Automatic Segmentation of Metastatic Breast Cancer Lesions on 18F-FDG PET/CT Longitudinal Acquisitions for Treatment Response Assessment
Abstract
Simple Summary
Abstract
1. Introduction
- Development of a deep learning network to segment breast cancer metastatic lesions on baseline acquisitions with whole-body PET/CT images as input. Our network achieved a mean dice score of 0.66.
- Development of a deep learning network to segment breast cancer metastatic lesions on follow-up acquisitions with whole-body PET/CT images as input. The difference of this network compared to the previous one lies in the use of baseline PET images and lesion segmentations as complementary inputs to the follow-up PET/CT images. This allows a better segmentation of the follow-up lesions that often present a lower contrast due to treatment response. Our network achieved a mean dice score of 0.58.
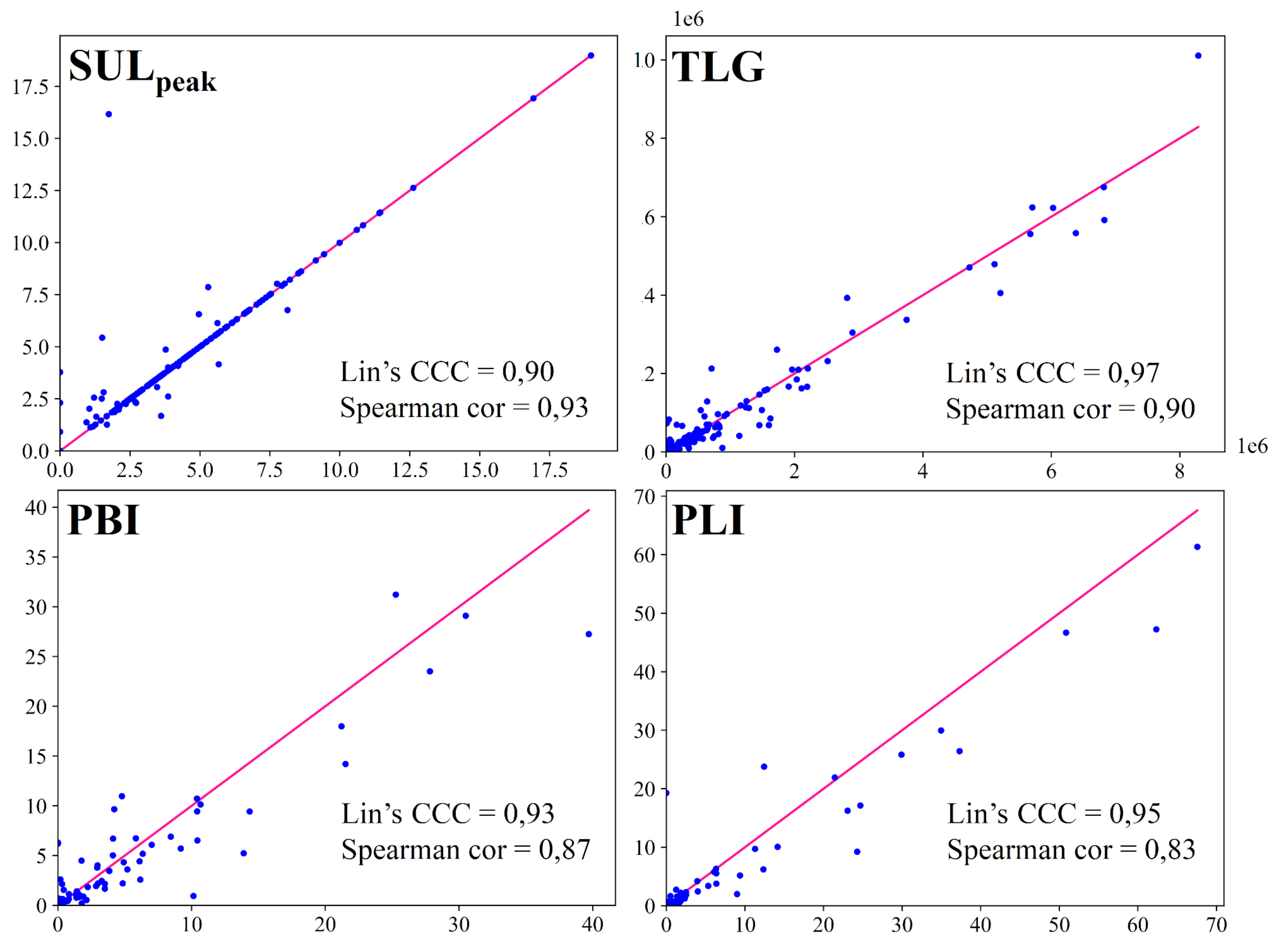
- Automatic computation of 4 biomarkers from the automatic segmentation: (1) SUL to assess metabolic changes, (2) TLG to determine metabolic and volume changes, (3) PET Bone Index (PBI) and (4) PET Liver Index (PLI), which estimates the lesion volume of the two sites most affected by metastatic breast cancer (bone and liver) [23]. We obtained good Lin’s concordance correlation coefficients (≥0.90) and Spearman’s rank correlation coefficients (≥0.80) between biomarkers computed on automatic segmentation and on manual segmentation.
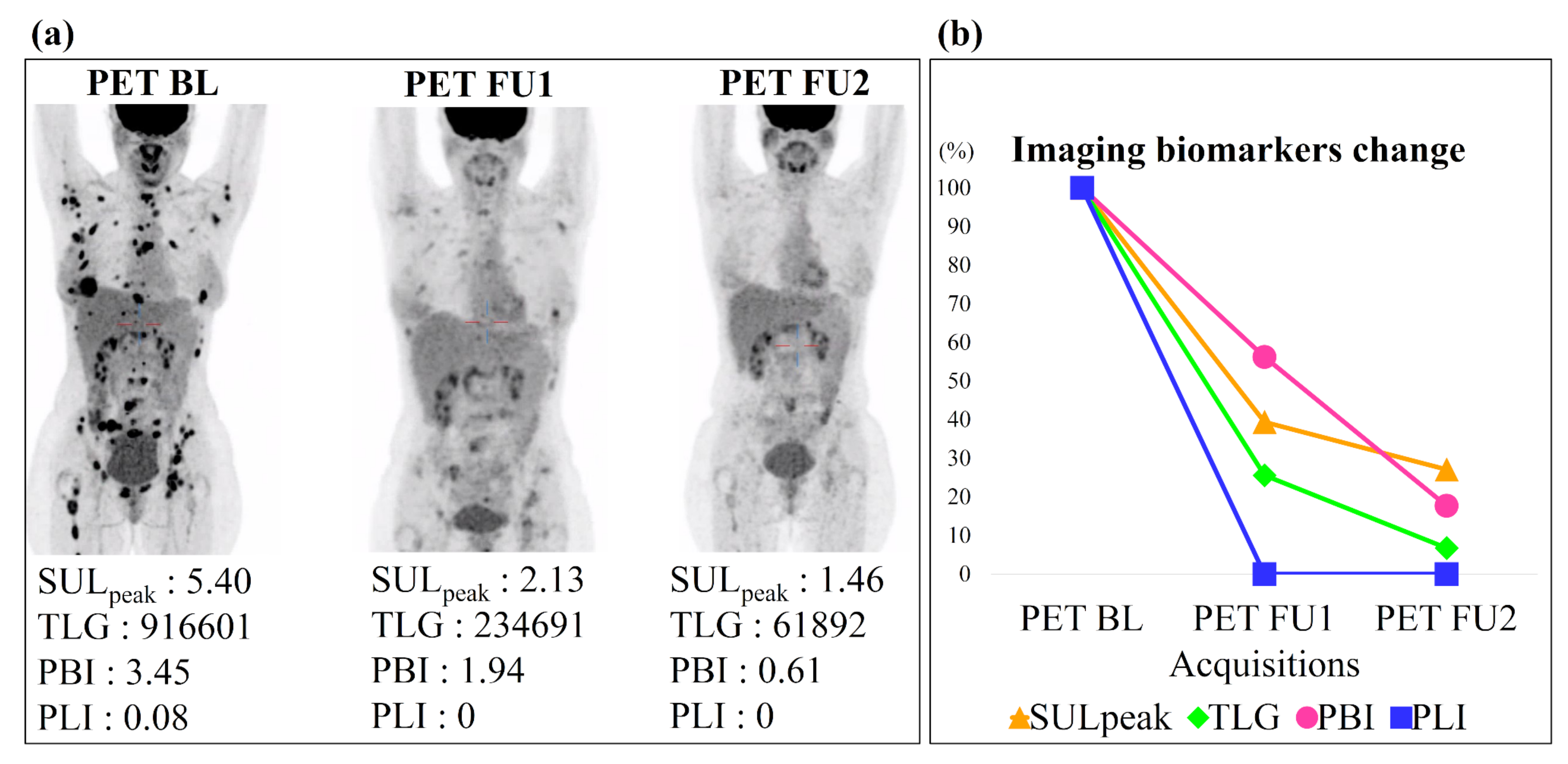
- Automatic assessment of patients’ treatment response using the previously defined biomarkers computed on the different PET/CT acquisitions. The SUL, with a 32% decrease between baseline and follow-up, was the biomarker best able to assess patients’ response (sensitivity 87%, specificity 87%).
2. Materials and Methods
2.1. Dataset
2.2. Metastatic Lesion Segmentation
2.3. Segmentation Evaluation
2.4. Imaging Biomarkers
2.5. Response Assessment
3. Results
3.1. Metastatic Lesion Segmentation
3.2. Imaging Biomarkers Measurements
3.3. Response Assessment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| RECIST | Response Evaluation Criteria in Solid Tumors |
| CT | Computed Tomography |
| MRI | Magnetic Resonance Imaging |
| PET | Positron Emission Tomography |
| PERCIST | PET Response Evaluation Criteria in Solid Tumors |
| OS | Overall Survival |
| PFS | Progression-Free Survival |
| MTV | Metabolic Tumor Volume |
| TLG | Total Leson Glycolysis |
| CNN | Convolutional Neural Networks |
| TP | True Positive |
| FN | False Negative |
| FP | False Positif |
| PBI | PET Bone Index |
| PLI | PET Liver Index |
| VOI | Volume of Interest |
| ROC | Receiver Operating Characteristic |
| AUC | Area Under the Curve |
| CR | Complete Response |
| PR | Partial Response |
| SD | Stable Disease |
| PD | Progressive Disease |
References
- O’Shaughnessy, J. Extending survival with chemotherapy in metastatic breast cancer. Oncologist 2005, 10, 20–29. [Google Scholar] [CrossRef]
- Sundquist, M.; Brudin, L.; Tejler, G. Improved survival in metastatic breast cancer 1985–2016. Breast 2017, 31, 46–50. [Google Scholar] [CrossRef] [PubMed]
- Eisenhauer, E.A.; Therasse, P.; Bogaerts, J.; Schwartz, L.H.; Sargent, D.; Ford, R.; Dancey, J.; Arbuck, S.; Gwyther, S.; Mooney, M.; et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur. J. Cancer 2009, 45, 228–247. [Google Scholar] [CrossRef]
- Schwartz, L.H.; Litière, S.; De Vries, E.; Ford, R.; Gwyther, S.; Mandrekar, S.; Shankar, L.; Bogaerts, J.; Chen, A.; Dancey, J.; et al. RECIST 1.1—Upyear and clarification: From the RECIST committee. Eur. J. Cancer 2016, 62, 132–137. [Google Scholar] [CrossRef]
- Yang, H.L.; Liu, T.; Wang, X.M.; Xu, Y.; Deng, S.M. Diagnosis of bone metastases: A meta-analysis comparing 18-FDG PET, CT, MRI and bone scintigraphy. Eur. Radiol. 2011, 21, 2604–2617. [Google Scholar] [CrossRef] [PubMed]
- Wahl, R.L.; Jacenne, H.; Kasamon, Y.; Lodge, M.A. From RECIST to PERCIST: Evolving Considerations for PET Response Criteria in Solid Tumors. J. Nucl. Med. 2009, 50 (Suppl. 1), 122S–150S. [Google Scholar] [CrossRef]
- Riedl, C.C.; Pinker, K.; Ulaner, G.A.; Ong, L.T.; Baltzer, P.; Jochelson, M.S.; McArthur, H.L.; Gönen, M.; Dickler, M.; Weber, W.A. Comparison of FDG-PET/CT and contrast-enhanced CT for monitoring therapy response in patients with metastatic breast cancer. Eur. J. Nucl. Med. Mol. Imaging 2017, 44, 1428–1437. [Google Scholar] [CrossRef]
- Sluis, J.V.; Heer, E.C.D.; Boellaard, M.; Jalving, M.; Brouwers, A.H.; Boellaard, R. Clinically feasible semi-automatic workflows for measuring metabolically active tumour volume in metastatic melanoma. Eur. J. Nucl. Med. Mol. Imaging 2021, 48, 1498–1510. [Google Scholar] [CrossRef]
- Kanoun, S.; Rossi, C.; Berriolo-Riedinger, A.; Dygai-Cochet, I.; Cochet, A.; Humbert, O.; Toubeau, M.; Ferrant, E.; Brunotte, F.; Casasnovas, R.O. Baseline metabolic tumour volume is an independent prognostic factor in Hodgkin lymphoma. Eur. J. Nucl. Med. Mol. Imaging 2014, 41, 1735–1743. [Google Scholar] [CrossRef]
- Choi, J.H.; Kim, H.A.; Kim, W.; Lim, I.; Lee, I.; Byun, B.H.; Noh, W.C.; Seong, M.K.; Lee, S.S.; Kim, B.I.; et al. Early prediction of neoadjuvant chemotherapy response for advanced breast cancer using PET/MRI image deep learning. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef]
- Foster, B.; Bagci, U.; Mansoor, A.; Xu, Z.; Mollura, D.J. A review on segmentation of positron emission tomography images. Comput. Biol. Med. 2014, 50, 76–96. [Google Scholar] [CrossRef] [PubMed]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; Van Der Laak, J.A.; Van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef]
- Zhou, T.; Ruan, S.; Canu, S. A review: Deep learning for medical image segmentation using multi-modality fusion. Array 2019, 3–4, 100004. [Google Scholar] [CrossRef]
- Zhao, X.; Li, L.; Lu, W.; Tan, S. Tumor co-segmentation in PET/CT using multi-modality fully convolutional neural network. Phys. Med. Biol. 2018, 64, 015011. [Google Scholar] [CrossRef]
- Xu, L.; Tetteh, G.; Lipkova, J.; Zhao, Y.; Li, H.; Christ, P.; Piraud, M.; Buck, A.; Shiand, K.; Menze, B.H. Automated Whole-Body Bone Lesion Detection for Multiple Myeloma on 68Ga-Pentixafor PET/CT Imaging Using Deep Learning Methods. Contrast Media Mol. Imaging 2018, 2018, 2391925. [Google Scholar] [CrossRef]
- Moreau, N.; Rousseau, C.; Fourcade, C.; Santini, G.; Ferrer, L.; Lacombe, M.; Guillerminet, C.; Campone, M.; Colombié, M.; Rubeaux, M.; et al. Deep learning approaches for bone and bone lesion segmentation on 18F-FDG PET/CT imaging in the context of metastatic breast cancer. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020. [Google Scholar] [CrossRef]
- Andrearczyk, V.; Oreiller, V.; Jreige, M.; Vallières, M.; Castelli, J.; Elhalawani, H.; Boughdad, S.; Prior, J.O.; Depeursinge, A. Overview of the HECKTOR challenge at MICCAI 2020: Automatic head and neck tumor segmentation in PET/CT. In 3D Head and Neck Tumor Segmentation in PET/CT Challenge; Springer: Cham, Switzerland, 2020; pp. 1–21. [Google Scholar] [CrossRef]
- Sadaghiani, M.S.; Rowe, S.P.; Sheikhbahaei, S. Applications of artificial intelligence in oncologic 18F-FDG PET/CT imaging: A systematic review. Ann. Transl. Med. 2021, 9, 823. [Google Scholar] [CrossRef] [PubMed]
- Denner, S.; Khakzar, A.; Sajid, M.; Saleh, M.; Spiclin, Z.; Kim, S.T.; Navab, N. Spatio-temporal Learning from Longitudinal Data for Multiple Sclerosis Lesion Segmentation. arXiv 2020, arXiv:2004.03675. [Google Scholar]
- Krüger, J.; Opfer, R.; Gessert, N.; Ostwaldt, A.C.; Manogaran, P.; Kitzler, H.H.; Schlaefer, A.; Schippling, S. Fully automated longitudinal segmentation of new or enlarged multiple sclerosis lesions using 3D convolutional neural networks. NeuroImage Clin. 2020, 28, 102445. [Google Scholar] [CrossRef] [PubMed]
- Sepahvand, N.M.; Arnold, D.L.; Arbel, T. CNN Detection of New and Enlarging Multiple Sclerosis Lesions from Longitudinal Mri Using Subtraction Images. In Proceedings of the 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Iowa City, IA, USA, 3–7 April 2020; pp. 127–130. [Google Scholar] [CrossRef]
- Jin, C.; Yu, H.; Ke, J.; Ding, P.; Yi, Y.; Jiang, X.; Tang, J.; Chang, D.T.; Wu, X.; Gao, F.; et al. Predicting treatment response from longitudinal images using multi-task deep learning. Nat. Commun. 2021, 12, 1–11. [Google Scholar] [CrossRef]
- Coleman, R.; Rubens, R. The clinical course of bone metastases from breast cancer. Br. J. Cancer 1987, 55, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Colombié, M.; Jézéquel, P.; Rubeaux, M.; Frenel, J.S.; Bigot, F.; Seegers, V.; Campone, M. The EPICURE study: A pilot prospective cohort study of heterogeneous and massive data integration in metastatic breast cancer patients. BMC Cancer 2021, 21, 333. [Google Scholar] [CrossRef]
- Isensee, F.; Jaeger, P.F.; Kohl, S.; Petersen, J.; Maier-Hein, K.H. Automated Design of Deep Learning Methods for Biomedical Image Segmentation. arXiv 2020, arXiv:1904.08128. [Google Scholar] [CrossRef]
- Heller, N.; Isensee, F.; Maier-Hein, K.H.; Hou, X.; Xie, C.; Li, F.; Nan, Y.; Mu, G.; Lin, Z.; Han, M.; et al. The state of the art in kidney and kidney tumor segmentation in contrast-enhanced CT imaging: Results of the KiTS19 challenge. Med. Image Anal. 2021, 67, 101821. [Google Scholar] [CrossRef]
- Antonelli, M.; Reinke, A.; Bakas, S.; Farahani, K.; Landman, B.A.; Litjens, G.; Menze, B.; Ronneberger, O.; Summers, R.M.; van Ginneken, B.; et al. The Medical Segmentation Decathlon. arXiv 2021, arXiv:2106.05735. [Google Scholar]
- Isensee, F.; Petersen, J.; Kohl, S.A.A.; Jäger, P.F.; Maier-Hein, K.H. nnU-Net: Breaking the Spell on Successful Medical Image Segmentation. arXiv 2019, arXiv:1904.08128. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention—MICCAI; Springer: Cham, Switzerland, 2015; Volume 9351. [Google Scholar] [CrossRef]
- Tahari, A.K.; Chien, D.; Azadi, J.R.; Wahl, R.L. Optimum Lean Body Formulation for Correction of Standardized Uptake Value in PET Imaging. J. Nucl. Med. 2014, 55, 1481–1484. [Google Scholar] [CrossRef] [PubMed]
- Hong, R.; Halama, J.; Bova, D.; Sethi, A.; Emami, B. Correlation of PET standard uptake value and CT window-level thresholds for target delineation in CT-based radiation treatment planning. Int. J. Radiat. Oncol. Biol. Phys. 2007, 67, 720–726. [Google Scholar] [CrossRef] [PubMed]
- Avants, B.B.; Tustison, N.; Song, G. Advanced normalization tools (ANTS). Insight J. 2009, 2, 1–35. [Google Scholar] [CrossRef]
- O, J.H.; Lodge, M.A.; Wahl, R.L. Practical PERCIST: A Simplified Guide to PET Response Criteria in Solid Tumors 1.0. Radiology 2016, 280, 576–584. [Google Scholar] [CrossRef]
- Dennis, E.R.; Jia, X.; Mezheristskiy, I.S.; Stephenson, R.D.; Schoder, H.; Fox, J.J.; Helle, G.; Scher, H.I.; Larson, S.M.; Morris, M.J. Bone Scan Index: A Quantitative Treatment Response Biomarker for Castration-Resistant Metastatic Prostate Cancer. J. Clin. Oncol. 2012, 30, 519. [Google Scholar] [CrossRef] [PubMed]
- Idota, A.; Sawaki, M.; Yoshimura, A.; Inaba, Y.; Oze, I.; Kikumori, T.; Kodera, Y.; Iwata, H. Bone Scan Index predicts skeletal-related events in patients with metastatic breast cancer. SpringerPlus 2016, 5, 1–6. [Google Scholar] [CrossRef]
- Cook, G.J.; Azad, G.K.; Goh, V. Imaging Bone Metastases in Breast Cancer: Staging and Response Assessment. J. Nucl. Med. 2016, 57 (Suppl. 1), 27S–33S. [Google Scholar] [CrossRef]
- Moreau, N.; Rousseau, C.; Fourcade, C.; Santini, G.; Ferrer, L.; Lacombe, M.; Guillerminet, C.; Jezequel, P.; Campone, M.; Normand, N.; et al. Comparison between threshold-based and deep learning-based bone segmentation on whole-body CT images. In Medical Imaging 2021: Computer-Aided Diagnosis; International Society for Optics and Photonics: Bellingham, WA, USA, 2021; Volume 11597, pp. 661–667. [Google Scholar] [CrossRef]
- Eichbaum, M.H.R.; Kaltwasser, M.; Bruckner, T.; de Rossi, T.M.; Schneeweiss, A.; Sohn, C. Prognostic factors for patients with liver metastases from breast cancer. Breast Cancer Res. Treat. 2006, 96, 1735–1743. [Google Scholar] [CrossRef]
- Bilic, P.; Christ, P.F.; Vorontsov, E.; Chlebus, G.; Chen, H.; Dou, Q.; Fu, C.W.; Han, X.; Heng, P.A.; Hesser, J.; et al. The Liver Tumor Segmentation Benchmark (LiTS). arXiv 2019, arXiv:1901.04056. [Google Scholar]
- Lin, L.I.K. A Concordance Correlation Coefficient to Evaluate Reproducibility. Biometrics 1989, 45, 255–268. [Google Scholar] [CrossRef] [PubMed]
- DeLong, E.R.; DeLong, D.M.; Clarke-Pearson, D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics 1988, 837–845. [Google Scholar] [CrossRef]
- Youden, W.J. Index for rating diagnostic tests. Cancer 1950, 3, 32–35. [Google Scholar] [CrossRef]
- Choy, G.; Khalilzadeh, O.; Michalski, M.; Do, S.; Samir, A.E.; Pianykh, O.S.; Geis, J.R.; Pandharipande, P.V.; Brink, J.A.; Dreyer, K.J. Current Applications and Future Impact of Machine Learning in Radiology. Radiology 2018, 288, 318–328. [Google Scholar] [CrossRef]
- Iantsen, A.; Visvikis, D.; Hatt, M. Squeeze-and-excitation normalization for automated delineation of head and neck primary tumors in combined PET and CT images. In 3D Head and Neck Tumor Segmentation in PET/CT Challenge; Springer: Cham, Switzerland, 2020; pp. 37–43. [Google Scholar] [CrossRef]
- Xie, J.; Peng, Y. The head and neck tumor segmentation using nnU-Net with spatial and channel ‘squeeze & excitation’blocks. In 3D Head and Neck Tumor Segmentation in PET/CT Challenge; Springer: Cham, Switzerland, 2020; pp. 28–36. [Google Scholar] [CrossRef]
- Blanc-Durand, P.; Jégou, S.; Kanoun, S.; Berriolo-Riedinger, A.; Bodet-Milin, C.; Kraeber-Bodéré, F.; Carlier, T.; Le Gouill, S.; Casasnovas, R.O.; Meignan, M.; et al. Fully automatic segmentation of diffuse large B cell lymphoma lesions on 3D FDG-PET/CT for total metabolic tumour volume prediction using a convolutional neural network. Eur. J. Nucl. Med. Mol. Imaging 2021, 48, 1362–1370. [Google Scholar] [CrossRef] [PubMed]
- Gudi, S.; Ghosh-Laskar, S.; Agarwal, J.P.; Chaudhari, S.; Rangarajan, V.; Paul, S.N.; Upreti, R.; Murthy, V.; Budrukkar, A.; Gupta, T. Interobserver variability in the delineation of gross tumour volume and specified organs-at-risk during IMRT for head and neck cancers and the impact of FDG-PET/CT on such variability at the primary site. J. Med. Imaging Radiat. Sci. 2017, 48, 184–192. [Google Scholar] [CrossRef]
- Reinke, A.; Eisenmann, M.; Tizabi, M.D.; Sudre, C.H.; Rädsch, T.; Antonelli, M.; Arbel, T.; Bakas, S.; Cardoso, M.J.; Cheplygina, V.; et al. Common limitations of image processing metrics: A picture story. arXiv 2021, arXiv:2104.05642. [Google Scholar]
- Hatt, M.; Groheux, D.; Martineau, A.; Espié, M.; Hindié, E.; Giacchetti, S.; De Roquancourt, A.; Visvikis, D.; Cheze-Le Rest, C. Comparison between 18F-FDG PET image–derived indices for early prediction of response to neoadjuvant chemotherapy in breast cancer. J. Nucl. Med. 2013, 54, 341–349. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Goulon, D.; Necib, H.; Henaff, B.; Rousseau, C.; Carlier, T.; Kraeber-Bodere, F. Quantitative evaluation of Therapeutic response by FDg-PeT–cT in Metastatic Breast cancer. Front. Med. 2016, 3, 19. [Google Scholar] [CrossRef] [PubMed]
- Gerratana, L.; Fanotto, V.; Bonotto, M.; Bolzonello, S.; Minisini, A.; Fasola, G.; Puglisi, F. Pattern of metastasis and outcome in patients with breast cancer. Clin. Exp. Metastasis 2015, 32, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Alzubaidi, L.; Al-Amidie, M.; Al-Asadi, A.; Humaidi, A.J.; Al-Shamma, O.; Fadhel, M.A.; Zhang, J.; Santamaría, J.; Duan, Y. Novel Transfer Learning Approach for Medical Imaging with Limited Labeled Data. Cancers 2021, 13, 1590. [Google Scholar] [CrossRef] [PubMed]
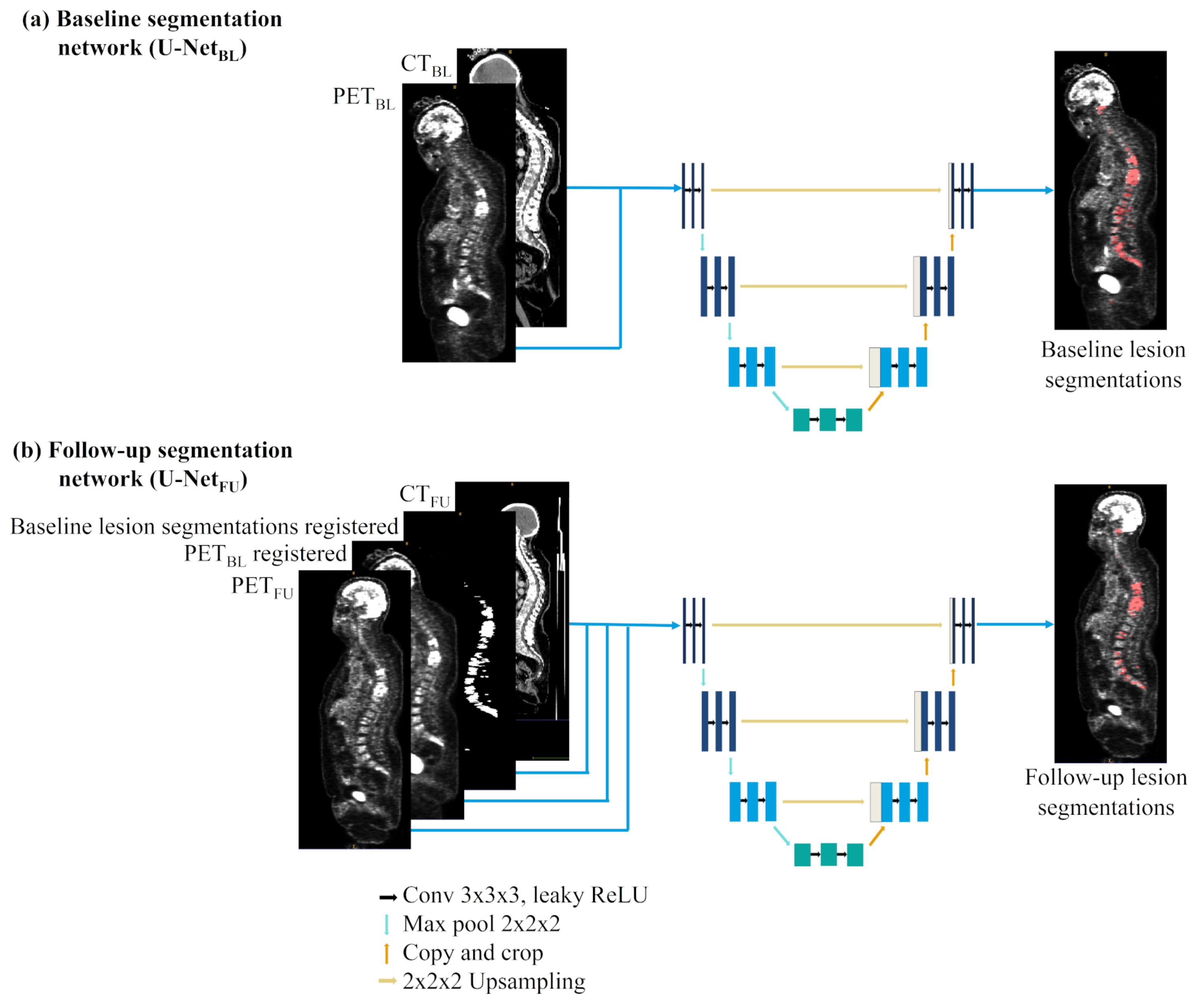



| Networks | Acquisitions | Mean Dice | Global Dice | Detection Recall | Detection Precision |
|---|---|---|---|---|---|
| U-Net | Baseline | 0.73 | 0.72 | 0.87 | |
| Follow-up | 0.53 | 0.43 | 0.75 | ||
| U-Net | Follow-up | 0.64 | 0.63 | 0.78 | |
| Networks | Acquisitions | Mean Dice | Global Dice | Detection Recall | Detection Precision |
| U-Net | Baseline | 0.84 | 0.67 | 0.92 | |
| Follow-up | 0.70 | 0.64 | 0.83 | ||
| U-Net | Follow-up | 0.77 | 0.75 | 0.88 |
| Biomarkers | AUC | Optimal Cutoff Value | Sensitivity | Specificity | p-Value |
|---|---|---|---|---|---|
| SUL | 0.89 | −32% | 87% | 87% | ≤0.001 * |
| TLG | 0.80 | −43% | 73% | 81% | ≤0.001 * |
| PBI | 0.72 | −8% | 69% | 69% | ≤0.001 * |
| PLI | 0.54 | 0% | 53% | 51% | ≤0.001 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moreau, N.; Rousseau, C.; Fourcade, C.; Santini, G.; Brennan, A.; Ferrer, L.; Lacombe, M.; Guillerminet, C.; Colombié, M.; Jézéquel, P.; et al. Automatic Segmentation of Metastatic Breast Cancer Lesions on 18F-FDG PET/CT Longitudinal Acquisitions for Treatment Response Assessment. Cancers 2022, 14, 101. https://doi.org/10.3390/cancers14010101
Moreau N, Rousseau C, Fourcade C, Santini G, Brennan A, Ferrer L, Lacombe M, Guillerminet C, Colombié M, Jézéquel P, et al. Automatic Segmentation of Metastatic Breast Cancer Lesions on 18F-FDG PET/CT Longitudinal Acquisitions for Treatment Response Assessment. Cancers. 2022; 14(1):101. https://doi.org/10.3390/cancers14010101
Chicago/Turabian StyleMoreau, Noémie, Caroline Rousseau, Constance Fourcade, Gianmarco Santini, Aislinn Brennan, Ludovic Ferrer, Marie Lacombe, Camille Guillerminet, Mathilde Colombié, Pascal Jézéquel, and et al. 2022. "Automatic Segmentation of Metastatic Breast Cancer Lesions on 18F-FDG PET/CT Longitudinal Acquisitions for Treatment Response Assessment" Cancers 14, no. 1: 101. https://doi.org/10.3390/cancers14010101
APA StyleMoreau, N., Rousseau, C., Fourcade, C., Santini, G., Brennan, A., Ferrer, L., Lacombe, M., Guillerminet, C., Colombié, M., Jézéquel, P., Campone, M., Normand, N., & Rubeaux, M. (2022). Automatic Segmentation of Metastatic Breast Cancer Lesions on 18F-FDG PET/CT Longitudinal Acquisitions for Treatment Response Assessment. Cancers, 14(1), 101. https://doi.org/10.3390/cancers14010101






