A Novel Multidrug-Resistant Cell Line from an Italian Intrahepatic Cholangiocarcinoma Patient
Abstract
:Simple Summary
Abstract
1. Introduction
2. Results
2.1. Generation and Immunophenotyping of ICC Cell Line
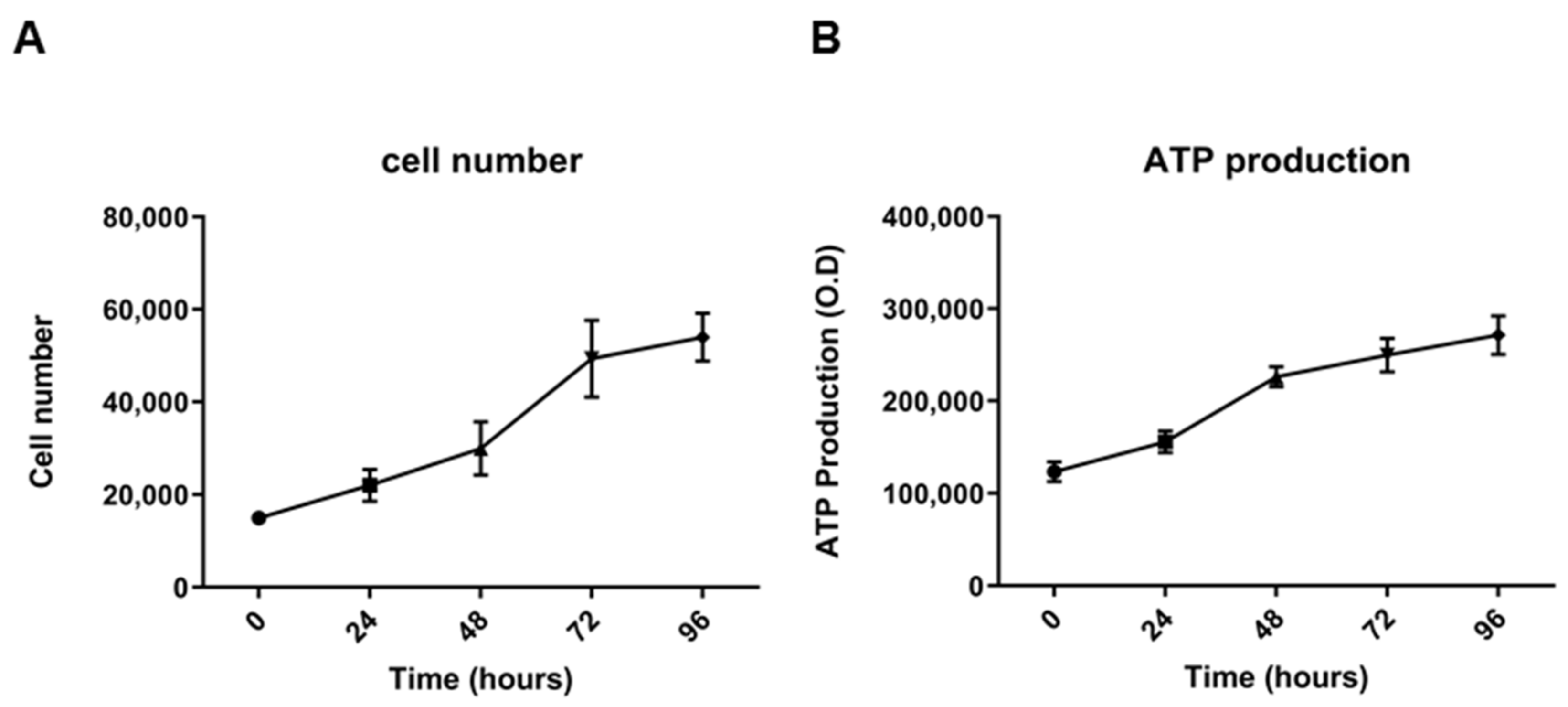
2.2. Morphological, Phenotypic, and Biological Characterization of 82.3 Cells
2.3. Genetic and Molecular Characterization of Cells 82.3
2.4. Multidrug Resistance of 82.3
2.5. Modulation of Genes Related to Epithelial to Mesenchymal Transition (EMT) and Drug Resistance (DR)
2.6. Transcriptomic Profile of 82.3 Cells
3. Discussion
4. Materials and Methods
4.1. Establishment of ICC Cell Line from a PDX
4.2. Cell Lines
4.3. Flow Cytometry Analysis
4.4. Immunofluorescence Staining
4.5. Population Doubling Time
4.6. Drugs
4.7. Cell Growth Assay
4.8. Apoptosis Assay
4.9. Sphere-Formation Assay
4.10. In Vitro Migration Assay
4.11. Tumorigenicity in NOD/SCID Mice
4.12. DNA Identify Analysis by Fingerprinting
4.13. Karyotype Analysis
4.14. RNA Extraction, RT2PCR Array for Epithelial to Mesenchymal Transition (EMT) and Drug Resistance and RNA Sequencing
4.15. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Alvaro, D.; Crocetti, E.; Ferretti, S.; Bragazzi, M.C.; Capocaccia, R.; the AISF “Cholangiocarcinoma” Committee. Descriptive epidemiology of cholangiocarcinoma in Italy. Dig. Liver. Dis. 2010, 42, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Valle, J.W.; Lamarca, A.; Goyal, L.; Barriuso, J.; Zhu, A.X. New Horizons for Precision Medicine in Biliary Tract Cancers. Cancer Discov. 2017, 7, 943–962. [Google Scholar] [CrossRef] [Green Version]
- Valle, J.W.; Furuse, J.; Jitlal, M.; Beare, S.; Mizuno, N.; Wasan, H.; Bridgewater, J.; Okusaka, T. Cisplatin and Gemcitabine for advanced biliary tract cancer: A meta-analysis of two randomised trials. Ann. Oncol. 2014, 25, 391–398. [Google Scholar] [CrossRef] [PubMed]
- Rueff, J.; Rodrigues, A.S. Cancer Drug Resistance: A Brief Overview from a Genetic Viewpoint. Methods Mol. Biol. 2016, 1395, 1–18. [Google Scholar] [PubMed]
- Schweitzer, N.; Kirstein, M.M.; Kratzel, A.M.; Mederacke, Y.S.; Fischer, M.; Manns, M.P.; Vogel, A. Second-line chemotherapy in biliary tract cancer: Outcome and prognostic factors. Liver Int. 2019, 39, 914–923. [Google Scholar] [CrossRef]
- Walter, T.; Horgan, A.M.; McNamara, M.; McKeever, L.; Min, T.; Hedley, D.; Serra, S.; Krzyzanowska, M.K.; Chen, E.; Mackay, H.; et al. Feasibility and benefits of second-line chemotherapy in advanced biliary tract cancer: A large retrospective study. Eur. J. Cancer 2013, 49, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Takahara, N.; Nakai, Y.; Isayama, H.; Sasaki, T.; Saito, K.; Oyama, H.; Kanai, S.; Suzuki, T.; Sato, T.; Hakuta, R.; et al. Second-line chemotherapy in patients with advanced or recurrent biliary tract cancer: A single center, retrospective analysis of 294 cases. Investig. New Drugs 2018, 36, 1093–1102. [Google Scholar] [CrossRef]
- Kim, B.J.; Yoo, C.; Kim, K.P.; Hyung, J.; Park, S.J.; Ryoo, B.Y.; Chang, H.M. Efficacy of fluoropyrimidine-based chemotherapy in patients with advanced biliary tract cancer after failure of Gemcitabine plus Cisplatin: Retrospective analysis of 321 patients. Br. J. Cancer 2017, 116, 561–567. [Google Scholar] [CrossRef] [Green Version]
- Wattanawongdon, W.; Hahnvajanawong, C.; Namwat, N.; Kanchanawat, S.; Boonmars, T.; Jearanaikoon, P.; Leelayuwat, C.; Techasen, A.; Seubwai, W. Establishment and characterization of Gemcitabine-resistant human cholangiocarcinoma cell lines with multidrug resistance and enhanced invasiveness. Int. J. Oncol. 2015, 47, 398–410. [Google Scholar] [CrossRef] [Green Version]
- Varamo, C.; Peraldo-Neia, C.; Ostano, P.; Basiricò, M.; Raggi, C.; Bernabei, P.; Venesio, T.; Berrino, E.; Aglietta, M.; Leone, F.; et al. Establishment and Characterization of a New Intrahepatic Cholangiocarcinoma Cell Line Resistant to Gemcitabine. Cancers 2019, 11, 519. [Google Scholar] [CrossRef] [Green Version]
- Cavalloni, G.; Peraldo-Neia, C.; Varamo, C.; Casorzo, L.; Dell’Aglio, C.; Bernabei, P.; Chiorino, G.; Aglietta, M.; Leone, F. Establishment and characterization of a human intrahepatic cholangiocarcinoma cell line derived from an Italian patient. Tumour Biol. 2015, 37, 4041–4052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiang, Z.; Zhang, W.; Jin, S.; Dai, K.; He, Y.; Tao, L.; Yu, H. Carcinoembryonic antigen, α-fetoprotein, and Ki67 as biomarkers and prognostic factors in intrahepatic cholangiocarcinoma: A retrospective cohort study. Ann. Hepatol. 2021, 20, 100242. [Google Scholar] [CrossRef]
- Ricci, F.; Fratelli, M.; Guffanti, F.; Porcu, L.; Spriano, F.; Dell’Anna, T.; Fruscio, R.; Damia, G. Patient-derived ovarian cancer xenografts re-growing after a Cisplatinum treatment are less responsive to a second drug re-challenge: A new experimental setting to study response to therapy. Oncotarget 2017, 8, 7441–7451. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Shen, W.; Peng, H.; Li, Y.; Chen, F.; Zheng, L.; Xu, J.; Jia, L. Fibronectin 1 promotes melanoma proliferation and metastasis by inhibiting apoptosis and regulating EMT. Onco Targets Ther. 2019, 12, 3207–3221. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Li, Y.; Kong, D.; Banerjee, S.; Ahmad, A.; Azmi, A.S.; Ali, S.; Abbruzzese, J.L.; Gallick, G.E.; Sarkar, F.H. Acquisition of epithelial-mesenchymal transition phenotype of Gemcitabine-resistant pancreatic cancer cells is linked with activation of the notch signaling pathway. Cancer Res. 2009, 69, 2400–2407. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Lee, J.; Jeon, M.; Nam, S.J.; Lee, J.E. Elevated TGF-β1 and -β2 expression accelerates the epithelial to mesenchymal transition in triple-negative breast cancer cells. Cytokine 2015, 75, 151–158. [Google Scholar] [CrossRef]
- Zhang, Y.; Zou, X.; Qian, W.; Weng, X.; Zhang, L.; Wang, S.; Cao, X.; Ma, L.; Wei, G.; Wu, Y.; et al. Enhanced PAPSS2/VCAN sulfation axis is essential for Snail-mediated breast cancer cell migration and metastasis. Cell Death Differ. 2019, 26, 565–579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singal, A.G.; Lampertico, P.; Nahon, P. Epidemiology and surveillance for hepatocellular carcinoma: New trends. J. Hepatol. 2020, 72, 250–261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, T.; Wang, H.; Wang, Y.; Lin, X.; Cui, Y.; Wang, Z. Clinical Significance of Preoperative Serum CEA, CA125, and CA19-9 Levels in Predicting the Resectability of Cholangiocarcinoma. Dis. Markers 2019, 2019, 6016931. [Google Scholar] [CrossRef]
- Ishii, T.; Yasuchika, K.; Suemori, H.; Nakatsuji, N.; Ikai, I.; Uemoto, S. Alpha-fetoprotein producing cells act as cancer progenitor cells in human cholangiocarcinoma. Cancer Lett. 2010, 294, 25–34. [Google Scholar] [CrossRef]
- Cavalloni, G.; Peraldo-Neia, C.; Sassi, F.; Chiorino, G.; Sarotto, I.; Aglietta, M.; Leone, F. Establishment of a patient-derived intrahepatic cholangiocarcinoma xenograft model with KRAS mutation. BMC Cancer 2016, 16, 90. [Google Scholar] [CrossRef] [Green Version]
- Adamska, A.; Elaskalani, O.; Emmanouilidi, A.; Kim, M.; Abdol Razak, N.B.; Metharom, P.; Falasca, M. Molecular and cellular mechanisms of chemoresistance in pancreatic cancer. Adv. Biol. Regul. 2018, 68, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Yu, S.; Tan, Q.; Guo, P.; Liu, H. Role of AhR in regulating cancer stem cell-like characteristics in choriocarcinoma. Cell Cycle 2018, 17, 2309–2320. [Google Scholar] [CrossRef] [Green Version]
- Asukai, K.; Kawamoto, K.; Eguchi, H.; Konno, M.; Asai, A.; Iwagami, Y.; Yamada, D.; Asaoka, T.; Noda, T.; Wada, H.; et al. Micro-RNA-130a-3p Regulates Gemcitabine Resistance via PPARG in Cholangiocarcinoma. Ann. Surg. Oncol. 2017, 24, 2344–2352. [Google Scholar] [CrossRef] [PubMed]
- Valencia-Cervantes, J.; Huerta-Yepez, S.; Aquino-Jarquín, G.; Rodríguez-Enríquez, S.; Martínez-Fong, D.; Arias-Montaño, J.A.; Dávila-Borja, V.M. Hypoxia increases chemoresistance in human medulloblastoma DAOY cells via hypoxia-inducible factor 1α-mediated downregulation of the CYP2B6, CYP3A4 and CYP3A5 enzymes and inhibition of cell proliferation. Oncol. Rep. 2019, 41, 178–190. [Google Scholar] [CrossRef] [PubMed]
- McGowan-Jordan, J.; Simons, A.; Schmid, M. ISCN. An International System for Human Cytogenetic Nomenclature; McGowan-Jordan, J., Simons, A., Schmid, M., Eds.; Karger Medical and Scientific Publishers: Basel, Switzerland, 2016; p. 149. [Google Scholar]











| Locus | 82.3 | |||
|---|---|---|---|---|
| Allele 1 | Allele 2 | >Allele 3 | >Allele 4 | |
| AMEL | X | Y | - | - |
| CSF1PO | 11 | - | - | - |
| D13S317 | 9 | 10 | - | - |
| D16S539 | 11 | - | - | - |
| D18S51 | 13 | 16 | - | - |
| D21S11 | 30 | - | - | - |
| D3S1358 | 14 | 15 | - | - |
| D5S818 | 12 | - | - | - |
| D7S820 | 8 | 9 | - | - |
| D8S1179 | 13 | 14 | - | - |
| FGA | 23 | - | - | - |
| Penta D | 9 | - | - | - |
| Penta E | 10 | 16 | - | - |
| TH01 | 9 | - | - | - |
| TPOX | 9 | - | - | - |
| vWA | 18 | - | - | - |
| Up-Regulated Genes | Average Fold Change | SD | Down-Regulated Genes | Average Fold Change | SD |
|---|---|---|---|---|---|
| AHR | 152.60 | 4.39 | ABCC1 | 0.11 | 0.04 |
| APC | 8.80 | 1.82 | ABCC2 | 0.04 | 0.01 |
| ARNT | 3.06 | 0.46 | ABCC3 | 0.33 | 0.08 |
| BCL2 | 2.22 | 0.49 | ABCC5 | 0.25 | 0.08 |
| BRCA2 | 11.43 | 1.41 | BAX | 0.46 | 0.07 |
| CCND1 | 1.67 | 0.44 | BCL2L1 | 0.09 | 0.03 |
| CDKN1A | 29.58 | 0.44 | CCNE1 | 0.26 | 0.03 |
| CYP2E1 | 6.37 | 1.99 | CDK2 | 0.41 | 0.07 |
| EGFR | 5.15 | 1.60 | CDK4 | 0.28 | 0.14 |
| EPHX1 | 1.69 | 0.51 | CDKN1B | 0.27 | 0.15 |
| HIF1A | 7.07 | 0.87 | CLPTM1L | 0.07 | 0.02 |
| MET | 3.16 | 0.40 | CYP3A5 | 0.11 | 0.03 |
| MSH2 | 5.66 | 1.25 | ELK1 | 0.01 | 0.01 |
| MYC | 1.74 | 0.19 | ERBB2 | 0.16 | 0.08 |
| NAT2 | 10.82 | 0.17 | GSK3A | 0.20 | 0.06 |
| PPARA | 2.19 | 0.31 | GSTP1 | 0.40 | 0.13 |
| PPARG | 9.29 | 1.41 | MVP | 0.15 | 0.07 |
| RB1 | 5.11 | 0.74 | NFKB2 | 0.22 | 0.00 |
| SOD1 | 1.69 | 0.17 | NFKBIB | 0.27 | 0.07 |
| TOP2A | 3.91 | 0.54 | RARA | 0.39 | 0.07 |
| TOP2B | 16.13 | 1.66 | RARG | 0.18 | 0.05 |
| TPMT | 6.25 | 0.01 | RELB | 0.36 | 0.08 |
| UGCG | 6.69 | 0.43 | RXRA | 0.18 | 0.04 |
| - | - | - | RXRB | 0.49 | 0.14 |
| - | - | - | SULT1E1 | 0.34 | 0.04 |
| - | - | - | TP53 | 0.07 | 0.02 |
| Up-Regulated Genes | Average Fold Change | SD | Down-Regulated Genes | Average Fold Change | SD |
|---|---|---|---|---|---|
| CALD1 | 3.51 | 0.12 | AKT1 | 0.17 | 0.03 |
| CAMK2N1 | 14.79 | 3.50 | BMP2 | 0.05 | 0.02 |
| CAV2 | 2.17 | 0.84 | ESR1 | 0.33 | 0.09 |
| CDH2 | 5.47 | 1.08 | F11R | 0.19 | 0.04 |
| COL5A2 | 3.62 | 1.07 | FOXC2 | 0.05 | 0.00 |
| DSC2 | 11.94 | 2.90 | IGFBP4 | 0.02 | 0.01 |
| DSP | 6.91 | 0.55 | ILK | 0.23 | 0.01 |
| EGFR | 5.92 | 0.59 | ITGA5 | 0.01 | 0.00 |
| FGFBP1 | 6.46 | 2.95 | KRT19 | 0.28 | 0.01 |
| FN1 | 31.61 | 6.18 | KRT7 | 0.37 | 0.13 |
| FZD7 | 2.10 | 0.41 | MAP1B | 0.13 | 0.01 |
| GSK3B | 5.43 | 0.34 | MSN | 0.15 | 0.04 |
| IL1RN | 4.31 | 0.50 | MST1R | 0.07 | 0.01 |
| ITGAV | 3.73 | 0.16 | NOTCH1 | 0.10 | 0.03 |
| ITGB1 | 4.55 | 0.29 | PPPDE2 | 0.36 | 0.00 |
| MMP9 | 29.40 | 0.13 | SPP1 | 0.16 | 0.01 |
| OCLN | 5.93 | 0.06 | - | - | - |
| PLEK2 | 2.19 | 0.29 | - | - | - |
| PTK2 | 2.02 | 0.51 | - | - | - |
| PTP4A1 | 2.89 | 0.03 | - | - | - |
| RAC1 | 13.17 | 1.75 | - | - | - |
| SIP1 | 2.58 | 0.35 | - | - | - |
| SMAD2 | 4.04 | 0.45 | - | - | - |
| STEAP1 | 2.78 | 0.47 | - | - | - |
| TCF4 | 6.10 | 0.26 | - | - | - |
| TGFB1 | 2.09 | 0.61 | - | - | - |
| TGFB2 | 89.35 | 16.11 | - | - | - |
| TGFB3 | 1.82 | 0.30 | - | - | - |
| TIMP1 | 1.67 | 0.14 | - | - | - |
| TMEFF1 | 12.51 | 4.12 | - | - | - |
| TSPAN13 | 5.72 | 0.59 | - | - | - |
| VCAN | 168.97 | 28.95 | - | - | - |
| VIM | 504.06 | 83.12 | - | - | - |
| VPS13A | 9.95 | 0.97 | - | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peraldo-Neia, C.; Massa, A.; Vita, F.; Basiricò, M.; Raggi, C.; Bernabei, P.; Ostano, P.; Casorzo, L.; Panero, M.; Leone, F.; et al. A Novel Multidrug-Resistant Cell Line from an Italian Intrahepatic Cholangiocarcinoma Patient. Cancers 2021, 13, 2051. https://doi.org/10.3390/cancers13092051
Peraldo-Neia C, Massa A, Vita F, Basiricò M, Raggi C, Bernabei P, Ostano P, Casorzo L, Panero M, Leone F, et al. A Novel Multidrug-Resistant Cell Line from an Italian Intrahepatic Cholangiocarcinoma Patient. Cancers. 2021; 13(9):2051. https://doi.org/10.3390/cancers13092051
Chicago/Turabian StylePeraldo-Neia, Caterina, Annamaria Massa, Francesca Vita, Marco Basiricò, Chiara Raggi, Paola Bernabei, Paola Ostano, Laura Casorzo, Mara Panero, Francesco Leone, and et al. 2021. "A Novel Multidrug-Resistant Cell Line from an Italian Intrahepatic Cholangiocarcinoma Patient" Cancers 13, no. 9: 2051. https://doi.org/10.3390/cancers13092051
APA StylePeraldo-Neia, C., Massa, A., Vita, F., Basiricò, M., Raggi, C., Bernabei, P., Ostano, P., Casorzo, L., Panero, M., Leone, F., Cavalloni, G., & Aglietta, M. (2021). A Novel Multidrug-Resistant Cell Line from an Italian Intrahepatic Cholangiocarcinoma Patient. Cancers, 13(9), 2051. https://doi.org/10.3390/cancers13092051






