Immune-Proteome Profiling in Classical Hodgkin Lymphoma Tumor Diagnostic Tissue
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
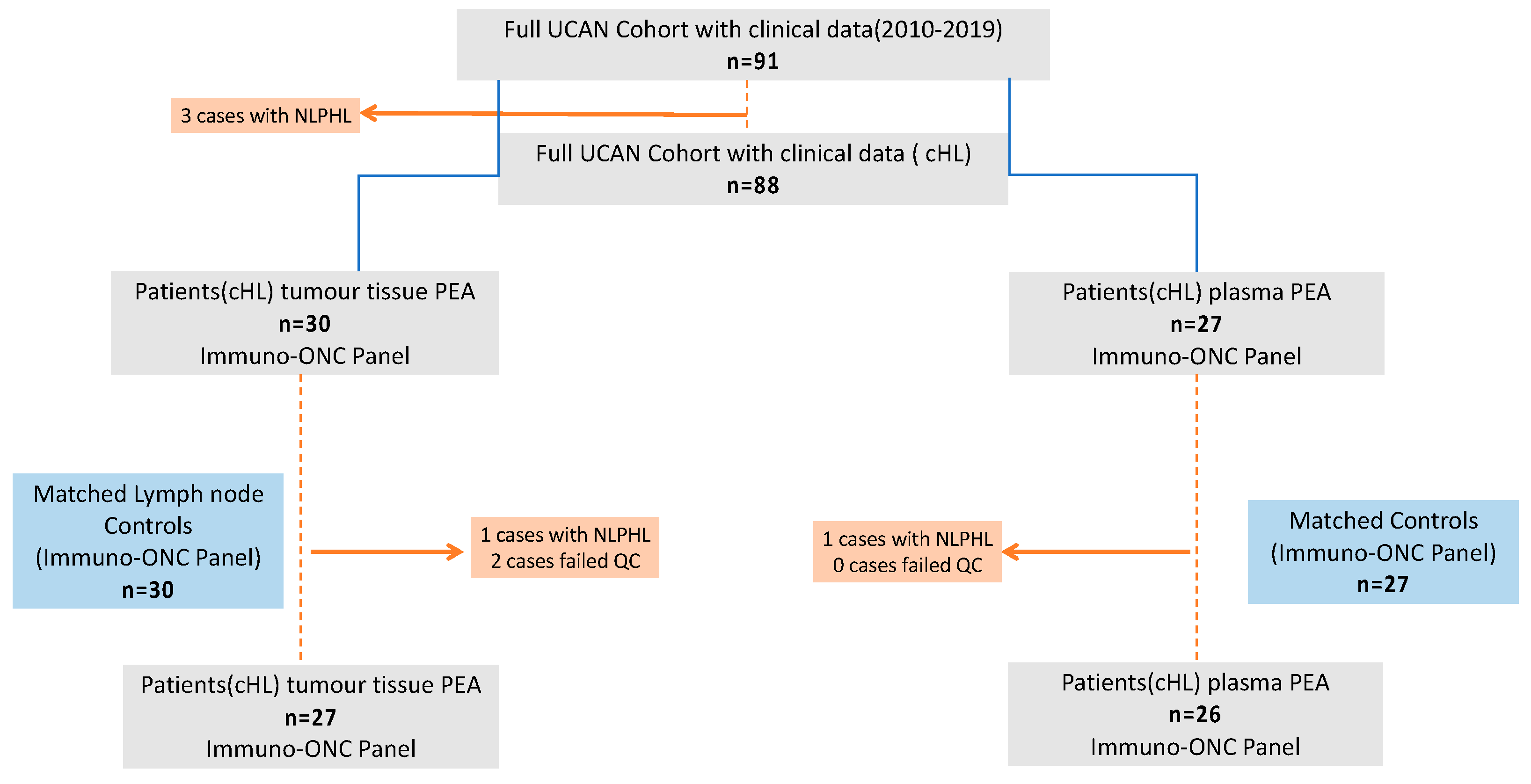
2.1. Study Cohort and Study Samples
2.2. Tissue Lysates Preparation and Plasma Samples
2.3. Proximity Extension Assay Overview, Plate Distribution, and Data Output
2.4. Statistics
3. Results
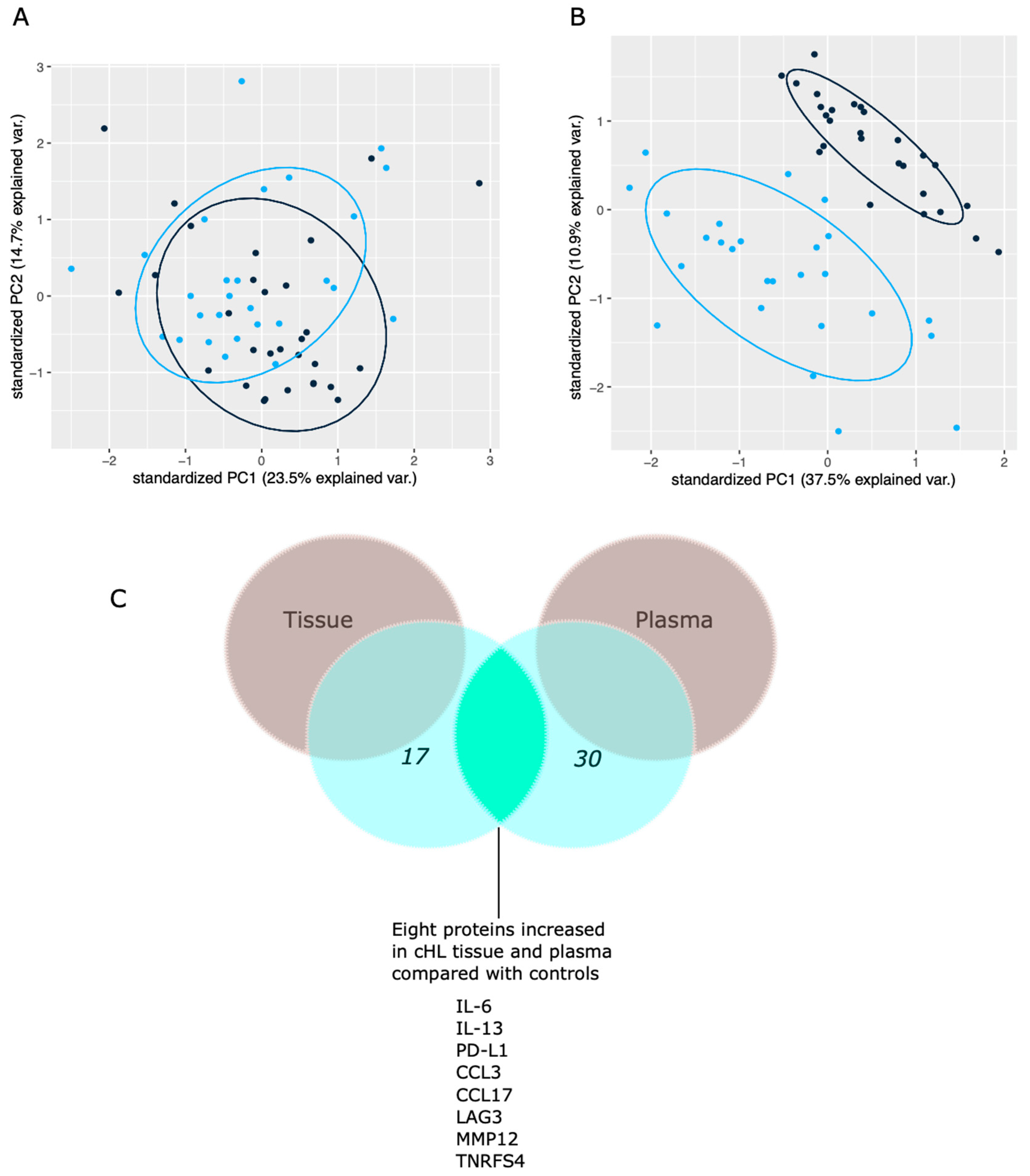
3.1. Proteins Distinguishing Patients from Controls
3.2. Clinicopathological Correlations
4. Discussion
4.1. Immunobiomarkers Elevated in cHL Tissues and Plasma Samples (n = 8)
4.2. Proteins with Decreased levels in cHL Tissues (n = 2)
4.3. Immunobiomarkers Elevated in cHL Tissues but Not Plasma (n = 7)
4.4. General Comments Regarding the Identified Proteome Profile
4.5. Study Limitations and Strengths
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Vardhana, S.; Younes, A. The Immune Microenvironment in Hodgkin Lymphoma: T Cells, B Cells, and Immune Checkpoints. Haematologica 2016, 101, 794–802. [Google Scholar] [CrossRef]
- Aldinucci, D.; Gloghini, A.; Pinto, A.; De Filippi, R.; Carbone, A. The Classical Hodgkin’s Lymphoma Microenvironment and Its Role in Promoting Tumour Growth and Immune Escape. J. Pathol. 2010, 221, 248–263. [Google Scholar] [CrossRef] [PubMed]
- De la Cruz-Merino, L.; Lejeune, M.; Nogales Fernández, E.; Henao Carrasco, F.; Grueso López, A.; Illescas Vacas, A.; Pulla, M.P.; Callau, C.; Álvaro, T. Role of Immune Escape Mechanisms in Hodgkin’s Lymphoma Development and Progression: A Whole New World with Therapeutic Implications. Clin. Dev. Immunol. 2012, 2012, 756353. [Google Scholar] [CrossRef] [PubMed]
- Evens, A.M.; Hutchings, M.; Diehl, V. Treatment of Hodgkin Lymphoma: The Past, Present, and Future. Nat. Clin. Pract. Oncol. 2008, 5, 543–556. [Google Scholar] [CrossRef]
- Collins, F.S.; Varmus, H. A New Initiative on Precision Medicine. N. Engl. J. Med. 2015, 372, 793–795. [Google Scholar] [CrossRef]
- Steidl, C.; Diepstra, A.; Lee, T.; Chan, F.C.; Farinha, P.; Tan, K.; Telenius, A.; Barclay, L.; Shah, S.P.; Connors, J.M.; et al. Gene Expression Profiling of Microdissected Hodgkin Reed-Sternberg Cells Correlates with Treatment Outcome in Classical Hodgkin Lymphoma. Blood 2012, 120, 3530–3540. [Google Scholar] [CrossRef]
- Maggio, E.M.; Van den Berg, A.; Visser, L.; Diepstra, A.; Kluiver, J.; Emmens, R.; Poppema, S. Common and Differential Chemokine Expression Patterns in Rs Cells of NLP, EBV Positive and Negative Classical Hodgkin Lymphomas. Int. J. Cancer 2002, 99, 665–672. [Google Scholar] [CrossRef] [PubMed]
- Skinnider, B.F.; Mak, T.W. The Role of Cytokines in Classical Hodgkin Lymphoma. Blood 2002, 99, 4283–4297. [Google Scholar] [CrossRef]
- Teruya-Feldstein, J.; Jaffe, E.S.; Burd, P.R.; Kingma, D.W.; Setsuda, J.E.; Tosato, G. Differential Chemokine Expression in Tissues Involved by Hodgkin’s Disease: Direct Correlation of Eotaxin Expression and Tissue Eosinophilia: Presented in Part at the 39th Annual Meeting of the American Society of Hematology, Held in San Diego, CA, December 5-9, 1997. Blood 1999, 93, 2463–2470. [Google Scholar] [CrossRef]
- Maggio, E.; van den Berg, A.; Diepstra, A.; Kluiver, J.; Visser, L.; Poppema, S. Chemokines, Cytokines and Their Receptors in Hodgkin’s Lymphoma Cell Lines and Tissues. Ann. Oncol. 2002, 13 (Suppl. 1), 52–56. [Google Scholar] [CrossRef]
- Fujii, K.; Kondo, T.; Yokoo, H.; Yamada, T.; Matsuno, Y.; Iwatsuki, K.; Hirohashi, S. Protein Expression Pattern Distinguishes Different Lymphoid Neoplasms. Proteomics 2005, 5, 4274–4286. [Google Scholar] [CrossRef] [PubMed]
- Wallentine, J.C.; Kim, K.K.; Seiler, C.E.; Vaughn, C.P.; Crockett, D.K.; Tripp, S.R.; Elenitoba-Johnson, K.S.J.; Lim, M.S. Comprehensive Identification of Proteins in Hodgkin Lymphoma-Derived Reed–Sternberg Cells by LC-MS/MS. Lab. Investig. 2007, 87, 1113–1124. [Google Scholar] [CrossRef][Green Version]
- Vergara, D.; Simeone, P.; De Matteis, S.; Carloni, S.; Lanuti, P.; Marchisio, M.; Miscia, S.; Rizzello, A.; Napolitano, R.; Agostinelli, C.; et al. Comparative Proteomic Profiling of Hodgkin Lymphoma Cell Lines. Mol. Biosyst. 2016, 12, 219–232. [Google Scholar] [CrossRef]
- Ma, Y.; Visser, L.; Roelofsen, H.; de Vries, M.; Diepstra, A.; van Imhoff, G.; van der Wal, T.; Luinge, M.; Alvarez-Llamas, G.; Vos, H.; et al. Proteomics Analysis of Hodgkin Lymphoma: Identification of New Players Involved in the Cross-Talk between HRS Cells and Infiltrating Lymphocytes. Blood 2008, 111, 2339–2346. [Google Scholar] [CrossRef] [PubMed]
- LaCasce, A.S. Treating Hodgkin Lymphoma in the New Millennium: Relapsed and Refractory Disease. Hematol. Oncol. 2019, 37 (Suppl. 1), 87–91. [Google Scholar] [CrossRef] [PubMed]
- Verma, M. Personalized Medicine and Cancer. J. Pers. Med. 2012, 2, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Shruthi, B.S.; Vinodhkumar, P. Selvamani Proteomics: A New Perspective for Cancer. Adv. Biomed. Res. 2016, 5, 67. [Google Scholar] [CrossRef]
- Assarsson, E.; Lundberg, M.; Holmquist, G.; Björkesten, J.; Thorsen, S.B.; Ekman, D.; Eriksson, A.; Rennel Dickens, E.; Ohlsson, S.; Edfeldt, G.; et al. Homogenous 96-Plex PEA Immunoassay Exhibiting High Sensitivity, Specificity, and Excellent Scalability. PLoS ONE 2014, 9, e95192. [Google Scholar] [CrossRef]
- Fredriksson, S.; Gullberg, M.; Jarvius, J.; Olsson, C.; Pietras, K.; Gústafsdóttir, S.M.; Ostman, A.; Landegren, U. Protein Detection Using Proximity-Dependent DNA Ligation Assays. Nat. Biotechnol. 2002, 20, 473–477. [Google Scholar] [CrossRef]
- Petrera, A.; von Toerne, C.; Behler, J.; Huth, C.; Thorand, B.; Hilgendorff, A.; Hauck, S.M. Multi-Platforms Approach for Plasma Proteomics: Complementarity of Olink PEA Technology to Mass Spectrometry-Based Protein Profiling. J. Proteome Res. 2021, 20, 751–762. [Google Scholar] [CrossRef]
- Carbone, P.P.; Kaplan, H.S.; Musshoff, K.; Smithers, D.W.; Tubiana, M. Report of the Committee on Hodgkin’s Disease Staging Classification. Cancer Res. 1971, 31, 1860–1861. [Google Scholar]
- Gobbi, P.G.; Cavalli, C.; Gendarini, A.; Crema, A.; Ricevuti, G.; Federico, M.; Di Prisco, U.; Ascari, E. Reevaluation of Prognostic Significance of Symptoms in Hodgkin’s Disease. Cancer 1985, 56, 2874–2880. [Google Scholar] [CrossRef]
- De Oliveira, F.M.S.; Mereiter, S.; Lönn, P.; Siart, B.; Shen, Q.; Heldin, J.; Raykova, D.; Karlsson, N.G.; Polom, K.; Roviello, F.; et al. Detection of Post-Translational Modifications Using Solid-Phase Proximity Ligation Assay. N. Biotechnol. 2018, 45, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Polom, K.; Williams, C.; de Oliveira, F.M.S.; Guergova-Kuras, M.; Lisacek, F.; Karlsson, N.G.; Roviello, F.; Kamali-Moghaddam, M. A Targeted Proteomics Approach Reveals a Serum Protein Signature as Diagnostic Biomarker for Resectable Gastric Cancer. EBioMedicine 2019, 44, 322–333. [Google Scholar] [CrossRef]
- Pla-Roca, M.; Leulmi, R.F.; Tourekhanova, S.; Bergeron, S.; Laforte, V.; Moreau, E.; Gosline, S.J.C.; Bertos, N.; Hallett, M.; Park, M.; et al. Antibody Colocalization Microarray: A Scalable Technology for Multiplex Protein Analysis in Complex Samples. Mol. Cell. Proteom. 2012, 11. [Google Scholar] [CrossRef] [PubMed]
- Olink-Article Number 95311. Available online: Https://Www.Olink.Com/Content/Uploads/2019/06/Olink-Immuno-Oncology-Validation-Data-v2.1.Pdf (accessed on 11 October 2021).
- El Halabi, L.; Adam, J.; Gravelle, P.; Marty, V.; Danu, A.; Lazarovici, J.; Ribrag, V.; Bosq, J.; Camara-Clayette, V.; Laurent, C.; et al. Expression of the Immune Checkpoint Regulators LAG-3 and TIM-3 in Classical Hodgkin Lymphoma. Clin. Lymphoma Myeloma Leuk. 2021, 21, 257–266. [Google Scholar] [CrossRef]
- Gandhi, M.K.; Lambley, E.; Duraiswamy, J.; Dua, U.; Smith, C.; Elliott, S.; Gill, D.; Marlton, P.; Seymour, J.; Khanna, R. Expression of LAG-3 by Tumor-Infiltrating Lymphocytes Is Coincident with the Suppression of Latent Membrane Antigen-Specific CD8+ T-Cell Function in Hodgkin Lymphoma Patients. Blood 2006, 108, 2280–2289. [Google Scholar] [CrossRef]
- Abro, E.U.; Law, S.C.; Keane, C.; Birch, S.; Sabdia, M.B.; Tobin, J.W.D.; Johnson, P.; Trotman, J.; Berkahn, L.; Fulham, M.; et al. A Critical Role for Intratumoral and Circulating LAG3 in Classical Hodgkin Lymphoma: Analysis from the Rathl Prospective Phase III International Clinical Trial. Blood 2018, 132, 1621. [Google Scholar] [CrossRef]
- Gholiha, A.R.; Hollander, P.; Glimelius, I.; Hedstrom, G.; Molin, D.; Hjalgrim, H.; Smedby, K.E.; Hashemi, J.; Amini, R.-M.; Enblad, G. Revisiting IL-6 Expression in the Tumor Microenvironment of Classical Hodgkin Lymphoma. Blood Adv. 2021, 5, 1671–1681. [Google Scholar] [CrossRef]
- Skinnider, B.F.; Kapp, U.; Mak, T.W. The Role of Interleukin 13 in Classical Hodgkin Lymphoma. Leuk. Lymphoma 2002, 43, 1203–1210. [Google Scholar] [CrossRef] [PubMed]
- Skinnider, B.F.; Elia, A.J.; Gascoyne, R.D.; Trümper, L.H.; von Bonin, F.; Kapp, U.; Patterson, B.; Snow, B.E.; Mak, T.W. Interleukin 13 and Interleukin 13 Receptor Are Frequently Expressed by Hodgkin and Reed-Sternberg Cells of Hodgkin Lymphoma. Blood 2001, 97, 250–255. [Google Scholar] [CrossRef] [PubMed]
- Kapp, U.; Yeh, W.C.; Patterson, B.; Elia, A.J.; Kägi, D.; Ho, A.; Hessel, A.; Tipsword, M.; Williams, A.; Mirtsos, C.; et al. Interleukin 13 Is Secreted by and Stimulates the Growth of Hodgkin and Reed-Sternberg Cells. J. Exp. Med. 1999, 189, 1939–1946. [Google Scholar] [CrossRef]
- Ohshima, K.; Akaiwa, M.; Umeshita, R.; Suzumiya, J.; Izuhara, K.; Kikuchi, M. Interleukin-13 and Interleukin-13 Receptor in Hodgkin’s Disease: Possible Autocrine Mechanism and Involvement in Fibrosis. Histopathology 2001, 38, 368–375. [Google Scholar] [CrossRef]
- Buri, C.; Körner, M.; Schärli, P.; Cefai, D.; Uguccioni, M.; Mueller, C.; Laissue, J.A.; Mazzucchelli, L. CC Chemokines and the Receptors CCR3 and CCR5 Are Differentially Expressed in the Nonneoplastic Leukocytic Infiltrates of Hodgkin Disease. Blood 2001, 97, 1543–1548. [Google Scholar] [CrossRef] [PubMed]
- Merz, H.; Fliedner, A.; Orscheschek, K.; Binder, T.; Sebald, W.; Müller-Hermelink, H.K.; Feller, A.C. Cytokine Expression in T-Cell Lymphomas and Hodgkin’s Disease. Its Possible Implication in Autocrine or Paracrine Production as a Potential Basis for Neoplastic Growth. Am. J. Pathol. 1991, 139, 1173–1180. [Google Scholar] [PubMed]
- Greaves, P.; Clear, A.; Owen, A.; Iqbal, S.; Lee, A.; Matthews, J.; Wilson, A.; Calaminici, M.; Gribben, J.G. Defining Characteristics of Classical Hodgkin Lymphoma Microenvironment T-Helper Cells. Blood 2013, 122, 2856–2863. [Google Scholar] [CrossRef]
- Ma, Y.; Visser, L.; Blokzijl, T.; Harms, G.; Atayar, C.; Poppema, S.; van den Berg, A. CD4+CD26-T Cell Population in Classical Hodgkin Lymphoma Displays a Distinctive Regulatory T Cell Population. Blood 2007, 110, 384. [Google Scholar] [CrossRef]
- Buglio, D.; Khaskhely, N.M.; Voo, K.S.; Martinez-Valdez, H.; Liu, Y.-J.; Younes, A. HDAC11 Plays an Essential Role in Regulating OX40 Ligand Expression in Hodgkin Lymphoma. Blood 2011, 117, 2910–2917. [Google Scholar] [CrossRef]
- Green, M.R.; Monti, S.; Rodig, S.J.; Juszczynski, P.; Currie, T.; O’Donnell, E.; Chapuy, B.; Takeyama, K.; Neuberg, D.; Golub, T.R.; et al. Integrative Analysis Reveals Selective 9p24.1 Amplification, Increased PD-1 Ligand Expression, and Further Induction via JAK2 in Nodular Sclerosing Hodgkin Lymphoma and Primary Mediastinal Large B-Cell Lymphoma. Blood 2010, 116, 3268–3277. [Google Scholar] [CrossRef]
- Hollander, P.; Kamper, P.; Smedby, K.E.; Enblad, G.; Ludvigsen, M.; Mortensen, J.; Amini, R.-M.; Hamilton-Dutoit, S.; d’Amore, F.; Molin, D.; et al. High Proportions of PD-1+ and PD-L1+ Leukocytes in Classical Hodgkin Lymphoma Microenvironment Are Associated with Inferior Outcome. Blood Adv. 2017, 1, 1427–1439. [Google Scholar] [CrossRef]
- Luciani, M.G.; Stoppacciaro, A.; Peri, G.; Mantovani, A.; Ruco, L.P. The Monocyte Chemotactic Protein a (MCP-1) and Interleukin 8 (IL-8) in Hodgkin’s Disease and in Solid Tumours. Mol. Pathol. 1998, 51, 273–276. [Google Scholar] [CrossRef] [PubMed]
- Foss, H.D.; Hummel, M.; Gottstein, S.; Ziemann, K.; Falini, B.; Herbst, H.; Stein, H. Frequent Expression of IL-7 Gene Transcripts in Tumor Cells of Classical Hodgkin’s Disease. Am. J. Pathol. 1995, 146, 33–39. [Google Scholar]
- Cattaruzza, L.; Gloghini, A.; Olivo, K.; Di Francia, R.; Lorenzon, D.; De Filippi, R.; Carbone, A.; Colombatti, A.; Pinto, A.; Aldinucci, D. Functional Coexpression of Interleukin (IL)-7 and Its Receptor (IL-7R) on Hodgkin and Reed-Sternberg Cells: Involvement of IL-7 in Tumor Cell Growth and Microenvironmental Interactions of Hodgkin’s Lymphoma. Int. J. Cancer 2009, 125, 1092–1101. [Google Scholar] [CrossRef]
- Gruss, H.J.; Kadin, M.E. Pathophysiology of Hodgkin’s Disease: Functional and Molecular Aspects. Baillieres Clin. Haematol. 1996, 9, 417–446. [Google Scholar] [CrossRef]
- McEarchern, J.A.; Smith, L.M.; McDonagh, C.F.; Klussman, K.; Gordon, K.A.; Morris-Tilden, C.A.; Duniho, S.; Ryan, M.; Boursalian, T.E.; Carter, P.J.; et al. Preclinical Characterization of SGN-70, a Humanized Antibody Directed against CD70. Clin. Cancer Res. 2008, 14, 7763–7772. [Google Scholar] [CrossRef]
- Oudejans, J.J.; Kummer, J.A.; Jiwa, M.; van der Valk, P.; Ossenkoppele, G.J.; Kluin, P.M.; Kluin-Nelemans, J.C.; Meijer, C.J. Granzyme B Expression in Reed-Sternberg Cells of Hodgkin’s Disease. Am. J. Pathol. 1996, 148, 233–240. [Google Scholar]
- Guo, Y.; Xu, F.; Lu, T.; Duan, Z.; Zhang, Z. Interleukin-6 Signaling Pathway in Targeted Therapy for Cancer. Cancer Treat. Rev. 2012, 38, 904–910. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Crabill, G.A.; Pritchard, T.S.; McMiller, T.L.; Wei, P.; Pardoll, D.M.; Pan, F.; Topalian, S.L. Mechanisms Regulating PD-L1 Expression on Tumor and Immune Cells. J. Immunother. Cancer 2019, 7, 305. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, Y.; Yan, Z.; Yang, H.; Sun, W.; Yao, Y.; Chen, Y.; Jiang, R. IL-6 Promotes PD-L1 Expression in Monocytes and Macrophages by Decreasing Protein Tyrosine Phosphatase Receptor Type O Expression in Human Hepatocellular Carcinoma. J. Immunother. Cancer 2020, 8, e000285. [Google Scholar] [CrossRef]
- Gorbachev, A.V.; Fairchild, R.L. Regulation of Chemokine Expression in the Tumor Microenvironment. Crit. Rev. Immunol. 2014, 34, 103–120. [Google Scholar] [CrossRef]
- Sauer, M.; Plütschow, A.; Jachimowicz, R.D.; Kleefisch, D.; Reiners, K.S.; Ponader, S.; Engert, A.; von Strandmann, E.P. Baseline Serum TARC Levels Predict Therapy Outcome in Patients with Hodgkin Lymphoma. Am. J. Hematol. 2013, 88, 113–115. [Google Scholar] [CrossRef]
- Niens, M.; Visser, L.; Nolte, I.M.; van der Steege, G.; Diepstra, A.; Cordano, P.; Jarrett, R.F.; Te Meerman, G.J.; Poppema, S.; van den Berg, A. Serum Chemokine Levels in Hodgkin Lymphoma Patients: Highly Increased Levels of CCL17 and CCL22. Br. J. Haematol. 2008, 140, 527–536. [Google Scholar] [CrossRef]
- Green, M.R.; Rodig, S.; Juszczynski, P.; Ouyang, J.; Sinha, P.; O’Donnell, E.; Neuberg, D.; Shipp, M.A. Constitutive AP-1 Activity and EBV Infection Induce PD-L1 in Hodgkin Lymphomas and Posttransplant Lymphoproliferative Disorders: Implications for Targeted Therapy. Clin. Cancer Res. 2012, 18, 1611–1618. [Google Scholar] [CrossRef]
- Anastasiadou, E.; Stroopinsky, D.; Alimperti, S.; Jiao, A.L.; Pyzer, A.R.; Cippitelli, C.; Pepe, G.; Severa, M.; Rosenblatt, J.; Etna, M.P.; et al. Epstein−Barr Virus-Encoded EBNA2 Alters Immune Checkpoint PD-L1 Expression by Downregulating MiR-34a in B-Cell Lymphomas. Leukemia 2019, 33, 132–147. [Google Scholar] [CrossRef]
- Petrelli, F.; Maltese, M.; Tomasello, G.; Conti, B.; Borgonovo, K.; Cabiddu, M.; Ghilardi, M.; Ghidini, M.; Passalacqua, R.; Barni, S.; et al. Clinical and Molecular Predictors of PD-L1 Expression in Non-Small-Cell Lung Cancer: Systematic Review and Meta-Analysis. Clin. Lung Cancer 2018, 19, 315–322. [Google Scholar] [CrossRef]
- Huard, B.; Gaulard, P.; Faure, F.; Hercend, T.; Triebel, F. Cellular Expression and Tissue Distribution of the Human LAG-3-Encoded Protein, an MHC Class II Ligand. Immunogenetics 1994, 39, 213–217. [Google Scholar] [CrossRef]
- Nagarsheth, N.; Wicha, M.S.; Zou, W. Chemokines in the Cancer Microenvironment and Their Relevance in Cancer Immunotherapy. Nat. Rev. Immunol. 2017, 17, 559–572. [Google Scholar] [CrossRef] [PubMed]
- Ntanasis-Stathopoulos, I.; Fotiou, D.; Terpos, E. CCL3 Signaling in the Tumor Microenvironment. Adv. Exp. Med. Biol. 2020, 1231, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Kubon, J.; Sikic, D.; Eckstein, M.; Weyerer, V.; Stöhr, R.; Neumann, A.; Keck, B.; Wullich, B.; Hartmann, A.; Wirtz, R.M.; et al. Analysis of CXCL9, PD1 and PD-L1 MRNA in Stage T1 Non-Muscle Invasive Bladder Cancer and Their Association with Prognosis. Cancers 2020, 12, 2794. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Zhang, S.-D.; McCrudden, C.; Chan, K.-W.; Lin, Y.; Kwok, H.-F. The Prognostic Significance of PD-L1 in Bladder Cancer. Oncol. Rep. 2015, 33, 3075–3084. [Google Scholar] [CrossRef] [PubMed]
- Lyu, J.; Joo, C.-K. Wnt-7a up-Regulates Matrix Metalloproteinase-12 Expression and Promotes Cell Proliferation in Corneal Epithelial Cells during Wound Healing. J. Biol. Chem. 2005, 280, 21653–21660. [Google Scholar] [CrossRef]
- Davis, S.; Aldrich, T.H.; Jones, P.F.; Acheson, A.; Compton, D.L.; Jain, V.; Ryan, T.E.; Bruno, J.; Radziejewski, C.; Maisonpierre, P.C.; et al. Isolation of Angiopoietin-1, a Ligand for the TIE2 Receptor, by Secretion-Trap Expression Cloning. Cell 1996, 87, 1161–1169. [Google Scholar] [CrossRef]
- Martin, V.; Liu, D.; Fueyo, J.; Gomez-Manzano, C. Tie2: A Journey from Normal Angiogenesis to Cancer and Beyond. Histol. Histopathol. 2008, 23, 773–780. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, R.G.; Lupton, S.; Schmierer, A.; Hjerrild, K.J.; Jerzy, R.; Clevenger, W.; Gillis, S.; Cosman, D.; Namen, A.E. Human Interleukin 7: Molecular Cloning and Growth Factor Activity on Human and Murine B-Lineage Cells. Proc. Natl. Acad. Sci. USA 1989, 86, 302–306. [Google Scholar] [CrossRef]
- Mazzucchelli, R.I.; Riva, A.; Durum, S.K. The Human IL-7 Receptor Gene: Deletions, Polymorphisms and Mutations. Semin. Immunol. 2012, 24, 225–230. [Google Scholar] [CrossRef]
- Gao, J.; Zhao, L.; Wan, Y.Y.; Zhu, B. Mechanism of Action of IL-7 and Its Potential Applications and Limitations in Cancer Immunotherapy. Int. J. Mol. Sci. 2015, 16, 10267–10280. [Google Scholar] [CrossRef]
- Goncharova, O.; Flinner, N.; Bein, J.; Döring, C.; Donnadieu, E.; Rikirsch, S.; Herling, M.; Küppers, R.; Hansmann, M.-L.; Hartmann, S. Migration Properties Distinguish Tumor Cells of Classical Hodgkin Lymphoma from Anaplastic Large Cell Lymphoma Cells. Cancers 2019, 11, 1484. [Google Scholar] [CrossRef] [PubMed]
- Jorgovanovic, D.; Song, M.; Wang, L.; Zhang, Y. Roles of IFN-γ in Tumor Progression and Regression: A Review. Biomark. Res. 2020, 8, 49. [Google Scholar] [CrossRef]
- Cárdenas-Mondragón, M.G.; Torres, J.; Sánchez-Zauco, N.; Gómez-Delgado, A.; Camorlinga-Ponce, M.; Maldonado-Bernal, C.; Fuentes-Pananá, E.M. Elevated Levels of Interferon-γ Are Associated with High Levels of Epstein-Barr Virus Reactivation in Patients with the Intestinal Type of Gastric Cancer. J. Immunol. Res. 2017, 2017, 7069242. [Google Scholar] [CrossRef]
- Mimura, K.; Teh, J.L.; Okayama, H.; Shiraishi, K.; Kua, L.-F.; Koh, V.; Smoot, D.T.; Ashktorab, H.; Oike, T.; Suzuki, Y.; et al. PD-L1 Expression Is Mainly Regulated by Interferon Gamma Associated with JAK-STAT Pathway in Gastric Cancer. Cancer Sci. 2018, 109, 43–53. [Google Scholar] [CrossRef]
- Garcia-Diaz, A.; Shin, D.S.; Moreno, B.H.; Saco, J.; Escuin-Ordinas, H.; Rodriguez, G.A.; Zaretsky, J.M.; Sun, L.; Hugo, W.; Wang, X.; et al. Interferon Receptor Signaling Pathways Regulating PD-L1 and PD-L2 Expression. Cell Rep. 2017, 19, 1189–1201. [Google Scholar] [CrossRef]
- Yoshimura, T. The Chemokine MCP-1 (CCL2) in the Host Interaction with Cancer: A Foe or Ally? Cell. Mol. Immunol. 2018, 15, 335–345. [Google Scholar] [CrossRef]
- Sabroe, I.; Hartnell, A.; Jopling, L.A.; Bel, S.; Ponath, P.D.; Pease, J.E.; Collins, P.D.; Williams, T.J. Differential Regulation of Eosinophil Chemokine Signaling via CCR3 and Non-CCR3 Pathways. J. Immunol. 1999, 162, 2946–2955. [Google Scholar]
- Steidl, C.; Lee, T.; Shah, S.P.; Farinha, P.; Han, G.; Nayar, T.; Delaney, A.; Jones, S.J.; Iqbal, J.; Weisenburger, D.D.; et al. Tumor-Associated Macrophages and Survival in Classic Hodgkin’s Lymphoma. N. Engl. J. Med. 2010, 362, 875–885. [Google Scholar] [CrossRef]
- Tan, K.L.; Scott, D.W.; Hong, F.; Kahl, B.S.; Fisher, R.I.; Bartlett, N.L.; Advani, R.H.; Buckstein, R.; Rimsza, L.M.; Connors, J.M.; et al. Tumor-Associated Macrophages Predict Inferior Outcomes in Classic Hodgkin Lymphoma: A Correlative Study from the E2496 Intergroup Trial. Blood 2012, 120, 3280–3287. [Google Scholar] [CrossRef]
- Touati, M.; Delage-Corre, M.; Monteil, J.; Abraham, J.; Moreau, S.; Remenieras, L.; Gourin, M.-P.; Dmytruk, N.; Olivrie, A.; Turlure, P.; et al. CD68-Positive Tumor-Associated Macrophages Predict Unfavorable Treatment Outcomes in Classical Hodgkin Lymphoma in Correlation with Interim Fluorodeoxyglucose-Positron Emission Tomography Assessment. Leuk. Lymphoma 2015, 56, 332–341. [Google Scholar] [CrossRef]
- Hughes, M.D. Analysis and Design Issues for Studies Using Censored Biomarker Measurements with an Example of Viral Load Measurements in HIV Clinical Trials. Stat. Med. 2000, 19, 3171–3191. [Google Scholar] [CrossRef]
| Entire Cohort (cHL) 2010–2019 n = 88 | Patient Plasma PEA n = 26 | Control: Plasma PEA n = 27 | Patient: Tissue PEA n = 27 | Controls: Tissue PEA n = 30 | |
|---|---|---|---|---|---|
| Age (y): Median (Range) | 41 (12–85) | 44 (21–85) | 45 (20–78) | 45 (21–85) | 45.50 (22–83) |
| Age ≥ 60 (n) | 23 (26%) | 7 (27%) | 8 (30%) | 8 (30%) | 7 (23%) |
| Male Sex (n) | 58 (66%) | 17 (65%) | 18 (67%) | 18 (67%) | 21 (70%) |
| Follow-up time (y); Median (range) | 4.50 (0.36–26.00) | 4.75 (0.66–9.26) | NA | 4.77 (0.66–26.00) | NA |
| 5 year OS probability | 85% | 87% | NA | 88% | NA |
| 2 year OS probability | 91% | 92% | NA | 93% | NA |
| 2 year EFS probability | 84% | 81% | NA | 82% | NA |
| Advanced stage (n) (IIB-IVA) | 56 (64%) | 12 (46%) | NA | 12 (57%) | NA |
| WHO 0–1 (n) | 74 (95%) Missing = 10 | 24 (92%) | NA | 25 (100%) Missing = 2 | NA |
| IPS ≥ 2 (n) | 44 (56%) Missing = 10 | 17 (68%) Missing = 1 | NA | 11 (49) Missing = 4 | NA |
| Treated with BEACOPP at first-line (n) | 14 (16%) Missing = 1 | n = 4 (15%) | NA | 4 (15%) Missing = 1 | NA |
| Treated with ABVD at first-line (n) | 55 (63%) Missing = 1 | n = 18 (69%) | NA | 17 (65%) Missing = 1 | NA |
| Univariate | Multivariate | Predictive | ||||
|---|---|---|---|---|---|---|
| Mean Difference. NPX | p | Padj | Padj | AUC | Pw (adj) | |
| TIE2 | −0.645 | <0.001 | <0.001 | <0.001 | 0.870 | <0.001 |
| IL7 | −0.585 | <0.001 | 0.001 | 0.002 | 0.833 | <0.001 |
| IL6 | 2.879 | <0.001 | <0.001 | <0.001 | 0.922 | <0.001 |
| MCP-1 | 0.985 | <0.001 | 0.001 | 0.002 | 0.825 | 0.001 |
| MCP-4 | 2.089 | <0.001 | 0.006 | 0.002 | 0.821 | 0.001 |
| MCP-2 | 1.688 | <0.001 | 0.001 | 0.001 | 0.823 | 0.001 |
| CCL4 | 1.728 | <0.001 | <0.001 | <0.001 | 0.878 | <0.001 |
| PD-L1 | 1.094 | <0.001 | 0.001 | <0.001 | 0.854 | <0.001 |
| CD70 | 0.799 | <0.001 | 0.003 | 0.001 | 0.799 | 0.005 |
| CCL3 | 1.291 | <0.001 | 0.003 | 0.001 | 0.805 | 0.003 |
| TNFRSF4 | 1.260 | <0.001 | <0.001 | <0.001 | 0.835 | <0.001 |
| CCL17 | 3.800 | <0.001 | <0.001 | <0.001 | 0.917 | <0.001 |
| IFN-gamma | 2.363 | <0.001 | <0.001 | <0.001 | 0.893 | <0.001 |
| MMP12 | 1.891 | <0.001 | <0.001 | 0.007 | 0.817 | 0.001 |
| LAG3 | 2.119 | <0.001 | <0.001 | <0.001 | 0.925 | <0.001 |
| IL13 | 2.382 | <0.001 | <0.001 | <0.001 | 0.909 | <0.001 |
| GZMB | 0.952 | 0.001 | 0.049 | 0.042 | 0.762 | 0.040 |
| Protein | Tissue | Plasma | Biological Annotation * | Cellular Annotation * | Studies in cHL |
|---|---|---|---|---|---|
| LAG3 | Increased in cHL cases | Increased in cHL cases | Suppressed tumor immunity: | Membrane | Mainly upregulated in Tregs adjacent to HRS cells but also observed in macrophages [27,28,29] |
| CCL17 | Increased in cHL cases | Increase in cHL cases | Chemotaxis: Produced by several leukocytes including M2 macrophages | Secretory | Confirmed in HRS cells, and monocytes in the TME [7,8,14] |
| IL6 | Increased in cHL cases | Increase in cHL cases | Inflammation/cell Survival signaling. Produced by several leukocytes including macrophages | Secretory | Confirmed in HRS cells, and various leukocytes [8,30] |
| IL13 | Increased in cHL cases | Increased in cHL cases | Inflammation/cell survival signaling | Secretory | Confirmed in HRS cells, and various lymphocytes [31,32,33,34] |
| CCL4 | Increased in cHL cases | Non-significantly increase in cHL cases | Chemotaxis. Recruit Tregs | Secretory | Higher levels in TAMs, HRS cells mainly negative [35] |
| IFN-gamma | Increased in cHL cases | Non-significantly increase in cHL | Inflammation, cell survival signaling | Secretory | Confirmed in HRS cells [8,9,36] |
| TIE2 | Decreased in cHL | Non-significantly decreased in cHL | Vascular remolding. Migration/permeability | Membrane. and secretory | No data available |
| TNFRSF4 | Increased in cHL cases | Increased in cHL cases | Chemotaxis/Cell signal survival. Induced host antitumor immunity | Membrane and intracellular | Confirmed in T-cells in cHL. Status in HRS cells limited [37,38,39] |
| PD-L1 | Increased in cHL cases | Increased in cHL cases | Suppressed host tumor immunity | Cell membrane | Confirmed in HRS cells and surrounding leukocytes [40,41] |
| MCP-1 | Increased in cHL cases | Non--significantly increase in cHL | Chemotaxis for monocytes | Secreted | Confirmed in Monocytes and HRS cells [42] |
| MCP-2 | Increased in cHL cases | Non-significantly increase in cHL | Chemotaxis for various leukocytes | Secreted | Confirmed in HRS cells [6] |
| IL7 | Decreased in cHL | Increased in cHL | Promoted immune host response | Secreted | Confirmed in HRS cells [43,44] |
| CD70 | Increased in cHL cases | Non-significantly increase in cHL | Cell survival signaling primarily for T-cells | Plasma membrane | Confirmed in HRS cells [45,46] |
| CCL3 | Increased in cHL cases | Increased in cHL cases | Inflammation, cell survival signaling, and chemotaxis | Secreted | Confirmed in HRS cells [9] |
| MCP-4 | Increased in cHL cases | Non-significantly increase in cHL | Chemotaxis and inflammation for monocytes and T-cells | Secreted | Confirmed in HRS cells [7] |
| MMP12 | Increased in cHL cases | Increased in cHL cases | Modulating extracellular matrix. Produced by macrophages | Secreted, mainly extracellular matrix | No data available |
| GZMB | Increased in cHL cases | Non-significantly increase in cHL | Apoptosis/cytotoxic | Secreted | Confirmed in HRS cells and cytotoxic lymphocytes [47] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gholiha, A.R.; Hollander, P.; Löf, L.; Larsson, A.; Hashemi, J.; Ulfstedt, J.M.; Molin, D.; Amini, R.-M.; Freyhult, E.; Kamali-Moghaddam, M.; et al. Immune-Proteome Profiling in Classical Hodgkin Lymphoma Tumor Diagnostic Tissue. Cancers 2022, 14, 9. https://doi.org/10.3390/cancers14010009
Gholiha AR, Hollander P, Löf L, Larsson A, Hashemi J, Ulfstedt JM, Molin D, Amini R-M, Freyhult E, Kamali-Moghaddam M, et al. Immune-Proteome Profiling in Classical Hodgkin Lymphoma Tumor Diagnostic Tissue. Cancers. 2022; 14(1):9. https://doi.org/10.3390/cancers14010009
Chicago/Turabian StyleGholiha, Alex Reza, Peter Hollander, Liza Löf, Anders Larsson, Jamileh Hashemi, Johan Mattsson Ulfstedt, Daniel Molin, Rose-Marie Amini, Eva Freyhult, Masood Kamali-Moghaddam, and et al. 2022. "Immune-Proteome Profiling in Classical Hodgkin Lymphoma Tumor Diagnostic Tissue" Cancers 14, no. 1: 9. https://doi.org/10.3390/cancers14010009
APA StyleGholiha, A. R., Hollander, P., Löf, L., Larsson, A., Hashemi, J., Ulfstedt, J. M., Molin, D., Amini, R.-M., Freyhult, E., Kamali-Moghaddam, M., & Enblad, G. (2022). Immune-Proteome Profiling in Classical Hodgkin Lymphoma Tumor Diagnostic Tissue. Cancers, 14(1), 9. https://doi.org/10.3390/cancers14010009







