM Group. The impact of age and gender on biology, clinical features and treatment outcome of non-hodgkin lymphoma in childhood and adolescence. Br. J. Haematol. 2005, 131, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Brugieres, L.; Le Deley, M.C.; Rosolen, A.; Williams, D.; Horibe, K.; Wrobel, G.; Mann, G.; Zsiros, J.; Uyttebroeck, A.; Marky, I.; et al. Impact of the methotrexate administration dose on the need for intrathecal treatment in children and adolescents with anaplastic large-cell lymphoma: Results of a randomized trial of the EICNHL group. J. Clin. Oncol. 2009, 27, 897–903. [Google Scholar] [CrossRef] [PubMed]
- Landmann, E.; Burkhardt, B.; Zimmermann, M.; Meyer, U.; Woessmann, W.; Klapper, W.; Wrobel, G.; Rosolen, A.; Pillon, M.; Escherich, G.; et al. Results and conclusions of the European intergroup euro-LB02 trial in children and adolescents with lymphoblastic lymphoma. Haematologica 2017, 102, 2086–2096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minard-Colin, V.; Auperin, A.; Pillon, M.; Burke, G.A.A.; Barkauskas, D.A.; Wheatley, K.; Delgado, R.F.; Alexander, S.; Uyttebroeck, A.; Bollard, C.M.; et al. Rituximab for high-risk, mature B-cell non-Hodgkin’s lymphoma in children. N. Engl. J. Med. 2020, 382, 2207–2219. [Google Scholar] [CrossRef]
- Minard-Colin, V.; Brugieres, L.; Reiter, A.; Cairo, M.S.; Gross, T.G.; Woessmann, W.; Burkhardt, B.; Sandlund, J.T.; Williams, D.; Pillon, M.; et al. Non-Hodgkin lymphoma in children and adolescents: Progress through effective collaboration, current knowledge, and challenges ahead. J. Clin. Oncol. 2015, 33, 2963–2974. [Google Scholar] [CrossRef]
- Burkhardt, B.; Hermiston, M.L. Lymphoblastic lymphoma in children and adolescents: Review of current challenges and future opportunities. Br. J. Haematol. 2019, 185, 1158–1170. [Google Scholar] [CrossRef]
- Woessmann, W.; Zimmermann, M.; Meinhardt, A.; Muller, S.; Hauch, H.; Knorr, F.; Oschlies, I.; Klapper, W.; Niggli, F.; Kabickova, E.; et al. Progressive or relapsed Burkitt lymphoma or leukemia in children and adolescents after BFM-type first-line therapy. Blood 2020, 135, 1124–1132. [Google Scholar] [CrossRef]
- Woessmann, W. XI. How to treat children and adolescents with relapsed non-Hodgkin lymphoma? Hematol. Oncol. 2013, 31, 64–68. [Google Scholar] [CrossRef]
- Knorr, F.; Brugieres, L.; Pillon, M.; Zimmermann, M.; Ruf, S.; Attarbaschi, A.; Mellgren, K.; Burke, G.A.A.; Uyttebroeck, A.; Wrobel, G.; et al. Stem cell transplantation and vinblastine monotherapy for relapsed pediatric anaplastic large cell lymphoma: Results of the international, prospective ALCL-relapse trial. J. Clin. Oncol. 2020, 38, 3999–4009. [Google Scholar] [CrossRef]
- Campana, D.; Pui, C.H. Minimal residual disease-guided therapy in childhood acute lymphoblastic leukemia. Blood 2017, 129, 1913–1918. [Google Scholar] [CrossRef] [Green Version]
- Schrappe, M. Detection and management of minimal residual disease in acute lymphoblastic leukemia. Hematol. Am. Soc. Hematol. Educ. Program. 2014, 2014, 244–249. [Google Scholar] [CrossRef] [Green Version]
- van Dongen, J.J.; van der Velden, V.H.; Bruggemann, M.; Orfao, A. Minimal residual disease diagnostics in acute lymphoblastic leukemia: Need for sensitive, fast, and standardized technologies. Blood 2015, 125, 3996–4009. [Google Scholar] [CrossRef] [Green Version]
- Chase, M.L.; Armand, P. Minimal residual disease in non-hodgkin lymphoma—Current applications and future directions. Br. J. Haematol. 2018, 180, 177–188. [Google Scholar] [CrossRef] [Green Version]
- Herrera, A.F.; Armand, P. Minimal residual disease assessment in lymphoma: Methods and applications. J. Clin. Oncol. 2017, 35, 3877–3887. [Google Scholar] [CrossRef]
- Rosolen, A.; Perkins, S.L.; Pinkerton, C.R.; Guillerman, R.P.; Sandlund, J.T.; Patte, C.; Reiter, A.; Cairo, M.S. Revised international pediatric non-Hodgkin lymphoma staging system. J. Clin. Oncol. 2015, 33, 2112–2118. [Google Scholar] [CrossRef]
- Kuroda, T.; Morikawa, N.; Matsuoka, K.; Fujino, A.; Honna, T.; Nakagawa, A.; Kumagai, M.; Masaki, H.; Saeki, M. Prognostic significance of circulating tumor cells and bone marrow micrometastasis in advanced neuroblastoma. J. Pediatr. Surg. 2008, 43, 2182–2185. [Google Scholar] [CrossRef]
- Kurtz, D.M.; Green, M.R.; Bratman, S.V.; Scherer, F.; Liu, C.L.; Kunder, C.A.; Takahashi, K.; Glover, C.; Keane, C.; Kihira, S.; et al. Noninvasive monitoring of diffuse large b-cell lymphoma by immunoglobulin high-throughput sequencing. Blood 2015, 125, 3679–3687. [Google Scholar] [CrossRef] [Green Version]
- Roschewski, M.; Dunleavy, K.; Pittaluga, S.; Moorhead, M.; Pepin, F.; Kong, K.; Shovlin, M.; Jaffe, E.S.; Staudt, L.M.; Lai, C.; et al. Circulating tumour DNA and CT monitoring in patients with untreated diffuse large B-cell lymphoma: A correlative biomarker study. Lancet Oncol. 2015, 16, 541–549. [Google Scholar] [CrossRef] [Green Version]
- Schleiermacher, G.; Peter, M.; Oberlin, O.; Philip, T.; Rubie, H.; Mechinaud, F.; Sommelet-Olive, D.; Landman-Parker, J.; Bours, D.; Michon, J.; et al. Increased risk of systemic relapses associated with bone marrow micrometastasis and circulating tumor cells in localized ewing tumor. J. Clin. Oncol. 2003, 21, 85–91. [Google Scholar] [CrossRef]
- Vo, K.T.; Edwards, J.V.; Epling, C.L.; Sinclair, E.; Hawkins, D.S.; Grier, H.E.; Janeway, K.A.; Barnette, P.; McIlvaine, E.; Krailo, M.D.; et al. Impact of two measures of micrometastatic disease on clinical outcomes in patients with newly diagnosed ewing sarcoma: A report from the children’s oncology group. Clin. Cancer Res. 2016, 22, 3643–3650. [Google Scholar] [CrossRef] [Green Version]
- Crist, W.M.; Shuster, J.J.; Falletta, J.; Pullen, D.J.; Berard, C.W.; Vietti, T.J.; Alvarado, C.S.; Roper, M.A.; Prasthofer, E.; Grossi, C.E. Clinical features and outcome in childhood T-cell leukemia-lymphoma according to stage of thymocyte differentiation: A pediatric oncology group study. Blood 1988, 72, 1891–1897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coustan-Smith, E.; Sandlund, J.T.; Perkins, S.L.; Chen, H.; Chang, M.; Abromowitch, M.; Campana, D. Minimal disseminated disease in childhood T-cell lymphoblastic lymphoma: A report from the children’s oncology group. J. Clin. Oncol. 2009, 27, 3533–3539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mussolin, L.; Buldini, B.; Lovisa, F.; Carraro, E.; Disaro, S.; Lo Nigro, L.; d’Amore, E.S.; Pillon, M.; Basso, G. Detection and role of minimal disseminated disease in children with lymphoblastic lymphoma: The AIEOP experience. Pediatr. Blood Cancer 2015, 62, 1906–1913. [Google Scholar] [CrossRef] [PubMed]
- Stark, B.; Avigad, S.; Luria, D.; Manor, S.; Reshef-Ronen, T.; Avrahami, G.; Yaniv, I. Bone marrow minimal disseminated disease (MDD) and minimal residual disease (MRD) in childhood T-cell lymphoblastic lymphoma stage III, detected by flow cytometry (FC) and real-time quantitative polymerase chain reaction (RQ-PCR). Pediatr. Blood Cancer 2009, 52, 20–25. [Google Scholar] [CrossRef]
- Balbach, S.T.; Makarova, O.; Bonn, B.R.; Zimmermann, M.; Rohde, M.; Oschlies, I.; Klapper, W.; Rossig, C.; Burkhardt, B. Proposal of a genetic classifier for risk group stratification in pediatric T-cell lymphoblastic lymphoma reveals differences from adult T-cell lymphoblastic leukemia. Leukemia 2016, 30, 970–973. [Google Scholar] [CrossRef]
- Bonn, B.R.; Rohde, M.; Zimmermann, M.; Krieger, D.; Oschlies, I.; Niggli, F.; Wrobel, G.; Attarbaschi, A.; Escherich, G.; Klapper, W.; et al. Incidence and prognostic relevance of genetic variations in T-cell lymphoblastic lymphoma in childhood and adolescence. Blood 2013, 121, 3153–3160. [Google Scholar] [CrossRef] [Green Version]
- Callens, C.; Baleydier, F.; Lengline, E.; Ben Abdelali, R.; Petit, A.; Villarese, P.; Cieslak, A.; Minard-Colin, V.; Rullier, A.; Moreau, A.; et al. Clinical impact of notch1 and/or fbxw7 mutations, flash deletion, and TCR status in pediatric T-cell lymphoblastic lymphoma. J. Clin. Oncol. 2012, 30, 1966–1973. [Google Scholar] [CrossRef]
- Dalla-Favera, R.; Bregni, M.; Erikson, J.; Patterson, D.; Gallo, R.C.; Croce, C.M. Human c-myc onc gene is located on the region of chromosome 8 that is translocated in Burkitt lymphoma cells. Proc. Natl. Acad. Sci. USA 1982, 79, 7824–7827. [Google Scholar] [CrossRef] [Green Version]
- Mussolin, L.; Basso, K.; Pillon, M.; D’Amore, E.S.; Lombardi, A.; Luzzatto, L.; Zanesco, L.; Rosolen, A. Prospective analysis of minimal bone marrow infiltration in pediatric Burkitt’s lymphomas by long-distance polymerase chain reaction for t(8;14)(q24;q32). Leukemia 2003, 17, 585–589. [Google Scholar] [CrossRef] [Green Version]
- Mussolin, L.; Pillon, M.; d’Amore, E.S.; Conter, V.; Piglione, M.; Lo Nigro, L.; Garaventa, A.; Buffardi, S.; Arico, M.; Rosolen, A. Minimal disseminated disease in high-risk Burkitt’s lymphoma identifies patients with different prognosis. J. Clin. Oncol. 2011, 29, 1779–1784. [Google Scholar] [CrossRef]
- Pillon, M.; Mussolin, L.; Carraro, E.; Conter, V.; Arico, M.; Vinti, L.; Garaventa, A.; Piglione, M.; Buffardi, S.; Sala, A.; et al. Detection of prognostic factors in children and adolescents with Burkitt and diffuse large B-cell lymphoma treated with the AIEOP LNH-97 protocol. Br. J. Haematol. 2016, 175, 467–475. [Google Scholar] [CrossRef]
- Busch, K.; Borkhardt, A.; Wossmann, W.; Reiter, A.; Harbott, J. Combined polymerase chain reaction methods to detect c-myc/igh rearrangement in childhood Burkitt’s lymphoma for minimal residual disease analysis. Haematologica 2004, 89, 818–825. [Google Scholar]
- Lovisa, F.; Mussolin, L.; Corral, L.; Pillon, M.; Cazzaniga, G.; Biondi, A.; Rosolen, A. IGH and IGK gene rearrangements as PCR targets for pediatric Burkitt’s lymphoma and mature B-all MRD analysis. Lab. Investig. 2009, 89, 1182–1186. [Google Scholar] [CrossRef]
- van Dongen, J.J.; Langerak, A.W.; Bruggemann, M.; Evans, P.A.; Hummel, M.; Lavender, F.L.; Delabesse, E.; Davi, F.; Schuuring, E.; Garcia-Sanz, R.; et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: Report of the biomed-2 concerted action BMH4-CT98-3936. Leukemia 2003, 17, 2257–2317. [Google Scholar] [CrossRef] [Green Version]
- Agsalda, M.; Kusao, I.; Troelstrup, D.; Shiramizu, B. Screening for residual disease in pediatric Burkitt lymphoma using consensus primer pools. Adv. Hematol. 2009, 2009, 412163. [Google Scholar] [CrossRef] [Green Version]
- Shiramizu, B.; Goldman, S.; Smith, L.; Agsalda-Garcia, M.; Galardy, P.; Perkins, S.L.; Frazer, J.K.; Sanger, W.; Anderson, J.R.; Gross, T.G.; et al. Impact of persistent minimal residual disease post-consolidation therapy in children and adolescents with advanced Burkitt leukaemia: A children’s oncology group pilot study report. Br. J. Haematol. 2015, 170, 367–371. [Google Scholar] [CrossRef]
- Mussolin, L.; Pillon, M.; Conter, V.; Piglione, M.; Lo Nigro, L.; Pierani, P.; Micalizzi, C.; Buffardi, S.; Basso, G.; Zanesco, L.; et al. Prognostic role of minimal residual disease in mature B-cell acute lymphoblastic leukemia of childhood. J. Clin. Oncol. 2007, 25, 5254–5261. [Google Scholar] [CrossRef]
- Mussolin, L.; Lovisa, F.; Gallingani, I.; Cavallaro, E.; Carraro, E.; Damanti, C.C.; Vinti, L.; Sala, A.; Micalizzi, C.; Santoro, N.; et al. Minimal residual disease analysis in childhood mature B-cell leukaemia/lymphoma treated with AIEOP LNH-97 protocol with/without anti-CD20 administration. Br. J. Haematol. 2020, 189, e108–e111. [Google Scholar] [CrossRef] [Green Version]
- Damm-Welk, C.; Klapper, W.; Oschlies, I.; Gesk, S.; Rottgers, S.; Bradtke, J.; Siebert, R.; Reiter, A.; Woessmann, W. Distribution of NPM1-ALK and X-ALK fusion transcripts in paediatric anaplastic large cell lymphoma: A molecular-histological correlation. Br. J. Haematol. 2009, 146, 306–309. [Google Scholar] [CrossRef]
- Lamant, L.; McCarthy, K.; d’Amore, E.; Klapper, W.; Nakagawa, A.; Fraga, M.; Maldyk, J.; Simonitsch-Klupp, I.; Oschlies, I.; Delsol, G.; et al. Prognostic impact of morphologic and phenotypic features of childhood alk-positive anaplastic large-cell lymphoma: Results of the ALCL99 study. J. Clin. Oncol. 2011, 29, 4669–4676. [Google Scholar] [CrossRef]
- Morris, S.W.; Kirstein, M.N.; Valentine, M.B.; Dittmer, K.G.; Shapiro, D.N.; Saltman, D.L.; Look, A.T. Fusion of a kinase gene, alk, to a nucleolar protein gene, NPM, in non-Hodgkin’s lymphoma. Science 1994, 263, 1281–1284. [Google Scholar] [CrossRef] [PubMed]
- Perkins, S.L.; Pickering, D.; Lowe, E.J.; Zwick, D.; Abromowitch, M.; Davenport, G.; Cairo, M.S.; Sanger, W.G. Childhood anaplastic large cell lymphoma has a high incidence of alk gene rearrangement as determined by immunohistochemical staining and fluorescent in situ hybridisation: A genetic and pathological correlation. Br. J. Haematol. 2005, 131, 624–627. [Google Scholar] [CrossRef] [PubMed]
- Montes-Mojarro, I.A.; Steinhilber, J.; Bonzheim, I.; Quintanilla-Martinez, L.; Fend, F. The pathological spectrum of systemic anaplastic large cell lymphoma (ALCL). Cancers (Basel) 2018, 10, 107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Damm-Welk, C.; Busch, K.; Burkhardt, B.; Schieferstein, J.; Viehmann, S.; Oschlies, I.; Klapper, W.; Zimmermann, M.; Harbott, J.; Reiter, A.; et al. Prognostic significance of circulating tumor cells in bone marrow or peripheral blood as detected by qualitative and quantitative PCR in pediatric NPM-ALK-positive anaplastic large-cell lymphoma. Blood 2007, 110, 670–677. [Google Scholar] [CrossRef]
- Mussolin, L.; Pillon, M.; d’Amore, E.S.; Santoro, N.; Lombardi, A.; Fagioli, F.; Zanesco, L.; Rosolen, A. Prevalence and clinical implications of bone marrow involvement in pediatric anaplastic large cell lymphoma. Leukemia 2005, 19, 1643–1647. [Google Scholar] [CrossRef] [Green Version]
- Damm-Welk, C.; Mussolin, L.; Zimmermann, M.; Pillon, M.; Klapper, W.; Oschlies, I.; d’Amore, E.S.; Reiter, A.; Woessmann, W.; Rosolen, A. Early assessment of minimal residual disease identifies patients at very high relapse risk in NPM-ALK-positive anaplastic large-cell lymphoma. Blood 2014, 123, 334–337. [Google Scholar] [CrossRef] [Green Version]
- Iijima-Yamashita, Y.; Mori, T.; Nakazawa, A.; Fukano, R.; Takimoto, T.; Tsurusawa, M.; Kobayashi, R.; Horibe, K. Prognostic impact of minimal disseminated disease and immune response to NPM-ALK in Japanese children with ALK-positive anaplastic large cell lymphoma. Int J. Hematol. 2018, 107, 244–250. [Google Scholar] [CrossRef]
- Mussolin, L.; Damm-Welk, C.; Pillon, M.; Zimmermann, M.; Franceschetto, G.; Pulford, K.; Reiter, A.; Rosolen, A.; Woessmann, W. Use of minimal disseminated disease and immunity to NPM-ALK antigen to stratify ALK-positive ALCL patients with different prognosis. Leukemia 2013, 27, 416–422. [Google Scholar] [CrossRef]
- Mussolin, L.; Le Deley, M.C.; Carraro, E.; Damm-Welk, C.; Attarbaschi, A.; Williams, D.; Burke, A.; Horibe, K.; Nakazawa, A.; Wrobel, G.; et al. Prognostic factors in childhood anaplastic large cell lymphoma: Long term results of the international ALCL99 trial. Cancers (Basel) 2020, 12, 2747. [Google Scholar] [CrossRef]
- Damm-Welk, C.; Kutscher, N.; Zimmermann, M.; Attarbaschi, A.; Schieferstein, J.; Knorr, F.; Oschlies, I.; Klapper, W.; Woessmann, W. Quantification of minimal disseminated disease by quantitative PCR and digital PCR for NPM-ALK as prognostic factor in children with anaplastic large cell lymphoma. Haematologica 2019, 105, 2141–2149. [Google Scholar] [CrossRef]
- Lowe, E.J.; Reilly, A.F.; Lim, M.S.; Gross, T.G.; Saguilig, L.; Barkauskas, D.A.; Wu, R.; Alexander, S.; Bollard, C.M. Brentuximab vedotin in combination with chemotherapy for pediatric patients with ALK+ALCL: Results of COG trial ANHL12P1. Blood 2021. online ahead of print. [Google Scholar] [CrossRef]
- Damm-Welk, C.; Schieferstein, J.; Schwalm, S.; Reiter, A.; Woessmann, W. Flow cytometric detection of circulating tumour cells in nucleophosmin/anaplastic lymphoma kinase-positive anaplastic large cell lymphoma: Comparison with quantitative polymerase chain reaction. Br. J. Haematol. 2007, 138, 459–466. [Google Scholar] [CrossRef]
- Rigaud, C.; Abbas, R.; Grand, D.; Minard-Colin, V.; Aladjidi, N.; Buchbinder, N.; Garniere, N.; Plat, G.; Couec, M.-L.; Duplan, M.; et al. Should treatment of ALK-positive anaplastic large cell lymphoma be stratified according to minimal residual disease? Pediatr. Blood Cancer 2021, e28982, Online ahead of print. [Google Scholar] [CrossRef]
- Gambacorti-Passerini, C.; Mussolin, L.; Brugieres, L. Abrupt relapse of ALK-positive lymphoma after discontinuation of crizotinib. N. Engl. J. Med. 2016, 374, 95–96. [Google Scholar] [CrossRef]
- Hebart, H.; Lang, P.; Woessmann, W. Nivolumab for refractory anaplastic large cell lymphoma: A case report. Ann. Intern. Med. 2016, 165, 607–608. [Google Scholar] [CrossRef]
- Mosse, Y.P.; Lim, M.S.; Voss, S.D.; Wilner, K.; Ruffner, K.; Laliberte, J.; Rolland, D.; Balis, F.M.; Maris, J.M.; Weigel, B.J.; et al. Safety and activity of crizotinib for paediatric patients with refractory solid tumours or anaplastic large-cell lymphoma: A children’s oncology group phase 1 consortium study. Lancet Oncol. 2013, 14, 472–480. [Google Scholar] [CrossRef] [Green Version]
- Mosse, Y.P.; Voss, S.D.; Lim, M.S.; Rolland, D.; Minard, C.G.; Fox, E.; Adamson, P.; Wilner, K.; Blaney, S.M.; Weigel, B.J. Targeting ALK with crizotinib in pediatric anaplastic large cell lymphoma and inflammatory myofibroblastic tumor: A children’s oncology group study. J. Clin. Oncol. 2017, 35, 3215–3221. [Google Scholar] [CrossRef]
- Branford, S.; Cross, N.C.; Hochhaus, A.; Radich, J.; Saglio, G.; Kaeda, J.; Goldman, J.; Hughes, T. Rationale for the recommendations for harmonizing current methodology for detecting BCR-ABL transcripts in patients with chronic myeloid leukaemia. Leukemia 2006, 20, 1925–1930. [Google Scholar] [CrossRef]
- Hughes, T.; Deininger, M.; Hochhaus, A.; Branford, S.; Radich, J.; Kaeda, J.; Baccarani, M.; Cortes, J.; Cross, N.C.; Druker, B.J.; et al. Monitoring CML patients responding to treatment with tyrosine kinase inhibitors: Review and recommendations for harmonizing current methodology for detecting BCR-ABL transcripts and kinase domain mutations and for expressing results. Blood 2006, 108, 28–37. [Google Scholar] [CrossRef] [Green Version]
- Hughes, T.P.; Kaeda, J.; Branford, S.; Rudzki, Z.; Hochhaus, A.; Hensley, M.L.; Gathmann, I.; Bolton, A.E.; van Hoomissen, I.C.; Goldman, J.M.; et al. Frequency of major molecular responses to imatinib or interferon alfa plus cytarabine in newly diagnosed chronic myeloid leukemia. N. Engl. J. Med. 2003, 349, 1423–1432. [Google Scholar] [CrossRef]
- Pfeifer, H.; Cazzaniga, G.; van der Velden, V.H.J.; Cayuela, J.M.; Schafer, B.; Spinelli, O.; Akiki, S.; Avigad, S.; Bendit, I.; Borg, K.; et al. Standardisation and consensus guidelines for minimal residual disease assessment in philadelphia-positive acute lymphoblastic leukemia (Ph + ALL) by real-time quantitative reverse transcriptase PCR of e1a2 BCR-ABL1. Leukemia 2019, 33, 1910–1922. [Google Scholar] [CrossRef] [PubMed]
- White, H.; Deprez, L.; Corbisier, P.; Hall, V.; Lin, F.; Mazoua, S.; Trapmann, S.; Aggerholm, A.; Andrikovics, H.; Akiki, S.; et al. A certified plasmid reference material for the standardisation of BCR-ABL1 MRNA quantification by real-time quantitative PCR. Leukemia 2015, 29, 369–376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dube, S.; Qin, J.; Ramakrishnan, R. Mathematical analysis of copy number variation in a DNA sample using digital PCR on a nanofluidic device. PLoS ONE 2008, 3, e2876. [Google Scholar] [CrossRef] [PubMed]
- Sanders, R.; Mason, D.J.; Foy, C.A.; Huggett, J.F. Evaluation of digital PCR for absolute RNA quantification. PLoS ONE 2013, 8, e75296. [Google Scholar] [CrossRef]
- Vogelstein, B.; Kinzler, K.W. Digital PCR. Proc. Natl. Acad. Sci. USA 1999, 96, 9236–9241. [Google Scholar] [CrossRef] [Green Version]
- Whale, A.S.; Cowen, S.; Foy, C.A.; Huggett, J.F. Methods for applying accurate digital PCR analysis on low copy DNA samples. PLoS ONE 2013, 8, e58177. [Google Scholar] [CrossRef] [Green Version]
- Quelen, C.; Grand, D.; Sarot, E.; Brugieres, L.; Sibon, D.; Pradines, A.; Laurent, C.; Brousset, P.; Lamant, L. Minimal residual disease monitoring using a 3’ALK universal probe assay in ALK-positive anaplastic large-cell lymphoma: DDPCR, an attractive alternative method to real-time quantitative PCR. J. Mol. Diagn. 2021, 23, 131–139. [Google Scholar] [CrossRef]
- Luthra, R.; Pugh, W.C.; Waasdorp, M.; Morris, W.; Cabanillas, F.; Chan, P.K.; Sarris, A.H. Mapping of genomic t(2;5)(p23;q35) break points in patients with anaplastic large cell lymphoma by sequencing long-range PCR products. Hematopathol. Mol. Hematol. 1998, 11, 173–183. [Google Scholar]
- Sarris, A.H.; Luthra, R.; Papadimitracopoulou, V.; Waasdorp, M.; Dimopoulos, M.A.; McBride, J.A.; Cabanillas, F.; Duvic, M.; Deisseroth, A.; Morris, S.W.; et al. Amplification of genomic DNA demonstrates the presence of the t(2;5) (p23;q35) in anaplastic large cell lymphoma, but not in other non-Hodgkin’s lymphomas, Hodgkin’s disease, or lymphomatoid papulosis. Blood 1996, 88, 1771–1779. [Google Scholar] [CrossRef] [Green Version]
- Krumbholz, M.; Woessmann, W.; Zierk, J.; Seniuk, D.; Ceppi, P.; Zimmermann, M.; Singh, V.K.; Metzler, M.; Damm-Welk, C. Characterization and diagnostic application of genomic NPM-ALK fusion sequences in anaplastic large-cell lymphoma. Oncotarget 2018, 9, 26543–26555. [Google Scholar] [CrossRef] [Green Version]
- Zur Stadt, U.; Alawi, M.; Adao, M.; Indenbirken, D.; Escherich, G.; Horstmann, M.A. Characterization of novel, recurrent genomic rearrangements as sensitive MRD targets in childhood B-cell precursor all. Blood Cancer J. 2019, 9, 96. [Google Scholar] [CrossRef]
- Cui, S.; Zhang, W.; Xiong, L.; Pan, F.; Niu, Y.; Chu, T.; Wang, H.; Zhao, Y.; Jiang, L. Use of capture-based next-generation sequencing to detect ALK fusion in plasma cell-free DNA of patients with non-small-cell lung cancer. Oncotarget 2017, 8, 2771–2780. [Google Scholar] [CrossRef] [Green Version]
- Kunimasa, K.; Kato, K.; Imamura, F.; Kukita, Y. Quantitative detection of ALK fusion breakpoints in plasma cell-free DNA from patients with non-small cell lung cancer using PCR-based target sequencing with a tiling primer set and two-step mapping/alignment. PLoS ONE 2019, 14, e0222233. [Google Scholar] [CrossRef]
- Wang, Y.; Tian, P.W.; Wang, W.Y.; Wang, K.; Zhang, Z.; Chen, B.J.; He, Y.Q.; Li, L.; Liu, H.; Chuai, S.; et al. Noninvasive genotyping and monitoring of anaplastic lymphoma kinase (ALK) rearranged non-small cell lung cancer by capture-based next-generation sequencing. Oncotarget 2016, 7, 65208–65217. [Google Scholar] [CrossRef] [Green Version]
- Scherer, F.; Kurtz, D.M.; Newman, A.M.; Stehr, H.; Craig, A.F.; Esfahani, M.S.; Lovejoy, A.F.; Chabon, J.J.; Klass, D.M.; Liu, C.L.; et al. Distinct biological subtypes and patterns of genome evolution in lymphoma revealed by circulating tumor DNA. Sci. Transl. Med. 2016, 8, 364ra155. [Google Scholar] [CrossRef] [Green Version]
- Kurtz, D.M.; Scherer, F.; Jin, M.C.; Soo, J.; Craig, A.F.M.; Esfahani, M.S.; Chabon, J.J.; Stehr, H.; Liu, C.L.; Tibshirani, R.; et al. Circulating tumor DNA measurements as early outcome predictors in diffuse large B-cell lymphoma. J. Clin. Oncol. 2018, 36, 2845–2853. [Google Scholar] [CrossRef]
- Rossi, D.; Diop, F.; Spaccarotella, E.; Monti, S.; Zanni, M.; Rasi, S.; Deambrogi, C.; Spina, V.; Bruscaggin, A.; Favini, C.; et al. Diffuse large B-cell lymphoma genotyping on the liquid biopsy. Blood 2017, 129, 1947–1957. [Google Scholar] [CrossRef]
- Darrah, J.M.; Herrera, A.F. Updates on circulating tumor DNA assessment in lymphoma. Curr. Hematol. Malig. Rep. 2018, 13, 348–355. [Google Scholar] [CrossRef]
- Gauthier, J.; Holmberg, L.; Wu, D.; Bensinger, W.; Gopal, A.K.; Press, O.; Maloney, D.; Green, D.J.; Till, B.G.; Byelykh, D.; et al. Minimal detectable disease confirmed by flow cytometry and poor outcome after autologous stem cell transplantation in peripheral T-cell lymphomas. Bone Marrow Transplant. 2016, 51, 1617–1619. [Google Scholar] [CrossRef]
- Attarbaschi, A.; Abla, O.; Arias Padilla, L.; Beishuizen, A.; Burke, G.A.A.; Brugieres, L.; Bruneau, J.; Burkhardt, B.; d’Amore, E.S.G.; Klapper, W.; et al. Rare non-Hodgkin lymphoma of childhood and adolescence: A consensus diagnostic and therapeutic approach to pediatric-type follicular lymphoma, marginal zone lymphoma, and nonanaplastic peripheral T-cell lymphoma. Pediatr. Blood Cancer 2020, 67, e28416. [Google Scholar] [CrossRef]
- Rzepiel, A.; Kutszegi, N.; Gezsi, A.; Sagi, J.C.; Egyed, B.; Peter, G.; Butz, H.; Nyiro, G.; Muller, J.; Kovacs, G.T.; et al. Circulating micrornas as minimal residual disease biomarkers in childhood acute lymphoblastic leukemia. J. Transl. Med. 2019, 17, 372. [Google Scholar] [CrossRef]
- Drandi, D.; Alcantara, M.; Benmaad, I.; Sohlbrandt, A.; Lhermitte, L.; Zaccaria, G.; Ferrante, M.; Genuardi, E.; Mantoan, B.; Villarese, P.; et al. Droplet digital PCR quantification of mantle cell lymphoma follow-up samples from four prospective trials of the European MCL network. Hemasphere 2020, 4, e347. [Google Scholar] [CrossRef]
- Damm-Welk, C.; Yamashita, Y.; Bench, A.; Turner, S.D.; Lamant, L.; Verge, V.; Tosato, E.; Schieferstein, J.; Mussolin, L. Quality control of standardized methods for NPM-ALK RT-PCR and anti-ALK-antibody measurement for anaplastic large cell lymphoma—A report of the EICNHL reference laboratory group. Br. J. Haematol. 2015, 171 (Suppl. 1), 56. [Google Scholar]

| Lymphoblastic Lymphoma | Burkitt Lymphoma | DLBCL, PMLBCL | PTCL | |||
|---|---|---|---|---|---|---|
| Marker | IG/TCR Rearrangement | Aberrant Marker Expression | IG Rearrangement | MYC–IGH Fusion Site (DNA) | IG Rearrangement | TCR Rearrangement |
| Techniques | Marker screening: PCR, NGS Quantification: RQ-PCR | Flow cytometry | Marker screening: PCR, NGS Quantification: RQ-PCR | Long-distance PCR | Marker screening: PCR, NGS Quantification: RQ-PCR | Marker screening: PCR, NGS Quantification: RQ-PCR |
| Applicability | Most patients with initial tumor | Most patients | Most patients with initial tumor | 65–70% | ? | ? |
| Sensitivity | 10−5 | 10−4 | 10−4 | 10−3–10−4 | 10−4–10−5 | 10−5 |
| Initial Tumor Material | needed | not necessarily | needed | needed | needed in most cases | needed |
| Clinical data on MDD | - | + | + | + | - | (+) |
| Clinical data on MRD | - | (+) | (+) | + | - | (+) |
| (A) Methods to measure minimal disease in ALK-positive anaplastic large cell lymphoma (ALCL). | ||||
| RT-PCR NPM–ALK Transcripts | RQ-PCR NPM–ALK Transcripts | dPCR NPM–ALK Transcripts | RQ-PCR for DNA Break | |
|---|---|---|---|---|
| Applicability | 85% | 85% | 85% | n.k. |
| Sensitivity | ≤10−5 | ≤10−5 | ≤10−5 | ≤10−5 |
| Advantage | easy QC, inexpensive | allows following response to therapies | High sensitivity QC easier compared to RQ-PCR | Patient-specific ctDNA detectable |
| Disadvantage | no quantitative response monitoring | difficult to harmonize, expensive | expensive | fresh tumor needed, expensive, laborious |
| (B) Established clinical applications for minimal disseminated (MDD) and minimal residual disease (MRD) in ALK-positive anaplastic large cell lymphoma (ALCL). | ||||
| Specific Marker | MDD/MRD | Patients Positive (%) | Clinical Relevance | Specific Marker |
| RT-PCR for NPM–ALK Transcripts (RNA) | MDD MRD | 50–60 25 | HR patients (50% EFS), validated [44,45,46,48,50] VHR patients (25% EFS), validated [46,53] | RT-PCR for NPM–ALK transcripts (RNA) |
| RQ-PCR or dPCR for NPM–ALK Transcripts (RNA) | MDD MRD | 20–25 | VHR patients (30% EFS) [44,50] Individual response to therapy [54,55,56,57,70] | RQ-PCR or dPCR for NPM–ALK transcripts (RNA) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mussolin, L.; Damm-Welk, C.; Pillon, M.; Woessmann, W. Minimal Disease Monitoring in Pediatric Non-Hodgkin’s Lymphoma: Current Clinical Application and Future Challenges. Cancers 2021, 13, 1907. https://doi.org/10.3390/cancers13081907
Mussolin L, Damm-Welk C, Pillon M, Woessmann W. Minimal Disease Monitoring in Pediatric Non-Hodgkin’s Lymphoma: Current Clinical Application and Future Challenges. Cancers. 2021; 13(8):1907. https://doi.org/10.3390/cancers13081907
Chicago/Turabian StyleMussolin, Lara, Christine Damm-Welk, Marta Pillon, and Wilhelm Woessmann. 2021. "Minimal Disease Monitoring in Pediatric Non-Hodgkin’s Lymphoma: Current Clinical Application and Future Challenges" Cancers 13, no. 8: 1907. https://doi.org/10.3390/cancers13081907
APA StyleMussolin, L., Damm-Welk, C., Pillon, M., & Woessmann, W. (2021). Minimal Disease Monitoring in Pediatric Non-Hodgkin’s Lymphoma: Current Clinical Application and Future Challenges. Cancers, 13(8), 1907. https://doi.org/10.3390/cancers13081907








